Abstract
Background
The emergence and spread of antibiotic resistant micro-organisms is a global concern, which is largely attributable to inaccurate prescribing of antibiotics to patients presenting with non-bacterial infections. The use of ‘omics’ technologies for discovery of novel infection related biomarkers combined with novel treatment algorithms offers possibilities for rapidly distinguishing between bacterial and viral infections. This distinction can be particularly important for patients suffering from lower respiratory tract infections (LRTI) and/or sepsis as they represent a significant burden to healthcare systems. Here we present the study details of the TAILORED-Treatment study, an observational, prospective, multi-centre study aiming to generate a multi-parametric model, combining host and pathogen data, for distinguishing between bacterial and viral aetiologies in children and adults with LRTI and/or sepsis.
Methods
A total number of 1200 paediatric and adult patients aged 1 month and older with LRTI and/or sepsis or a non-infectious disease are recruited from Emergency Departments and hospital wards of seven Dutch and Israeli medical centres. A panel of three experienced physicians adjudicate a reference standard diagnosis for all patients (i.e., bacterial or viral infection) using all available clinical and laboratory information, including a 28-day follow-up assessment. Nasal swabs and blood samples are collected for multi-omics investigations including host RNA and protein biomarkers, nasal microbiota profiling, host genomic profiling and bacterial proteomics. Simplified data is entered into a custom-built database in order to develop a multi-parametric model and diagnostic tools for differentiating between bacterial and viral infections. The predictions from the model will be compared with the consensus diagnosis in order to determine its accuracy.
Discussion
The TAILORED-Treatment study will provide new insights into the interplay between the host and micro-organisms. New host- or pathogen-related biomarkers will be used to generate a multi-parametric model for distinguishing between bacterial and viral infections. This model will be helpful to better guide antimicrobial therapy for patients with LRTI and sepsis. This study has the potential to improve patient care, reduce unnecessary antibiotic prescribing and will contribute positively to institutional, national and international healthcare economics.
Trial Registration
NCT02025699. Registration Date: January, 1, 2014.
Similar content being viewed by others
Background
The burden of antibiotic misuse
Antibiotics are the most prescribed class of drugs worldwide [1]. However, 30–50% of antibiotics are estimated to be prescribed inappropriately, making antibiotics also the most misused drug class [1,2,3,4,5]. Antibiotic overuse, i.e. prescribing antibiotics to treat a non-bacterial disease, is a serious problem. For example, in the US over 80 million antibiotic prescriptions are given annually for viral infections in the outpatient setting [6]. This may cause the emergence of adverse events (AEs) such as allergic reactions and antibiotic-associated diarrhoea [7]. Importantly, antibiotic overuse has been linked to the emergence of resistant strains of bacteria, by the Center for Disease Control considered as “one of the world’s most pressing health problems in the 21st century” [8, 9]. Annually, 23,000 deaths due to resistant bacteria are expected in the US alone [8, 10]. Resistant bacteria are projected to cause over 10 M annual deaths worldwide by 2050, surpassing cancer as the leading cause of death [8, 11]. A second type of antibiotic misuse is underuse, i.e. delayed or no antibiotic treatment in case of a bacterial disease, from which a patient could have benefited [12,13,14,15,16,17]. Although reducing the risk of antibiotic-related AEs [18, 19], underuse of antibiotics may lead to a prolonged disease duration and an increased rate of complications that could have been avoided with early antimicrobial treatment [19,20,21]. For example, up to 15–40% of adult patients hospitalized for bacterial pneumonia in the US receive delayed or no antibiotic treatment [14, 15]. Finally, applying the wrong antibiotic spectrum to treat a bacterial disease is the third type of antibiotic misuse. For example, administering third generation cephalosporins instead of macrolides to treat a respiratory infection caused by atypical bacteria.
Health economics consequences
Antibiotic overuse has also several health economics consequences. For instance, the annual cost of unnecessary antibiotic prescriptions for adult respiratory infections in the US is estimated to be $1.1 billion [22]. In addition, there are also indirect costs, including the costs of treating preventable antibiotic-related AEs and the costs of prolonged hospital stay as a result of AEs. Lastly, are the costs related to the emergence of antibiotic-resistant bacterial strains. The cost of treating patients with antibiotic-resistant bacteria is estimated at $16–26 billion annually in the US [23,24,25] and over €1.5 billion in the EU [26]. The health economics consequences of underuse of antibiotics include the costs of treating preventable complications and a prolonged disease duration resulting from a delayed or no antibiotic treatment [19,20,21].
Antibiotics in respiratory tract infections and sepsis
Respiratory tract infections are one of the most common causes of hospital and outpatient visits in the EU, comprising 1 out of 3 admissions annually [9]. In patients with mild respiratory tract infections, over prescription of antibiotics in general and prescription of broad spectrum antibiotics are large problems. Broad spectrum antibiotics account for approximately half of all antibiotic prescribing for children with acute RTI [27], with the above mentioned consequences [8]. Sepsis-related hospital admissions are less frequent but represent a significant burden to healthcare systems in terms of adverse patient outcomes and the need for rapid, but effective, intervention strategies [28]. In septic patients the key clinical challenge is to ensure that the correct antimicrobial treatment is administered, and that this therapy is administered as soon as possible [29].
Limitations of current diagnostic tools
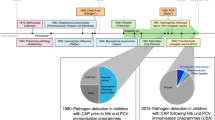
Although the current diagnostic tools for facilitating appropriate use of antibiotics (such as culture-, PCR-, and immunoassay-based) may be valuable in some clinical situations, they have major limitations (Fig. 1). First, existing diagnostic tools often require hours to days to provide information, whereas physicians need to decide whether to prescribe antibiotics within shorter time periods. Second, available diagnostic technologies usually require direct sampling of the pathogen. Such sampling is often not feasible if the infection site is inaccessible (e.g., pneumonia, sinusitis, middle-ear infection, etc.). A third limitation is that available technologies usually search for the presence of specific bacteria or viruses. However, many types of bacteria and viruses could be present as part of the natural flora without causing an infection. For example, the important respiratory tract pathogenic bacteria S. pneumoniae is also part of the upper respiratory tract natural flora in up to 68% and 15% of healthy children and adults, respectively [30] and respiratory viruses were detected in 35.4% of a-symptomatic children [31]. Finally, diagnostic solutions that were developed for detection of a specific pathogenic strain, often fail to detect the constantly evolving and emerging strains of the same family of pathogens, for example rapid Influenza tests showed low sensitivity, 45%, during the 2009 H1N1 influenza A viral outbreak [32]. These limitations lead to a ‘diagnostic gap’, which in turn often leads physicians to either over-prescribe (e.g., ‘just-in-case’ approach) or under-prescribe (e.g., ‘watchful waiting’ approach) antibiotics [18,19,20], both of which adversely influence patient care and health economics.
The TAILORED-treatment project
The health and health economics consequences of antibiotic misuse highlight the need for a diagnostic tool that would help physicians to use antibiotics appropriately. Ideally, this diagnostic tool should provide a diagnosis that can accurately and rapidly differentiate between bacterial and viral infections. However, as previously described, current diagnostic tools are often inadequate (Fig. 1). Therefore, the TAILORED-Treatment consortium was established to develop new tools aimed at increasing the effectiveness of antibiotic therapy, reduce adverse events, and limit the emergence of antimicrobial resistance in children and adults. In reality, targeted antimicrobial therapy can most effectively be achieved by utilizing personalized data to facilitate a tailored and optimized approach to individual patient treatment. This can best be achieved by utilizing knowledge gained from both host-centred and pathogen-centred parameters during health and disease. Unfortunately, these parameters have traditionally, tended to be measured independently, and used on an ad hoc basis without careful integration for the best treatment of the patient. However, recent advances in the development of high-throughput and sensitive molecular-based technologies, improved databases, and bioinformatics analysis tools, rapidly enabling that the goal of personalized medicine and treatment can be reached. Unfortunately, however, there currently exists a ‘technological gap’ between recent state-of-the-art methodologies (for example with respect to gaining new insights into novel host-pathogen interactions) and laboratory-to-bedside results to benefit patients, physicians and society as a whole [33]. The observational TAILORED-Treatment project is designed to bridge this technological gap in order to generate novel insights and innovations that are readily exploitable in the field of personalized medicine and infectious diseases.
Study objectives
Primary objective
To develop a novel multi-parametric diagnostic model for the management of patients with LRTI and/or sepsis that is based on novel pathogen- and host-related factors.
Secondary objectives
-
1.
Identify individual as well as sets of blood host biomarkers for differentiating between bacterial, and viral aetiologies and determine the sensitivity and specificity of these biomarkers in distinguishing between the different infection types;
-
2.
Identify individual as well as sets of blood host biomarkers for differentiating between bacterial subtypes, namely Gram positive, Gram negative and atypical bacteria;
-
3.
Characterize the temporal dynamics of the host-pathogen interactions based on multiple sampling before and during antimicrobial treatment;
-
4.
Characterize the respiratory microbiome in patients presenting with LRTI and/or sepsis;
-
5.
Develop a mass spectrometry based, rapid detection technique for identification of microbial pathogens and antimicrobial resistances in clinical samples;
-
6.
Develop a personalized treatment algorithm for tailored antimicrobial therapy that integrates clinical, molecular and biochemical data;
-
7.
Define genetic mechanisms underlying the differential host response between patients with viral versus bacterial infection.
Methods
General study design
In this prospective, multi-centre, observational study, an expected total number of 1200 paediatric and adult patients aged 1 month and older with suspected LRTI and/or sepsis or a non-infectious disease are recruited, in order to develop a diagnostic model to differentiate between bacterial and viral infections. The study will generate a model for patients with LRTI and a separate model for patients with sepsis. The prediction of the model obtained from each patient will be compared with a consensus diagnosis by a panel of three experts in order to determine its accuracy (Fig. 2). The study is a non-interventional study, and therefore, the diagnostic prediction of the model will not be used for any clinical or diagnostics decision making. Local data monitor plans were used and executed by local, independent monitors.
Study participants
Patients aged 1 month and older that attend the Emergency Department (ED) and hospital wards of seven Dutch and Israel medical centres (secondary and tertiary) due to a suspected LRTI and/or sepsis that agree (or their legal guardians agrees) to sign an informed consent and meet the inclusion criteria and none of the exclusion criteria (Table 1), are eligible to participate in the study. Informed consent is asked by the study team (i.e. research nurses or medical doctor). Sepsis criteria are defined according to published criteria [34]. Patients receive care as usual. The non-infectious disease group include patients with clinical signs of a non-infectious disease.
Data/sample collection
Clinical data
Clinical evaluation and management is performed according to standard institutional procedures (Fig. 2). Collected clinical data, includes demographics, medical history, clinical symptoms, physical examination, disease course, laboratory measurements, as well as more advanced diagnostic information including microbiological (including an additional multiplex PCR), and serological investigations. A follow-up phone call 28-day after recruitment, defines the end of follow-up and generates data to be used by the expert panel (e.g. maximum disease severity, total use of antibiotics, Additional file 1).
Sample collection
For study purposes two blood samples, two nasal swabs and (in case of diarrhoea) a stool sample are collected (Fig. 2). Blood and nasal swabs sampling at two sampling points (at inclusion and after 3–5 days) is performed in hospitalized Dutch children in order to monitor the temporal dynamics of the host-pathogen interactions. One nasal swab sample (Universal Transport Medium, Copan, USA) is used for PCR based microbiological investigations performed for the establishment of patient diagnosis (only at inclusion). The PCR results are included in the eCRF and are available to the TAILORED-Treatment expert panel while determining the diagnosis of the patient (Fig. 2). The second nasal swab is collected in Transport Medium suitable for bacterial culturing (e-swab, Liquid Amies Medium, Copan), this sample is used for respiratory microbiome investigations and to develop a rapid detection technique for identification of microbial pathogens and antimicrobial resistances, based on mass spectrometry and proteomics for discovery of peptide biomarkers. Serum samples are used for different proteomic measurements, leukocytes are isolated for discovery of RNA-based biomarkers. Following RNA isolation, remaining DNA is used for GWAS studies. GWAS analyses is performed in the Netherlands, as Institutional review board (IRB) approval for genetic testing is not obtained in Israel. DNA collection is mentioned explicitly in the patient information. Blood samples are collected from children with a non-infectious disease only if blood sampling is required as part of their routine care. In cases of diarrhoea on enrolment, stool samples are collected. These samples will be evaluated to identify Clostridium difficile infections.
‘Proteotyping’
Shotgun proteomic detection and identification of microorganisms, or ‘proteotyping’, relies on recognition of species-unique peptides by tandem mass spectrometry (MS), allowing discrimination of species facilitated at amino acid level resolution. The recent evolution of mass spectrometers, having high sensitivity, accuracy and resolution, enables detection of almost the complete proteome of a microorganism. With such analytical means, it is feasible to determine directly, within a clinical sample, the species identity, the sub-species strain type and the factors expressing virulence and antimicrobial resistance [35]. Clinical samples are processed by first removing human biomass and subsequently collecting bacteria. The proteins of the collected bacteria were processed by the LPI™ FlowCell or in-solution digestion protocols followed by liquid chromatography tandem MS (LC-MS/MS) analysis. The recently developed technology called Lipid-based Protein Immobilization (LPI™) is based on immobilization of biological material within a flow cell, followed by digestion of exposed proteins by an enzyme, such as trypsin [36,37,38]. The MS-based proteomics methodology has been applied successfully, without prior cultivations to the analyses of clinical nasal swab samples from the Sahlgrenska University Hospital that have been confirmed positive for a respiratory infectious bacterial species, i.e., S. pneumoniae, H. influenza, M. catarrhalis and S. aureus (unpublished data).
Central database
All clinical data (eCRFs) and research data (host biomarkers, host microbiome, pathogen identity and resistance) is coded and uploaded to a central, secured database (HoPOIT database) to which access is granted only to research partners (Fig. 3). The centralized clinical, microbiological, molecular and biochemical data is used to develop a multi-parametric model and diagnostic tools for differentiating between bacterial and viral infections.
Aetiology determination
Currently, no single reference standard exists for differentiating between bacterial and viral aetiology in an acute infectious disease. Therefore, we follow the Standards for Reporting of Diagnostic Accuracy (STARD) recommendation [39], and create a highly rigorous reference standard based on expert panel adjudication (Fig. 2). The eCRF of each patient is available to a panel of three independent physicians that are affiliated to the country of recruitment. Based on the information included in the eCRF, each member of the panel assigns one of the following diagnostic labels to each one of the patients: (i) bacterial infection; (ii) viral infection; (iii) mixed infection (i.e., bacterial and viral co-infection); (iv) non-infectious disease; or (v) undetermined. Of note, each expert is blinded to the research results and to the labels of his peers on the expert panel. For this purpose, mixed infection (bacterial and viral co-infection) is considered as bacterial infections, as they often elicit similar patient management. A final diagnosis is determined based on consensus agreement between all three experts (Fig. 2). The final diagnosis is used for the purpose of calculating the accuracy of the newly developed diagnostic tools. Cases with only majority agreement (2 out of 3 experts) are analysed as a separate sub-group. The remaining cases are labelled as undetermined. Specifically, this label is assigned in cases where each of the experts assigns a different diagnosis or in cases where 2 or more of the experts assign an “undetermined” diagnosis. These patients are used for data analysis and are expected to comprise roughly 10% of all patients.
Study stages
The study is conducted in two consecutive stages. Stage A is used to collect molecular, biochemical, microbiological and clinical data on a group of approximately 900 patients. Collected data is used to develop unique diagnostic signatures able to distinguish between bacterial and viral aetiologies, a mass spectrometry based rapid detection technique for identification of microbial pathogens and antimicrobial resistances and a personalized treatment algorithm for tailored antimicrobial therapy that integrates clinical, molecular and biochemical data. In stage B, a second, independent cohort of approximately 300 patients is recruited for the purpose of testing and validating the performances of the multi-parametric model.
Outcomes
Primary endpoint
Sensitivity and specificity for a multi-parametric diagnostic model, incorporating different pathogen- and host-related factors, in differentiating between bacterial and viral aetiology in patients with LRTI and/or sepsis.
Secondary endpoints
-
1.
Sensitivity and specificity for host-related individual blood biomarkers, in differentiating between bacterial and viral aetiology from other aetiologist in patients with LRTI and/or sepsis;
-
2.
Sensitivity and specificity for host-related sets of blood biomarkers, in differentiating Gram positive or Gram negative or atypical aetiology from other disease aetiologist in patients with LRTI and/or sepsis;
-
3.
Monitoring the temporal dynamics of host-related blood biomarkers levels during the course of disease in patients with LRTI and/or sepsis;
-
4.
A list of significant bacterial microbiome components that are associated with poor or favourable clinical outcome in patients with LRTI and/or sepsis;
-
5.
Sensitivity and specificity for liquid chromatography-mass spectrometry (LC-MS/MS) and lipid-based Protein Immobilization (LPI) proteomics-based rapid detection technique in identifying pathogens in clinical samples of patients with LRTI and/or sepsis;
-
6.
A web-based application that recommends physicians with a preferred antimicrobial treatment based on patients clinical, molecular and biochemical data;
-
7.
To define genetic mechanisms underlying the different host response between patients with viral versus bacterial infection.
Sample size calculation
The following power analysis aims to achieve the primary study objective of developing a new diagnostic model for classifying patients with viral and bacterial aetiologist and treatment algorithms to guide antimicrobial prescribing.
Paediatric population analysis
First, we estimate the sample size required to reject the null hypothesis that the sensitivity over the entire population, P, is lower than P0 = 70% (H0: P ≤ P0, H1: P > P0) with significance level of 1% (α = 0.01), power of 80% (=1-β) for a true sensitivity P1 of 90% (P1 - P0 ≥ 0.2). For sufficiently large sample sizes (n > ~ 30) the sample distribution of the sensitivity, is approximately normally distributed, (\( \widehat{p} \) ~ N{P, P(1-P)/n}). Thus, the number of bacterial patients that are required is:
We estimate that the ratio between bacterial and viral paediatric patients is 1:3 and that expert consensus is achieved in 65% of the cases. Thus, the number of patients with an acute infection required in order to reach 76 bacterial patients is 468 = (76 / 25% / 65%). Using similar considerations for computing specificity the sample size for viral patients is also 76. Of note, we anticipate that the above mentioned 468 patients will include 229 (= 468 × 75% × 65%) viral patients, and thus there is no need for additional patients. To assess the host-response based diagnostics accuracy in differentiating patients with an acute infection (both viral and bacterial) and non-infectious patients, assuming P0 = 70% (H0: P ≤ P0, H1: P > P0) with significance level of 1% (α = 0.01), power of 80% (=1-β) for a true sensitivity P1 of 90% (P1 - P0 ≥ 0.2) requires 76 non-infectious patients. Overall, the paediatric cohort should include at least 468 patients with a suspected infectious disease and 76 patients with non-infectious.
Adult population analysis
Using the same model as for the paediatric populations (assuming α = 0.01, 1-β = 0.2, P1 = 90%, P0 = 70% and a consensus rate of 65%) and assuming a viral to bacterial rate of 3:7 yields 390 patients with an infectious disease. Additionally, 76 non-infectious patients will be enrolled (based on similar calculations as for the paediatric population).
Methods for computing the accuracy of a multi-parametric model
The multi-parametric model integrates various host-related and pathogen-related parameters into a single score that determines the likelihood of bacterial and viral aetiology (Fig. 3). The score is computed using a multinomial logistic regression formula that is developed during Stage A of the study. Using predetermined cut-offs, each of the patients is classified as bacterial, viral, or inconclusive (i.e., a marginal immune response that is neither clearly bacterial nor viral). The model diagnosis will be compared with the consensus diagnosis in order to determine its accuracy. The diagnostic accuracy is quantified by computing the following measures: sensitivity, specificity, positive predictive value, negative predictive value, total accuracy, positive likelihood ratio, negative likelihood ratio and diagnostic odds ratio. The area under the receiver operation curve is computed to perform cut-off independent comparisons of different diagnostic methods.
Discussion and expected results
The inappropriate use of antibiotics has severe global health and economic consequences, including the emergence of antibiotic-resistant bacteria [33]. A major driver of antibiotic misuse is the inability to accurately distinguish between bacterial and viral infections based on currently available diagnostic solutions [33]. Many different biomarkers have been investigated in previous studies, none of them being accurate enough to establish or rule out bacterial infections [40]. The TAILORED-Treatment study is a novel collaboration between (university) hospitals, academic institutions and (diagnostic) industry, aimed to collect and analyse ‘multi-omics’ data from patients suffering from LRTI and/or sepsis. Univariate and multivariate analysis of the data will generate new biomarkers to distinguish between bacterial, viral, and non-infectious disease. The multi-omics approach to generate infectious disease algorithms utilised by this study is currently unique within this area of medicine and provides a template for follow-up combined infectious disease data studies in the future.
Some challenges remain for this type of study. First of all, as described previously, in a subset of patients, sampling of blood and nasal swabs at two sampling points (at presentation and after 3–5 days) should allow to measure data on the temporal dynamics of the host-pathogen interaction. However, if patients are not admitted to the hospital or are already discharged after 1 or 2 days, it is hard to collect a second sample. However, consecutive samples are not necessarily needed in order to achieve the primary objective. A second challenge is how to best interpret and efficiently act on the wealth of data available. Therefore, dedicated bioinformatics professionals are part of the TAILORED-Treatment consortium, for building the unique (HoPOIT) database and to develop the treatment algorithms. Finally, the study is a non-interventional study, and therefore, the diagnostic prediction of the model is not used for any clinical, diagnostic or prognostic decision making. After completing the study, utility studies will be needed to establish the clinical impact of any TAILORED-Treatment algorithms developed.
In summary, the aim of this prospective international (TAILORED-Treatment) study is to develop algorithms to differentiate between bacterial and viral infections in children and adults with LRTI and/or sepsis. The study has the potential to improve patient care, reduce unnecessary antibiotic prescribing (thereby reducing the spread of antibiotic resistances) and will contribute positively to institutional, regional and national healthcare economics.
Abbreviations
- AE:
-
Adverse events
- eCRF:
-
Electronic case record form
- ED:
-
Emergency Department
- EMC:
-
Erasmus University Medical Centre Rotterdam
- HAD:
-
Hadassah University Hospital
- LRTI:
-
Lower respiratory tract infection
- PCR:
-
Polymerase chain reaction
- SIRS:
-
Systemic inflammatory response syndrome
- UMCU:
-
University Medical Centre Utrecht
References
Get Smart: Fast facts about antibiotic resistance. [https://www.cdc.gov/antibiotic-use/community/about/fast-facts.html].
Davey P, Brown E, Fenelon L, Finch R, Gould I, Holmes A, Ramsay C, Taylor E, Wiffen P, Wilcox M. Systematic review of antimicrobial drug prescribing in hospitals. Emerg Infect Dis. 2006;12(2):211–6.
Linder JA, Stafford RS. Antibiotic treatment of adults with sore throat by community primary care physicians: a national survey, 1989-1999. JAMA. 2001;286(10):1181–6.
Pulcini C, Cua E, Lieutier F, Landraud L, Dellamonica P, Roger PM. Antibiotic misuse: a prospective clinical audit in a French university hospital. Eur J Clin Microbiol Infect Dis. 2007;26(4):277–80.
Cadieux G, Tamblyn R, Dauphinee D, Libman M. Predictors of inappropriate antibiotic prescribing among primary care physicians. CMAJ. 2007;177(8):877–83.
Antibiotic Use in the United States, 2017: Progress and Opportunities [https://www.cdc.gov/antibiotic-use/stewardship-report/outpatient.html ].
Bartlett JG. Clinical practice. Antibiotic-associated diarrhea. N Engl J Med. 2002;346(5):334–9.
About Antimicrobial Resistance [https://www.cdc.gov/drugresistance/about.html].
Arias CA, Murray BE. Antibiotic-resistant bugs in the 21st century--a clinical super-challenge. N Engl J Med. 2009;360(5):439–43.
Battle of the Bugs: Fighting Antibiotic Resistance [https://www.fda.gov/drugs/resourcesforyou/consumers/ucm143568.htm].
Review on Antimicrobial Resistance: Tackling drug-resistant infections globally: final report and recommendations. [https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf].
Battleman DS, Callahan M, Thaler HT. Rapid antibiotic delivery and appropriate antibiotic selection reduce length of hospital stay of patients with community-acquired pneumonia: link between quality of care and resource utilization. Arch Intern Med. 2002;162(6):682–8.
Craig JC, Williams GJ, Jones M, Codarini M, Macaskill P, Hayen A, Irwig L, Fitzgerald DA, Isaacs D, McCaskill M. The accuracy of clinical symptoms and signs for the diagnosis of serious bacterial infection in young febrile children: prospective cohort study of 15 781 febrile illnesses. BMJ. 2010;340:c1594.
Houck PM, Bratzler DW, Nsa W, Ma A, Bartlett JG. Timing of antibiotic administration and outcomes for Medicare patients hospitalized with community-acquired pneumonia. Arch Intern Med. 2004;164(6):637–44.
Houck PM, Bratzler DW, Niederman M, Bartlett JG. Pneumonia treatment process and quality. Arch Intern Med. 2002;162(7):843–4.
De Miguel-Yanes JM, Andueza-Lillo JA, Gonzalez-Ramallo VJ, Pastor L, Munoz J. Failure to implement evidence-based clinical guidelines for sepsis at the ED. Am J Emerg Med. 2006;24(5):553–9.
Metersky ML, Sweeney TA, Getzow MB, Siddiqui F, Nsa W, Bratzler DW. Antibiotic timing and diagnostic uncertainty in Medicare patients with pneumonia: is it reasonable to expect all patients to receive antibiotics within 4 hours? Chest. 2006;130(1):16–21.
Little PS, Williamson I. Are antibiotics appropriate for sore throats? Costs outweigh the benefits. Bmj. 1994;309(6960):1010–1.
Spiro DM, Tay KY, Arnold DH, Dziura JD, Baker MD, Shapiro ED. Wait-and-see prescription for the treatment of acute otitis media: a randomized controlled trial. Jama. 2006;296(10):1235–41.
Little P. Delayed prescribing of antibiotics for upper respiratory tract infection. Bmj. 2005;331(7512):301–2.
Zwart S, Sachs AP, Ruijs GJ, Gubbels JW, Hoes AW, de Melker RA. Penicillin for acute sore throat: randomised double blind trial of seven days versus three days treatment or placebo in adults. Bmj. 2000;320(7228):150–4.
Fendrick AM, Monto AS, Nightengale B, Sarnes M. The economic burden of non-influenza-related viral respiratory tract infection in the United States. Arch Intern Med. 2003;163(4):487–94.
John JF Jr, Fishman NO. Programmatic role of the infectious diseases physician in controlling antimicrobial costs in the hospital. Clin Infect Dis. 1997;24(3):471–85.
Study: Antibiotics problems cost U.S. between $17B and $26B a year [http://www.bizjournals.com/triangle/stories/2009/10/19/daily4.html].
Roberts RR, Hota B, Ahmad I, Scott RD 2nd, Foster SD, Abbasi F, Schabowski S, Kampe LM, Ciavarella GG, Supino M, et al. Hospital and societal costs of antimicrobial-resistant infections in a Chicago teaching hospital: implications for antibiotic stewardship. Clin Infect Dis. 2009;49(8):1175–84.
Antibiotic resistance - Frequently asked question [http://www.euro.who.int/en/health-topics/disease-prevention/antimicrobial-resistance/antibiotic-resistance/frequently-asked-questions].
Gerber JS, Ross RK, Bryan M, Localio AR, Szymczak JE, Wasserman R, Barkman D, Odeniyi F, Conaboy K, Bell L, et al. Association of Broad- vs narrow-Spectrum antibiotics with treatment failure, adverse events, and quality of life in children with acute respiratory tract infections. Jama. 2017;318(23):2325–36.
Sterling SA, Miller WR, Pryor J, Puskarich MA, Jones AE. The impact of timing of antibiotics on outcomes in severe sepsis and septic shock: a systematic review and meta-analysis. Crit Care Med. 2015;43(9):1907–15.
Dellinger RP, Levy MM, Rhodes A, Annane D, Gerlach H, Opal SM, Sevransky JE, Sprung CL, Douglas IS, Jaeschke R, et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2012. Crit Care Med. 2013;41(2):580–637.
Greenberg D, Broides A, Blancovich I, Peled N, Givon-Lavi N, Dagan R. Relative importance of nasopharyngeal versus oropharyngeal sampling for isolation of Streptococcus pneumoniae and Haemophilus influenzae from healthy and sick individuals varies with age. J Clin Microbiol. 2004;42(10):4604–9.
Rhedin S, Lindstrand A, Rotzen-Ostlund M, Tolfvenstam T, Ohrmalm L, Rinder MR, Zweygberg-Wirgart B, Ortqvist A, Henriques-Normark B, Broliden K, et al. Clinical utility of PCR for common viruses in acute respiratory illness. Pediatrics. 2014;133(3):e538–45.
Cruz AT, Demmler-Harrison GJ, Caviness AC, Buffone GJ, Revell PA. Performance of a rapid influenza test in children during the H1N1 2009 influenza a outbreak. Pediatrics. 2010;125(3):e645–50.
Cohen A, Bont L, Engelhard D, Moore E, Fernandez D, Kreisberg-Greenblatt R, Oved K, Eden E, Hays JP. A multifaceted ‘omics’ approach for addressing the challenge of antimicrobial resistance. Future Microbiol. 2015;10(3):365–76.
Levy MM, Fink MP, Marshall JC, Abraham E, Angus D, Cook D, Cohen J, Opal SM, Vincent JL, Ramsay G. 2001 SCCM/ESICM/ACCP/ATS/SIS international Sepsis definitions conference. Intensive Care Med. 2003;29(4):530–8.
Karlsson R, Gonzales-Siles L, Boulund F, Svensson-Stadler L, Skovbjerg S, Karlsson A, Davidson M, Hulth S, Kristiansson E, Moore ER. Proteotyping: proteomic characterization, classification and identification of microorganisms--a prospectus. Syst Appl Microbiol. 2015;38(4):246–57.
Karlsson R, Davidson M, Svensson-Stadler L, Karlsson A, Olesen K, Carlsohn E, Moore ER. Strain-level typing and identification of bacteria using mass spectrometry-based proteomics. J Proteome Res. 2012;11(5):2710–20.
Gonzales-Siles L, Karlsson R, Kenny D, Karlsson A, Sjoling A. Proteomic analysis of enterotoxigenic Escherichia coli (ETEC) in neutral and alkaline conditions. BMC Microbiol. 2017;17(1):11.
Karlsson R, Thorell K, Hosseini S, Kenny D, Sihlbom C, Sjoling A, Karlsson A, Nookaew I. Comparative analysis of two helicobacter pylori strains using genomics and mass spectrometry-based proteomics. Front Microbiol. 2016;7:1757.
Cohen JF, Korevaar DA, Altman DG, Bruns DE, Gatsonis CA, Hooft L, Irwig L, Levine D, Reitsma JB, de Vet HC, et al. STARD 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration. BMJ Open. 2016;6(11):e012799.
Kapasi AJ, Dittrich S, Gonzalez IJ, Rodwell TC. Host biomarkers for distinguishing bacterial from non-bacterial causes of acute febrile illness: a comprehensive review. PLoS One. 2016;11(8):e0160278.
Organization WH. Pocket book of Hospital Care for Children. Geneva, Switzerland: World Health Organization; 2005.
Goldstein B, Giroir B, Randolph A. International pediatric sepsis consensus conference: definitions for sepsis and organ dysfunction in pediatrics. Pediatr Crit Care Med. 2005;6(1):2–8.
Acknowledgements
The authors would like to acknowledge the following co-workers on the TAILORED-Treatment project, who together with the authors are helping to realize the project goals: Racheli Kreisberg, Vincent Collins, Hans Hoogeveen, Erik Kristiansson, Anders Karlsson, Astrid Heikema, Deborah Horst-Kreft and Julio Font Perez.
Funding
The TAILORED-Treatment study received funding from the EU’s Seventh Framework Programme FP7 under REA grant agreement No. HEALTH-F3–602860-2013 (TAILORED-Treatment; www.tailored-treatment.eu). The EU peer-reviewed the protocol and had no further role in the study.
Availability of data and materials
The datasets generated and/or analysed during the current study will be available to clinicians via a dedicated website (www.TAILORED-Treatment.eu) and upon request to the study coordinator (J. P. Hays one of the authors of this article). Datasets may also be available via individual publications and repositories as results are published by respective TAILORED-Treatment partners.
Author information
Authors and Affiliations
Contributions
All named authors in this article participated in the TAILORED-Treatment study and were responsible for formulating the clinical protocol outlined in this publication, as well as data analysis for the study. All authors have read and approved this manuscript. JH leaded the consortium and was responsible for the communication regarding the EU. CH, KO, EE, AC, DE and LB were responsible for the clinical aspects of the protocol, including patient recruitment. SB, RK, RK, YL, AS, EM and JH provided advice on laboratory testing and helped shape the protocol, especially the sample collection section. DF and EG developed the central database.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Protocol version 4, date 08–08-2014. Approval for this protocol was obtained prior to study start from the Medical Ethical Committee of UMCU (14–104, approval date: 09–09-2014) and the Institutional Review Boards of Hillel Yaffe Medical Centre (HYMC-0108-13 and HYMC-0107-13), Bnai Zion Medical Centre (BNZ-0107-14 and BNZ-0011-14) and Hadassah University Medical Centre (HMO-0007-14 and HMO-0006-14). Written, informed consent will be obtained from each patient-participant by research staff (by research nurse, research fellow or the principal investigator) prior to enrolment in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Questionnaire follow-up. (DOCX 33 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
van Houten, C.B., Oved, K., Eden, E. et al. Observational multi-centre, prospective study to characterize novel pathogen-and host-related factors in hospitalized patients with lower respiratory tract infections and/or sepsis - the “TAILORED-Treatment” study. BMC Infect Dis 18, 377 (2018). https://doi.org/10.1186/s12879-018-3300-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-018-3300-9