Abstract
Background
Routine culture-based diagnosis of Pseudomonas aeruginosa lung infection in Cystic Fibrosis (CF) patients can be hampered by the phenotypic variability of the microorganism, including its transition to a Viable But Non-Culturable (VBNC) state. The aim of this study was to validate an ecfX-targeting qPCR protocol developed to detect all viable P. aeruginosa bacteria and to identify VBNC forms in CF sputum samples.
Methods
The study involved 115 P. aeruginosa strains of different origins and 10 non-P. aeruginosa strains and 88 CF sputum samples, 41 Culture-Positive (CP) and 47 Culture-Negative (CN). Spiking assays were performed using scalar dilutions of a mixture of live and dead P. aeruginosa ATCC 9027 and a pooled P. aeruginosa-free sputum batch. Total DNA from sputum samples was extracted by a commercial kit, whereas a crude extract was obtained from the broth cultures. Extracellular DNA (eDNA) interference was evaluated by comparing the qPCR counts obtained from DNase-treated and untreated aliquots of the same samples. The statistical significance of the results was assessed by the Wilcoxon test and Student’s t test.
Results
The newly-developed qPCR protocol identified 96.6% of the P. aeruginosa isolates; no amplification was obtained with strains belonging to different species. Spiking assays supported protocol reliability, since counts always matched the amount of live bacteria, thus excluding the interference of dead cells and eDNA. The protocol sensitivity threshold was 70 cells/ml of the original sample. Moreover, qPCR detected P. aeruginosa in 9/47 CN samples and showed higher bacterial counts compared with the culture method in 10/41 CP samples.
Conclusions
Our findings demonstrate the reliability of the newly-developed qPCR protocol and further highlight the need for harnessing a non-culture approach to achieve an accurate microbiological diagnosis of P. aeruginosa CF lung infection and a greater understanding of its evolution.
Similar content being viewed by others
Background
Pseudomonas aeruginosa lung infection represents the main risk of morbidity and mortality in Cystic Fibrosis (CF) patients [1]. After lung colonisation, a wide range of strategies – including biofilm development [2], hypermutation, multidrug resistance, metabolic changes [3], and atypical phenotypes including mucoid forms, small colony variants and slow-growing and even Viable But Non-Culturable (VBNC) forms [4, 5] – promotes pathogen survival and infection persistence. Routine culture-based diagnosis of P. aeruginosa lung infection may fail to detect these bacterial populations in CF patients, providing false negative results or underestimating bacterial abundance [5]. A variety of alternative detection approaches have been described [6]. At present, real-time PCR is considered as the best technique to replace culture, and several protocols targeting different species–specific genes have been developed [7,8,9,10]. However, most of them seem to possess limited sensitivity and ability to discriminate live from dead cells or extracellular DNA (eDNA) [11]. In this study, we report the diagnostic performance of a newly-developed qPCR protocol targeting ecfX, which proved capable of detecting all viable P. aeruginosa subpopulations, including VBNC forms, and showed the specificity, sensitivity, ease of use, and cost-effectiveness required for routine microbiological use.
Methods
Sample collection and bacterial strains
A total number of 88 sputum samples – 41 Culture-Positive (CP) and 47 Culture-Negative (CN) – from CF patients were randomly and anonymously collected from September 2014 to May 2016 among those sent by the Marche regional Cystic Fibrosis Centre to the Microbiology lab of “Ospedali Riuniti” Hospital (Ancona, Italy). Samples were supplemented 1:1 with Sputasol (Oxoid, Basingstoke, UK) before processing. The study also included 115 P. aeruginosa isolates of different origins (51 clinical CF, 55 clinical non-CF, and 9 environmental); 9 recently isolated strains of different bacterial species (1 Klebsiella pneumoniae, 1 Proteus mirabilis 1 Achromobacter xylosoxidans, 1 Burkholderia cepacia, 2 Stenotrophomonas maltophilia, 2 Serratia marcescens and 1 Morganella morganii) provided the Microbiology labs of “Ospedali Riuniti” (Ancona, Italy), “Ospedale A. Murri” (Fermo, Italy), and “Arpam” (Pesaro, Italy); and Escherichia coli ATCC 25922. P. aeruginosa ATCC 9027 was the reference strain.
Culture-based detection of P. aeruginosa in sputum samples
Routine culture assays of CF sputum were performed according to the guidelines described by Gilligan [12] and reported in the UK Consensus Document [13]. P. aeruginosa abundance (CFU/ml) was reported as falling in one of the following ranges: < 102 (CN), 102–103; 103–104; 104–105; 105–106.
The 51 clinical CF P. aeruginosa isolates were identified by MALDI TOF (VITEK-MS, BioMèrieux, Marcy-l’Étoile, France), whereas the non-CF clinical and environmental strains and the non-P. aeruginosa Gram-negative isolates were identified by different systems (MiscroScan Walk Away 40, Beckman Coulter, CA, USA and API 20NE, BioMérieux).
Spiking assays
Spiking assays were performed by inoculating 10-fold (from 3 × 106 to 3 × 102 cells/ml) or 2-fold (from 1.5 × 102 to 16 cells/ml) scalar dilutions of a mid-log phase culture or of a 1:10 mixture of live (log phase) and dead cells (heat-killed at 75 °C for 15 min) of P. aeruginosa ATCC 9027 in a 1 ml aliquot of a pooled sputum batch that was negative for P. aeruginosa both by culture and by real-time PCR.
DNase I digestion
One ml of each sample (sputum or broth culture) was centrifuged at 15,000×g at 4 °C for 15 min, resuspended in 500 μl Phosphate Buffered Saline (PBS), and digested with 18 U DNase I (Ambion-Thermofisher, Carlsbad, CA, USA) at 37 °C for 30 min. After enzyme inactivation at 75 °C for 10 min, samples were washed in 1 ml PBS and resuspended in 100 μl of the same buffer.
DNA extraction
DNA was extracted from sputum samples using the QIAamp DNA Mini Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions, using 80 μl of elution solution to obtain a DNA concentration between 10 and 102 ng/μl.
DNA amount and purity were verified using an ND-1000 Nanodrop spectrophotometer (Thermo Scientific, Wilmington, NC, USA) and agarose gel electrophoresis. DNA samples were stored at − 20 °C until use.
DNA extraction from broth cultures was performed by a rough extraction protocol as previously described [14].
PCR assays
The primer combination ecfX-F 5’-AGCGTTCGTCCTGCACAAGT-3′ [15] and ecfX-R 5’-TCATCCTTCGCCTCCCTG-3′ [7] (amplicon size 145 bp) and the reference strain P. aeruginosa ATCC 9027 were used.
Classic PCRs were performed with 5 μl of DNA, 0.5 μM of each primer, and Dream-Taq Polymerase (Thermo Fisher Scientific, Waltham, MA, USA). Amplification conditions were as follows: 10 min at 95 °C, followed by 35 cycles of 30 s at 94 °C, 30 s at 61 °C, 45 s at 72 °C, and a final elongation step of 5 min at 72 °C.
Real-time PCR assays were performed using 0.2 μM of each primer, 10 μl of 2 × Rotor-Gene SYBR Green PCR master mix (Qiagen), and 2 μl DNA. Cycling conditions were 95 °C for 5 min, followed by 33 cycles of 95 °C for 10 s, 61 °C for 30 s, and 72 °C for 20 s. A melting curve was obtained by ramping the temperature from 59 °C to 95 °C (0.5 °C/10 s) and analysed with Qiagen’s Rotor-Gene Q MDx software. Each reaction was run in triplicate. P. aeruginosa ATCC 9027 DNA and RNase-free water were the positive and negative control, respectively. The amplicons obtained from P. aeruginosa ATCC 9027 and a CF sputum sample were purified with the Gene Elute PCR Clean-up kit (Sigma-Aldrich, Saint Louis, MO, USA) and sequenced bidirectionally (BMR Genomics service (www.bmr-genomics.it/), to confirm their identity. To quantify P. aeruginosa cells, a calibration curve was constructed using scalar dilutions (from 10− 5 to 10− 9 ng/reaction) of an ecfX amplicon of P. aeruginosa ATCC 9027 DNA; intra- and interassay Coefficients of Variation (CV) were calculated as described previously [16]. Data were analysed using Qiagen’s Rotor Gene Q Series software. P. aeruginosa abundance was calculated as reported previously [16] based on amplicon size (145 bp), the weight of 1 bp (1.095 × 10− 12 ng), and the number of ecfX copies found in the P. aeruginosa genome (n = 19). The value thus obtained was then doubled considering the 1:1 sample dilution with Sputasol. qPCR and CFU counts differing by < 0.5 log were considered similar.
The Limit Of Detection (LOD) of the qPCR protocol was determined as described previously [17].
Statistical analysis
The Wilcoxon test and Student’s t test were used for statistical analyses. A p value < 0.05 was considered significant.
Results
Primer pair sensitivity and specificity
The primer pair targeting ecfX was tested by standard PCR assays on 115 P. aeruginosa strains of different origins. It produced a correct amplicon in 111/115 (96.6%) isolates; no amplicon was obtained from strains belonging to species other than P. aeruginosa. The inability of the protocol to detect 4/115 P. aeruginosa strains was not statistically significant (p > 0.1), demonstrating that the target sequence is largely conserved (Table 1).
Validation of the real-time PCR protocol
The reliability of the real-time PCR protocol was first assessed by testing for the presence of inhibitors in direct DNA extracts from sputum and by measuring its sensitivity threshold.
Purity of DNA extracts
Ten-fold and 100-fold dilutions of the DNA extracts obtained from spiked samples were used to evaluate the presence of PCR inhibitors. The progressive threshold cycle (Ct) average delay of 3.3 cycles for each 10-fold dilution excluded their recovery [18].
Accuracy and sensitivity of real-time PCR P. aeruginosa direct detection
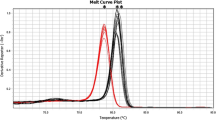
The accuracy and sensitivity of the real-time PCR protocol were determined by spiking assays using serial dilutions of P. aeruginosa ATCC 9027. The melting peaks were always similar to that of a DNA extract from a broth culture of P. aeruginosa ATCC 9027. The sensitivity threshold was 70 cells/ml.
The calibration curve obtained from a purified ecfX amplicon of P. aeruginosa ATCC 9027 yielded regression coefficients close to 1 (R2: 0.998); mean qPCR efficiency was 0.97 ± 0.02. The intra- and interassay CV are reported in Table 2. The LOD was 5.2 × 10− 9 ng/reaction, corresponding to about 138 cells/ml of the original sample. The similarity of the qPCR counts and the amount of P. aeruginosa cells used for spiking demonstrated that the qPCR protocol reliably detected the microorganism in sputum samples.
eDNA interference in the qPCR counts
The possible interference of eDNA in the qPCR counts was evaluated using 3 types of samples (Fig. 1): a crude extract of a P. aeruginosa ATCC 9027 broth culture containing a 1:10 mixture of live (log phase) and dead (heat-killed) cells; a DNA extract from a sputum sample spiked with a mid-log phase culture of P. aeruginosa ATCC 9027; and a DNA extract of a sputum sample spiked with a 1:10 mixture of live and dead P. aeruginosa ATCC 9027 cells. Each sample was divided into two aliquots, of which one was digested with DNase I and the other was not treated. Although a lower amount of total DNA was consistently recovered from treated compared with untreated aliquots (data not shown), similar qPCR counts were always found in untreated and DNase-treated aliquots of spiked sputum samples; these counts consistently matched the live portion of the whole inoculum. The bacterial counts of DNase-digested aliquots were lower (by 1 log) than those of untreated aliquots only in the crude extract of the P. aeruginosa broth culture, suggesting that the commercial kit used for direct DNA extraction from sputum samples efficiently removed the interference due to dead cells or eDNA. When the same analysis was performed in 26 CF sputum samples of clinical origin, no significant (p > 0.05) differences in qPCR counts were obtained in both untreated and DNase I-treated aliquots, confirming the absence of eDNA in the direct DNA extract.
Effect of DNase I treatment on P. aeruginosa quantification by qPCR. The effect of DNase I treatment was assessed by comparing the qPCR counts of two aliquots of the same sample, one digested and one undigested. L/D-BC: P. aeruginosa ATCC 9027 broth culture containing live and dead cells. L/D-SP: sputum sample spiked with 3 × 106 cells of a broth culture containing live and dead cells. Log-SP: sputum sample spiked with 3 × 106 cells of a log phase culture
Comparison of culture and qPCR results
Eighty-eight CF sputum samples – 41 CP and 47 CN – were analysed for their P. aeruginosa content by the newly-developed qPCR protocol. The results were compared to those obtained by the routine culture method (Table 3). The qPCR results were similar to the CFU/ml counts in 31 CP samples and were higher in 10 (see Additional file 1: Table S1). Moreover, in 9 CN samples qPCR detected P. aeruginosa with abundances up to 106 cells/ml (see Additional file 2: Table S2).
qPCR was significantly more efficient (p < 0.05) than routine culture in detecting P. aeruginosa in CN samples.
Influence of sample storage at 4 °C on culture and qPCR P. aeruginosa counts
To investigate whether the sputum samples were affected by storage for a week before analysis, we investigated the influence of storage at 4 °C on plate and qPCR P. aeruginosa counts of 5 sputum samples (CF43–47). Counts were performed on the day of sampling (T0) and after sample storage at 4 °C for a week (T1). Plate counts results were comparable (< 0.5 log difference) at T0 and T1 for 4 samples (CF43, CF44, CF45, and CF47), whereas the count was slightly lower (T0, 1.04 × 105 vs T1, 4.00 × 104 CFU/ml) at T1 for the fifth (CF46). qPCR counts did not show marked differences (< 0.5 log difference) between T0 and T1 in 3 samples (CF43, CF44 and CF45), whereas in the remaining 2 samples (CF46 and CF47) they were higher at T1 (Fig. 2). Whereas at T0 the qPCR and plate counts did not show clear differences, the qPCR counts were uniformly higher at T1 than at T0 and also than the plate counts, suggesting the induction of non-culturable forms during storage at 4 °C, after initial multiplication.
Discussion
In CF lung infection, the possible shift of P. aeruginosa to a dormant, non-culturable form [4] can hamper pathogen eradication, leading to recurrent episodes of relapse [19]. The urgent need for a reliable routine technique capable of detecting all the different forms of P. aeruginosa is thus widely acknowledged. However, none of the protocols devised to date has proved entirely satisfactory, due to suboptimal specificity and sensitivity [20] and to their inability to discriminate between live and dead cells or to exclude the possible interference of eDNA in bacterial counts [11].
In this study, we described a new real-time PCR targeting ecfX, which amplifies a 145 bp sequence, and demonstrated that it reliably detected and quantified P. aeruginosa in CF sputum samples. Notably, none of the previously described ecfX-targeting qPCR protocols [7, 15, 21], used alone, has provided the desired specificity and sensitivity, possibly because they targeted different sequences in the same gene. The SYBR Green ecfX-targeting PCR described herein provided several major advantages compared with routine culture, including faster (a few hours) and reliable pathogen detection and a very high sensitivity threshold (70 cells/ml vs. 100 CFU/ml) that was comparable to the one reported for probe-based assays [22]. With regard to DNA purity and abundance, spiking assays performed with live and dead cells after excluding the presence of PCR inhibitors, showed that qPCR counts matched the amount of the live inoculum. Conservation of the target sequence was supported by its detection in all the CP samples analysed and in 96.6% of the P. aeruginosa strains isolated from samples of different origins; on the other hand, the 19 ecfX copies contained in the P. aeruginosa genome make mutational events affecting all of them unlikely. In addition, the absence of cross-reaction with similar bacterial species supports protocol specificity. These data, together with our previous report of real-time PCRs targeting the P. aeruginosa genes gyrB and oprL [23], highlight the efficiency of the qPCR protocol described here, which reliably detected P. aeruginosa by targeting a single gene. Experiments are currently under way to achieve further improvements, both by designing new primer pairs inside the 145 bp amplicon and by developing an ecfX Taqman probe; sequencing of 23S rDNA amplicons is also being performed to confirm P. aeruginosa identification by the newly-developed PCR protocol [24].
eDNA detection is a longstanding limitation in qPCR assays. Treatment of biofilm samples with propidium or ethidium monoazide before DNA extraction [25] has proved unsatisfactory [26]. The DNase I digestion assays performed in the present study provided similar qPCR counts of both digested and undigested aliquots of spiked sputum samples and demonstrated that the counts corresponded with the live bacterial population. These findings, coupled with the results obtained in untreated and DNase-treated aliquots of 26 clinical CF sputum samples, suggest that the DNA extraction procedure from sputum samples efficiently excluded eDNA as well as DNA from dead bacterial cells, which may be too small and/or too damaged to be retained by the DNA purification column.
The discrepancy between the culture-based and the qPCR approach, i.e. P. aeruginosa detection in 9/47 CN samples and the greater bacterial abundance found in 10/41 CP samples, supports the ability of the proposed PCR protocol to provide a fast and reliable diagnosis and suggests that VBNC forms of P. aeruginosa may be found in CF samples, in line with earlier works [4, 5, 23, 27]. Moreover, the ability of antibiotics to induce a VBNC state in biofilm-growing Staphylococcus aureus [28] further supports the notion that a VBNC P. aeruginosa population may survive in the CF pulmonary biofilm subjected to extended and repeated antibiotic treatment [19].
Conclusions
The pivotal role of P. aeruginosa lung infections in CF patients stresses the urgent need for efficient methods to diagnose P. aeruginosa infection. Compared to routine culture methods, the qPCR-based procedure described herein allows faster and more reliable pathogen detection. Moreover, by detecting all P. aeruginosa phenotypes, including dormant forms, it enables monitoring the effectiveness of antibiotic treatment, providing new insight into the actual evolution of the infection.
Abbreviations
- CF:
-
Cystic Fibrosis
- CN:
-
Culture-Negative
- CP:
-
Culture-Positive
- Ct:
-
Threshold Cycle
- CV:
-
Coefficients of Variation
- eDNA:
-
extracellular DNA
- LOD:
-
Limit of Detection
- PBS:
-
Phosphate Buffered Saline
- VBNC:
-
Viable But Non-Culturable
References
Greipel L, Fischer S, Klockgether J, Dorda M, Mielke S, Wiehlmann L, et al. Molecular epidemiology of mutations in antimicrobial resistance loci of Pseudomonas aeruginosa isolates from airways of cystic fibrosis patients. Antimicrob Agents Chemother. 2016;60(11):6726–34.
Høiby N, Ciofu O, Bjarnsholt T. Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol. 2010;5(11):1663–74.
Hauser AR, Jain M, Bar-Meir M, McColley SA. Clinical significance of microbial infection and adaptation in cystic fibrosis. Clin Microbiol Rev. 2011;24(1):29–70.
Deschaght P, Schelstraete P, Van Simaey L, Vanderkercken M, Raman A, Mahieu L, et al. Is the improvement of CF patients, hospitalized for pulmonary exacerbation correlated to a decrease in bacterial load? PLoS One. 2013;8(11):e79010.
Héry-Arnaud G, Nowak E, Caillon J, David V, Dirou A, Revert K, et al. Evaluation of quantitative PCR for early diagnosis of Pseudomonas aeruginosa infection in cystic fibrosis: a prospective cohort study. Clin Microbiol Infect. 2017;23(3):203–7.
Høiby N, Bjarnsholt T, Moser C, Jensen PØ, Kolpen M, Qvist T, et al. Diagnosis of biofilm infections in cystic fibrosis patients. APMIS. 2017;125(4):339–43.
Lavenir R, Jocktane D, Laurent F, Nazaret S, Cournoyer B. Improved reliability of Pseudomonas aeruginosa PCR detection by the use of the species-specific ecfX gene target. J Microbiol Methods. 2007;70(1):20–9.
Motoshima M, Yanagihara K, Fukushima K, Matsuda J, Sugahara K, Hirakata Y, et al. Rapid and accurate detection of Pseudomonas aeruginosa by real-time polymerase chain reaction with melting curve analysis targeting gyrB gene. Diagn Microbiol Infect Dis. 2007;58(1):53–8.
Le Gall F, Le Berre R, Rosec S, Hardy J, Gouriou S, Boisramé-Gastrin S, et al. Proposal of a quantitative PCR-based protocol for an optimal Pseudomonas aeruginosa detection in patients with cystic fibrosis. BMC Microbiol. 2013;13:143.
Lagares A Jr, Agaras B, Bettiol MP, Gatti BM, Valverde C. A cultivation-independent PCR-RFLP assay targeting oprF gene for detection and identification of Pseudomonas spp. in samples from fibrocystic pediatric patients. J Microbiol Methods. 2015;114(Jul):66–74.
Pattison SH, Rogers GB, Crockard M, Elborn JS, Tunney MM. Molecular detection of CF lung pathogens: current status and future potential. J Cyst Fibros. 2013;12(3):194–205.
Gilligan P. Respiratory cultures from cystic fibrosis patients. In: Garcia LS, Isenberg HD, editors. Clinical microbiology procedures handbook. Washington DC: ASM press; 2007. p. 311–9.
UK Cystic Fibrosis Trust Microbiology Laboratory Standards Working Group. Laboratory standards consensus document. Laboratory standards for processing microbiological samples from people with cystic fibrosis. (2010).
Hynes WL, Ferretti JJ, Gilmore MS, Segarra RA. PCR amplification of streptococcal DNA using crude cell lysate. FEMS Microbiol Lett. 1992;73(1–2):139–42.
Clifford RJ, Milillo M, Prestwood J, Quintero R, Zurawski DV, Kwak YI, et al. Detection of bacterial 16S rRNA and identification of four clinically important bacteria by real-time PCR. PLoS One. 2012;7(11):e48558.
Di Cesare A, Luna GM, Vignaroli C, Pasquaroli S, Tota S, Paroncini P, et al. Aquaculture can promote the presence and spread of antibiotic-resistant enterococci in marine sediments. PLoS One. 2013;8(4):e62838.
Bustin S, Benes V, Garson J, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–22.
Gerriets JE, Greiner TC, Gebhart CL. Implementation of a T4 extraction control for molecular assays of cerebrospinal fluid and stool specimens. J Mol Diagn. 2008;10(1):28–32.
Yum HK, Park IN, Shin BM, Choi SJ. Recurrent Pseudomonas aeruginosa infection in chronic lung disease: relapse or reinfection? Tuberc Respir Dis. 2014;77(4):172–7.
Blanchard AC, Rooney AM, Yau Y, Zhang Y, Stapleton PJ, Horton E, et al. Early detection using qPCR of Pseudomonas aeruginosa infection in children with cystic fibrosis undergoing eradication treatment. JCyst Fibros. 2018. https://doi.org/10.1016/j.jcf.2018.02.008.
Shi H, Trinh Q, Xu W, Zhai B, Luo Y, Huang K. A universal primer multiplex PCR method for typing of toxinogenic Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2012;95(6):1579–87.
Deschaght P, De Baere T, Van Simaey L, Van Daele S, De Baets F, De Vos D, et al. Comparison of the sensitivity of culture, PCR and quantitative real-time PCR for the detection of Pseudomonas aeruginosa in sputum of cystic fibrosis patients. BMC Microbiol. 2009;9:244.
Amiri M, Mangiaterra G, Pasquaroli S, Vignaroli C, Cirilli N, Manso E, et al. Molecular diagnosis of Pseudomonas aeruginosa infection in culture-negative samples from cystic fibrosis patients. J Cys Fibros. 2015;14:S72.
McCulloch E, Lucas C, Ramage G, Williams C. Improved early diagnosis of Pseudomonas aeruginosa by real-time PCR to prevent chronic colonisation in a paediatric cystic fibrosis population. J Cyst Fibros. 2011;10(1):21–4.
Nocker A, Camper AK. Novel approaches toward preferential detection of viable cells using nucleic acid amplification techniques. FEMS Microbiol Lett. 2009;291(2):137–42.
Taylor MJ, Bentham RH, Ross KE. Limitations of using propidium monoazide with qPCR to discriminate between live and dead Legionella in biofilm samples. Microbiol Insights. 2014;7:5–24.
Boutin S, Weitnauer M, Hassel S, Graeber SY, Stahl M, Dittrich AS, et al. One time quantitative PCR detection of Pseudomonas aeruginosa to discriminate intermittent from chronic infection in cystic fibrosis. J Cyst Fibros. 2018;17(3):348–55.
Pasquaroli S, Zandri G, Vignaroli C, Vuotto C, Donelli G, Biavasco F. Antibiotic pressure can induce the viable but non-culturable state in Staphylococcus aureus growing in biofilms. J Antimicrob Chemother. 2013;68:1812–7.
Acknowledgements
We are grateful to Dr. Rolando Gagliardini for his clinical advice, to Prof. Luigi Ferrante for the statistical analysis, and to Marina Lombardi and Natascia Gracciotti for their technical assistance.
We also thank Dr. Marisiana Granatelli for providing the non-CF P. aeruginosa clinical strains, Dr. Silvia Magi for providing environmental P. aeruginosa strains, and Prof. Gianfranco Donelli for providing the reference strain P. aeruginosa ATCC9027.
Funding
This work was supported by FFC grant # 13/2017 of the Italian Cystic Fibrosis Research Foundation through a scholarship contract and financial support in the purchase of laboratory materials.
Availability of data and materials
Not applicable.
Author information
Authors and Affiliations
Contributions
GM was involved in all phases of the experimental plan, with particular regard to the evaluation of the specificity and sensitivity of the developed PCR protocol and to the effectiveness of the bacterial DNA extraction from CF samples; MA in the development of the qPCR and in its application to CF sputum samples; ADC and SP in the qPCR counts and in the general planning and critical discussion of the results; EM in the collection and cultural analysis of CF samples; NC in the discussion about P. aeruginosa CF lung colonization and the need of improved diagnostic approaches; BC in the collection and amplification of the P. aeruginosa and non-P. aeruginosa strain; CV in the methods improvement; FB planned, organized and coordinated the whole work. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. Comparison of plate and qPCR quantification of P. aeruginosa in 41 CP samples from CF patients. Description: P. aeruginosa abundance in the 41 CP samples quantified by both plate count and qPCR. Higher qPCR than plate counts suggest the presence of non-culturable forms and are indicated by a star. (DOCX 19 kb)
Additional file 2:
Table S2. qPCR quantification of 9 CN sputum samples from CF patients. Description: P. aeruginosa quantification by qPCR in 9 CN sputum samples; the samples which were shown to contain a P. aeruginosa amount under the LOD of the developed qPCR were indicated as <LOD. (DOCX 18 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Mangiaterra, G., Amiri, M., Di Cesare, A. et al. Detection of viable but non-culturable Pseudomonas aeruginosa in cystic fibrosis by qPCR: a validation study. BMC Infect Dis 18, 701 (2018). https://doi.org/10.1186/s12879-018-3612-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-018-3612-9