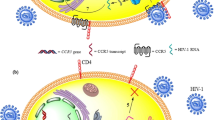
Abstract
Background
Cysteine-cysteine chemokine receptor 5 is the main HIV co-receptor involved in the virus and cell-to-cell spread. A variant of the CCR5 gene known as CCR5-Δ32 which is a product of 32 base pair deletion in the gene plays critical role in the infection and progression to AIDS. The study was carried out to determine the CCR5 genotype of HIV-infected subjects attending University of Calabar Teaching Hospital, Calabar.
Methods
A total of 100 subjects attending HIV clinic, University of Calabar Teaching Hospital were purposively recruited for this study. DNA was extracted from each sample using the Quick gDNA miniprep DNA extraction kit, Zymo Research. Polymerase chain reaction (PCR) was used in the amplification of CCR5 gene in each DNA in a 9700 ABI Thermo cycler and then resolved on 4% agarose gel electrophoresis.
Result
Out of the 100 samples assessed, 100 (100%) were homozygous for the CCR5 wild type gene (CCR5-wt), while none (0%) was homozygous for the CCR5-Δ32 (mutant type), and heterozygosity was not observed.
Conclusion
This study observed absence of CCR5-Δ32 deletion gene among the studied subjects in Calabar, implying lack of genetic advantage in HIV infection and possible rapid progression towards AIDS if other precautions are not checked.
Similar content being viewed by others
Background
Human immunodeficiency virus (HIV)/ Acquired immunodeficiency syndrome (AIDS) despite the campaigns has remained a public health concern [1] with approximately 36.7 million people living with HIV globally at the end of 2016. The burden of the epidemic varies between regions and countries with sub-Saharan Africa being the most affected having nearly 1 in every 25 adult living with HIV accounting for two-third of people living with HIV globally [2,3,4]. According to the 2014 Gap report, 9% of those living with HIV reside in Nigeria [5]. With 3.2 million (9%) people living with HIV in Nigeria as of 2016, she has ranked the second largest HIV global disease burden behind South Africa that housed 7.1 million (19%) people living with HIV [6, 7]. Infection with HIV is associated with progressive loss of cellular immunity which results to life threatening opportunistic infections and progressive development of acquired immunodeficiency syndrome [8].
The major event in HIV infection is the continuous decrease in CD4+T cells that leads to deficient immune system that is unable to withstand the actions of opportunistic pathogens. The more interesting part is the significant disparity of individuals in the susceptibility to the infection, the time lag for depletion of CD4+ T – lymphocytes, as well as time lag to progress to AIDS-defining state [9,10,11]. Host genes collectively known as AIDS restriction genes (ARGs); C-C type chemokine receptor 5 (CCR5) and C-X-C chemokine receptor type 4 (CXCR4) have been known to modify individual response to HIV-1 exposure, infection and pathogenesis markedly [12,13,14]. The human C-C type chemokine receptor (CCR5) belongs to the G-protein-coupled receptor family and is found mostly on the cell surface of some white blood cells such as T4 cells, monocytes and macrophages [15, 16]. The CCR5 wild type protein (CCR5-wt) is made up of 352 amino acids folded into 7 membrane-spanning domains linked by 3 intracellular loops. Aside the wild type, there is CCR5 allele with a 32 base pair deletion (CCR5-Δ32) in the coding region. This mutation results in a truncated protein that cannot be detected on the cell surface [14, 15]. The CCR5-Δ32 mutation has been shown to significantly affect the entry of the HIV and also the progression to AIDS [14]. Individuals who are homozygous for this defect (polymorphism) appear to be resistant to HIV-1 infection [12, 14] and same retards disease progression in heterozygotes [17]. HIV gains entry into the host’s cell through its glycoprotein 120 receptor (gp120) which binds to the hosts cell receptor cluster of differentiation four (CD4+) on T lymphocytes. For the virus to enter the host cell, it requires membrane fusion. This occurs through the virus glycoprotein 41 (gp41) which catalyzes the fusion of the virus to a host co -receptor being either CCR5 or CXCR4, and anchors the virus to the host membrance [17]. In the case of the mutated protein (in this case CCR5-Δ32) prevents the fusion of the virus to the host’s cell surface, thereby denying if entry into the cell, hence, rendering the cell resistant to HIV [12, 17]. The R5-tropic virus strain of HIV-1 prevail using CCR5 as co-receptor for fusion with the cell in the early stage of infection [18,19,20], while the X4-tropic strain of HIV-1 utilizes CXCR4 as co-receptor [21,22,23]. Probably due to the emergence of the X4-tropic HIV strains, the protective effect of the homozygous deletion in the CCR5 gene is not entire [24]. The CCR5- ∆ 32 mutatiion is predominantly found in European population and rare or no occurrence in the Asians and African population [12, 14, 25].
Considering the lack of information of CCR5 profile in Calabar, Cross River State, this study set off to bridge the gap in literature.
Method
Study location
This cross sectional study was carried out in University of Calabar Teaching Hospital, Cross River State, Nigeria. Cross River State is located in Southern Nigeria [26] in the Niger Delta Region. It is bounded in the north by Benue State, the west by Ebonyi and Abia State, the east by Cameroon Republic and the south by Akwa Ibom State and the Atlantic Ocean [27]. Cross River State has an area of 21,787km2 and a population of 2,892,988 (2006 census) [28]. University of Calabar is sited in Calabar Metropolis which is a fusion of Calabar south local government and Calabar municipality. Calabar has a geographical coordinates of 8019′37.02E with an estimated population of 375,196 (2006 census) [28, 29]. The map showing the study area is in indicated in Fig. 1.
Study population
One hundred HIV-infected clients attending antiretroviral therapy (ART) clinic of University of Calabar Teaching Hospital were purposively recruited for this study. Positive subjects who have been confirmed and were on ART treatment who gave their consent were recruited. HIV negative and HIV positive subject who declined were excluded.
DNA extraction
DNA was extracted using gDNA mini prep DNA extraction kit, Zymo Research supplied by Inqaba Biotechnological South Africa. One hundred microliter (100 μL) of whole blood was pipetted into a 1.5 ml. Four hundred microliters (400 μL) of the genomic lysis buffer were added. The samples were mixed by votexing for 5 s and were allowed to stand at room temperature for 10 min. The mixtures were transferred to a zymo-spin column in a collection tube. It was then centrifuged at 12,000 rpm for 1 min. The flow through and collection tubes were discarded. The zymo-spin columns were transferred to a new collection tube and 200 μL of DNA pre-wash buffer was added and centrifuged at 12,000 rpm for 1 min. Five hundred microliter (500 μL) of gDNA was buffered and was added to the spin column and centrifuged at 12,000 rpm for 1 min. The spin column was then transferred into another micro-centrifuge tube and 50 μL of DNA elution buffer added to the spin column and incubated at room temperature for 5 min followed by centrifugation for 30 s to elute the DNA. The eluted DNA was then used for PCR.
CCR5 gene amplification
The CCR5 gene was amplified by polymerase chain reaction (PCR) utilizing primers that flank the 32-base pair deletion; primary: P1: P1:5’CAAAAGGTCTTCATTACACC3′ and secondary: P2: P2:5’CCTGTGCCTCTTCTCATTTC-3’ on a 9700 ABI thermal cycler. The PCR mixture included 1X master mix (which contains Tq polymerase, DNTPS, MgCl2), forward and reverse primers at concentration of 0.4 M and 3 μL of the extracted DNA. DNAse free water was used to make up the PCR to a final volume of 20 μL. The PCR conditions used for the amplification of the gene was as follows: 5 cycles of 94 °C for 3 min (initial denaturation), 94 °C for 45 s (denaturation), 55 °C for 45 s (annealing), 72 °C for 1.5 s (extension) and and a final extension of 72 °C for 5 min and 30 cycles at 94 °C for 30 s (denaturation), 60 °C for 30 s (annealing) and 72 °C for 30 s (extension) and a final extension of 72 °C for 3 min. A 5 μl aliquot of each amplicon were resolved on 4% agarose gel at 120 V for 20 min and visualized in an ultraviolet trans-illuminator. The presence of a single fragment of 185 bp was considered to represent homozygote for CCR5 gene.
HIV-V3 region amplification
A two-step PCR amplification, first with outer primers and then with nested or inner primers, was performed to detect the presence of HIV-1 in infected patients’ PBMC. The DNA oligonucleotide primers HIV-19: 5′-AATGTCAGCACAGTACAATGTACA-3′ and HIV-20: 5′-CAGTAGAAAAAATTCCCCTCCACAATT-3′ and HIV-21: 5′-CTGCTGTTAAATGGCAGTCTAGC-3′, and HIV-22: 5′-TCTGGGTCCCCTCCTGAGGA-3′ were used The PCR components comprised of 5 PCR buffer (100 mM Tris-HCl [pH 8.3], 500 mM KCl, 15 mM MgCl2, 0.01% gelatin), 200 mM each dATP, dCTP, dGTP, and TTP, 0.2 to 1.0 mM each HIV-1 outer primer pair. The reactions were carried out at 94 °C for 1.5 min, 45 °C for 2 min, and 72 °C for 3 min for 35 cycles. The amplified DNA products were analyzed by electrophoresis on a 1.2% agarose gel. Negative controls consisting of DNA from PBMC of seronegative individuals were included in each set of reactions, which were negative in all assays. After the first round PCR, 1 ml of the product was amplified for 25 cycles with the corresponding inner primers at 94 °C for 1.5 min, 50 °C for 2 min, and 72 °C for 3 min. The PCR products were analyzed by electrophoresis on a 1.2% agarose gel.
Sequencing of CCR5 and HIV-V3 gene
The sequencing of the V3 gene and CCR5 was done using the Big Dye Terminator kit on ABI 3500 sequence by Inquaba, South Africa.
Phylogenic analysis
Obtained sequences were edited using the Bioinformatic software Bioedit. Similar sequences were downloaded from the National Centre for Bioinformatics Information (NCBI) using Blast N. Downloaded sequence were aligned using the alignment software MAFT and the evolutionary relationship was eliminated at 500 bootstrap using MEGA.
Results
Table 1 shows the virological and clinical details of the studied population. Majority (65%) of the studied subjects were females. Subjects with CD4 < 200 cells/μL, CD4 200–499 cells/μL and CD4 ≥ 500 cells/μL consisted 14, 41 and 45% of the studied population. Majority of the studied subjects fell into the CDC category A. The studied participants weren’t sure of their mode of acquisition of HIV (Table 1).
Table 2 shows the sequencing of CCR5 receptor protein among the HIV-infected subjects and reveals a similar protein sequence without any mutation among amino acid sequence with high concentration of Leucine 7 (15.99%) followed by Lysine 5 (11.6%).
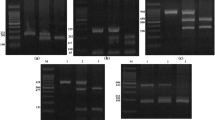
Figure 2 shows the profiling of CCR5 gene among the HIV-infected participants using PCR. The amplified CCR5 gene resolved on agarose at 120 V for 20 min showed that CCR5 DNA bands were slightly ahead of 200 bp of the ladder and the sizes were estimated at 189 bp. The gel revealed that all he subjects (100%) were homozygote for CCR5-wt gene, as no band was detected at 157 bp indicating a 32 bp deletion.
Table 3 shows the frequency of CCR5 gene genotype in the studied population. All the assessed subjects (100%) were homozygous for the wild type CCR5 (CCR5-wt). No homozygous mutant type (CCR5-Δ32) nor heterozygote was observed.
The amplified V3 gene was resolved on agarose at 120 V for 20 min and visualized on UV trans-illuminator. The subjects amplicons were ran along a 100 bp Biolabs molecular ladder. The V3 DNA bands aligned with the 300 bp of the ladder and the sized were estimated to be 300 bp as expected (Fig. 3).
Figure 4 shows sequence alignment of the V3 gene showing the various nucleotide polymorphisms, while Fig. 5 shows the phylogenic tree showing the genetic relationship between the HIV strains. The obtained V3 sequences from the patients produced an exact match during the megablast search for highly similar sequences from the NCBI non-redundant nucleotide (nr/nt) database. The V3 sequence showed a percentage similarity to other V3 region ranging from 98 to 100%. The evolutionary distances computed using the Jukes-Cantor method were in agreement with the phylogenetic placement of the V3 sequences within the HIV and revealed a closely relatedness to HIV-1 isolate 97CIRMF09 (gi\24,061,974) from Senegal, HIV-1 HC 19 (gi/5932488) from Gabon and HIV-1 (gi/918567001) from Cameroon. Furthermore, the gene2pheno software revealed that all the strains were HIV1 subtypes C (Table 4) though the sequence alignment showed some variations in the nucleotides sequence of the V3 gene of various strains (Fig. 4).
Discussion
The CCR5 gene profiling among the HIV-infected subjects in Calabar showed that all (100%) subjects that participated in the study were homozygotes for the normal allele/wild type (CCR5-wt). There was no detection of CCR5 ∆ 32 mutation. The absence of CCR5 ∆ 32 mutation observed in this study is consistent with 1% earlier report by Martinson and colleagues [25] for Nigeria and a more recent report of Nil value by Kenneth et al., 2016 [30] in Bayelsa State, Nigeria. However, higher frequencies of CCR5- ∆ 32 mutation has been reported in northern Europe, particularly the Baltic region as represented by data by Roy and Chakrabarti [9] in Sweden, Belarus, Finland, Lithuania and Estonia. Other areas with reported high frequency of CCR5- ∆ 32 polymorphism included northern coast of France, the Russian cities of Moscow and Ryazan, portions of Volga-Ural region of Russia [31]. Population based studies reported the phenotype of the wild type gene as 75.56% in Rusia [32], 87.5% in Turkey [25], 98.21% in Cyprus Greek [32], 91.22% in Jordan [32], 97.16% in Syria [32], 97.96% in Kuwait [31], 100% in Yemen [25] being studies in Euro - Asia while 100% has been reported in Kenya [25] and Sudan [33]. The relatively higher frequency of the wild type gene of the CCR5 in African region is suggestive that the mutant variant (CCR5- ∆ 32) is fairly recent in terms of human evolution [34, 35].
The implication of the absence of the CCR5- ∆ 32 gene in the studied population revealed a poor resistance to HIV and above all, underlined the fact that our HIV infected subjects are likely rapid progressors. The V3 sequencing showed genetic relationship between HIV strains isolated in this study and those isolated from Senegal P3764 (V3e), Garbon (V3d and Veg), V3b from Cameroon (V3b) and (V3e) from Senegal, this relatedness among strains could be due to trans-border movement of the population which may facilitate the transfer of one stain from a region to the other,
Conclusion
This study has revealed the absence of the mutant gene type of CCR5 (CCR5-Δ32) in the studied subjects implying that the studied population lack one of the prominent genetic advantage against HIV-infection as well as possible rapid progression to AIDS, hence the need to strengthen management and diagnostic strategies.
Availability of data and materials
Datasets generated and analysed in this study are available from the corresponding author on request.
Abbreviations
- AIDS:
-
Acquired immunodeficiency syndrome
- ART:
-
Antiretroviral therapy
- CCR5:
-
C-C- type chemokine receptor
- CD:
-
Cluster of differentiation
- CXCR4:
-
C-X-C chemokine receptor 4
- DNA:
-
Deoxyrebonucleic acid
- HIV:
-
Human Immunodeficiency virus
- PCR:
-
Polymerase chain reaction
References
Asemota EA, Okafor IM, Okoroiwu HU, Ekong ER, Anyanwu SO, Effiong EE, et al. Zinc, copper, CD4 T – cell count and some hematological parameters of HIV – infected subjects in southern Nigeria. Integr Med Res. 2018;7:53–60.
World Health Organization (WHO). Global Health Observation (GHO) data: HIV/AIDS. Available at: www.who.gho/hiv/en. Accessed 26 May 2019.
Okoroiwu HU, Okafor IM, Asemota EA, Okpokam DC. Seroprevalence of transfusion-transmissible infections (HBV, HCV, syphilis and HIV) among prospective blood donors in a tertiary health care facility in Calabar, Nigeria; an eleven years evaluation. BMC Public Health. 2018;18:645.
Coco-Bassey SB, Asemota EA, Okoroiwu HU, Etura JE, Efiong EE, Inyang IJ, et al. Glutathione, glutathione peroxidase and some hematological parameters of HIV-seropositive subjects attending clinic in University of Calabar Teaching Hospital, Calabar, Nigeria. BMC Infect Dis. 2019;19:944.
Joint United Nations Programme on HIV and AIDs (UNAIDS). The Gap report: children and pregnant women living with HIV. Geneva: UNAIDS; 2014.
Joint United Nations programme on HIV and AIDs (UNAIDS). Country fact sheets; Nigeria. 2017. Available at: www.unaids.org/en/regionscountries/nigeria. Accessed 26 May 2019.
Joint United Nations Programme on HIV and AIDs (UNAIDS). Country fast sheets; South Africa. 2017. Available at: www.unaids.org/en/regionscountries/southafrica. Accessed 26 May 2019.
Nsonwu-Anyanwu AC, Ighodalo EV, King D, Agu CE, Jeremiah S, Solomon OT, et al. Biomarken of oxidative stress in HIV seropositive individuals on highly active antiretroviral therapy. Reactive Oxygen Species. 2017;3(9):1–11.
Roy CP, Chakrabarti S. Mutation in AIDS restriction gene affecting the HIV infection and disease progression in a high risk group from northeastern India. Med J Armed Forces India. 2016;722:111–5.
Buchbinder SP, Katz MH, Hessol NA, O’Malley PM, Holmberg SD. Long-term HIV-1 infection without immunologic progression. AIDS. 1994;8:1123–8.
Winkler C, Ping AN, Stephen J, Brien O. Patterns of ethnic diversity among the genes that influence AIDS. Hum Mol Gent. 2004;13(1):R1–R19.
Samson M, Libert F, Doranz BJ, et al. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–5.
O’Brien SJ, Nelson GW. Human genes that limit AIDS. Nat Genet. 2004;36:565–74.
Liu R, Paxton WA, Choe S, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply – exposed individuals to HIV-1 infection. Cell. 1996;86:367–717.
Solloch UV, Lang K, Lang V, Bohne I, Shmidt AH. Frequencies of gene variant CCR5-∆32 in 87 countries based on next generation sequencing of 1.3 million individuals sampled from 3 national DKMS donor centres. Hum Immunol. 2017;78:710–7.
Wu L, Paxton WA, Kassam N, Ruffin N, Rottman JB, Sullivan N, et al. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp Med. 1997;185:1681–91.
Barmania F, Potgiefer M, Pepper MS. Mutation in C-C chemoreceptor type 5 (CCR5) in south African individuals. Int J Infect Dis. 2013;17:e1148–53.
Connor RI, Sheridan KE, Ceradini S, Choe S, Landau NR. Change in coreceptor use correlates with disease progression in HIV-1 infected individuals. J Exp Med. 1997;185:621–8.
Dragic T, Litwin V, Allaway G, Martin SR, Huang Y, Nagashima KA, et al. HIV-1 entry into CD4+ cells in mediated by the chemokine receptor CCR5. Nature. 1996;381:667–73.
Deng H, Lin R, Ellmeier W, Choe S, Unutrnaz D, Burkhart M, et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–6.
Feng Y, Broder CC, Kennedy PI, Berger EA. HIV-1 entry co-factor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;872:877.
Khorramdelazad H, Bagheri V, Hassanshahi G, Zeinali M, Vakilian A. New insight into the role of stromal cell-derived factor 1 (SDF-1/CXCL12) in the pathophysiology of multiple schlerosis.
Karimbadad MN, Falahati-Pour SK, Hassanshabi G. Significant role(s) of CXCL12 and SDF-1 31A genetic variant in the pathophysiology of multiple sclerosis. Neuroimmunomodulation. 2016;23:197–208.
Michael NI, Nelson JA, Kewal Ramani VN, Chang G, O’Brien SJ, Mascola JR, et al. Exclusive and persistent use of the entry co-rector CxCR4 by human immunodeficiency virus type 1 form a subject homozygous for CCR5 delta 32. J Virol. 1998;72:6040–7.
Martinson JJ, Chapman NH, Rees DC, Liu YT, Clegg JB. Global distribution of the CCR5 gene 32 base pair deletion. Nat Genet. 1997;16:100–3.
Nwabueze BO. A constitutional history of Nigeria. United Kingdom: C. Hurst and Co. LTD.; 1982.
Wikipedia, the free encyclopedia. Geography of Cross River State. Available at: https//en.wikipedia.org/wiki/Cross_River_State . Accessed 27 May 2019.
National Bureau of Statistics. Annual abstract of statistics 2011. NBS Federal Republic of Nigeria 2011.
Okoroiwu HU, Asemota EA. Blood donors deferral prevalence and causes in a tertiary health care hospital, southern Nigeria. BMC Health Serv Res. 2019;19:510.
Kenneth OZ, Opuata S, Tatfeng YM TCA, Zaccheaus AJ. Null single nucleotide polymorphism in Cysteine – cysteine chemokine receptor 5 genes among Ijaw Etnic Population of Nigeria. Open J Blood Dis. 2016;2(3):1–6.
Novembre J, Galvani AP, Slatkin M. The geographic spread of the CCR5∆32 HIV-resistance allele. Plos Biol. 2005;3(11):e339.
Salem AH, Batzer MA. Distribution of the HIV resistance CCR5-delta 32 allele among Egytians and Syrians. Mutat Res. 2007;616:175–80.
Su B, Sun G, Lu D, Xiao J, Hu F, Chakraborty R, Deka R, Jin L. Distribution of three HIV-1 resistance – conferring Polymorphisms (SDF1–3’A, CCR2–641 and CCR5 -∆32) in global populations. Eur J Hum Genet. 2000;8:975–9.
Galvani AP, November J. The evolutionary history of the CCR5-Delta 32 HIV-resistance mutation. Microbes Infect. 2005;7:302–9.
Picton ACP, Shalekoff S, Paximadis M, Tiemessen CT. Marked differences in CCR5 expression and activation levels in two South African Populations. Immunology. 2012;136:397–407.
Acknowledgements
Not applicable.
Funding
This study was funded by the authors.
Author information
Authors and Affiliations
Contributions
EFE conceived the study, designed the study, performed analytical procedures, analysed data and edited the initial manuscript; MFU designed the study, analysed data, supervised the study. HUO analysed data, performed statistical analysis, performed literature search, wrote the initial manuscript draft. TYM participated in the analytical procedure, analysed data. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by Health Research Ethical Committee (HREC) of the University of Calabar Teaching Hospital. Informed consent (written) was obtained from the study participants before commencing the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Ekere, E.F., Useh, M.F., Okoroiwu, H.U. et al. Cysteine-cysteine chemokine receptor 5 (CCR5) profile of HIV-infected subjects attending University of Calabar Teaching Hospital, Calabar, Southern Nigeria. BMC Infect Dis 20, 5 (2020). https://doi.org/10.1186/s12879-019-4737-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-019-4737-1