Abstract
Background
Maternal vaginal colonization with antibiotic resistant organisms is a growing concern in countries with high antibiotic resistance rates.
Methods
A low vaginal swab was collected from mothers on admission, on discharge and a peri-rectal swab was collected from the neonates born to these mothers on discharge. Routine microbiological methods were used to identify the colonization rates for Escherichia coli, Klebsiella spp. and Streptococcus agalactiae.
Results
The pre-delivery colonization rate among the 250 participants for total Enterobacteriaceae was 18.8%. The colonization rates for Klebsiella spp., E. coli and S. agalactiae were, 12.4, 5.6 and 14.8% respectively. Two Klebsiella spp. and two E. coli isolates were confirmed to be exentend spectrum β lactamase (ESBL) producers with the commonest resistant determinant being blaCTX-M. Post-delivery swabs were collected from 130 participants and the colonization rates were 41.5% for Enterobacteriaceae, 25.4% for Klebsiella spp., 10.8% for E. coli, and 10.8% for S. agalacteiae. Three Klebsiella isolates and one E. coli isolate were confirmed to be ESBL producers with the commonest resistant determinant being blaCTX-M. Considering the 130 participants with both samples, there was a significant increase in the colonization with any Enterobacteriaceae and Klebsiella spp. (p < 0.05). Peri-rectal swabs were collected from neonates in 159 instances. The isolation rates for Enterobacteriaceae was 34%. The genus specific isolation rate for Klebsiella was 21.4% while the rates for E. coli and S.agalactiae were 10.1 and 5.7% respectively. Two of the E. coli were confirmed to be ESBL producers while none of the klebsiellae were identified to be so. Considering these 159 instances where both the mother and baby were sampled, random amplification of polymorphic DNA (RAPD) analysis revealed that Enterbacteriaceae with same strain type was present in 6.9% of the instances, indicating possible transfer between the mother and neonate. The transfer rate for ESBL producers were 0.6%.
Conclusions
The lower level of antimicrobial resistance among these potentially community acquired isolates is encouraging. However, in view of the increasing level of resistance reported elsewhere in the region, regular monitoring is warranted.
Similar content being viewed by others
Background
Antimicrobial resistance is a major global health issue of the current era. Resistant organisms, initially found in health care environments are now being found commonly in community settings. Asymptomatic colonization with resistant organisms is an area that needs to be studied further as these organisms may subsequently cause serious infections in other sites of the given individual or in susceptible individuals [1,2,3].
Neonatal sepsis with antibiotic resistant organisms is an emerging issue. Neonatal sepsis associated with antibiotic resistant Gram negatives is a recognized problem in the Indian Subcontinent [4]. While there are different risk factors for early onset neonatal sepsis (EOS), defined as infections occurring within the first 7 days of birth, majority of the organisms causing EOS are colonizers of the maternal birth canal [5, 6]. Therefore studying the resistance pattern among the maternal vaginal colonizers during late pregnancy could be used as a proxy to predict the potential burden of EOS with antibiotic resistant organisms.
Studies on maternal colonization with Streptococcus agalactiae are frequently reported while studies on Gram negative colonization are not that common. Colonization rate with extended spectrum β lactamase (ESBL) producing Enterobacteriaecae among pregnant women being admitted for delivery in Madagascar has been found to be 18.5% [7]. The isolation rate for ampicillin resistant Escherichia coli was found to increase from 25 to 36% between pre and post-partum periods in a study conducted in Florida, USA while the isolation rates for ampicillin resistant Enterobacteriaciaea was found to increase from 38% in the antenatal period to 51% in the post-partum period [8].
There is an association between bacteria colonizing mothers and their babies [9]. Neonatal colonization with antibiotic resistant organisms also has multiple risk factors. However parental maternal colonization with resistant organisms has been identified as a key risk factor for neonatal colonization in some studies while not in others [10, 11].
Neonatal colonization with antibiotic resistant bacteria may be a transient phenomenon, or it may result in neonatal sepsis. This may also lead to outbreaks associated with resistant organisms [3]. Furthermore, evidence also suggest that those who are colonized may remain carriers for a long period [12].
Despite its obvious importance, research in to the epidemiology of perinatal transmission of resistant organisms remains a neglected area [13]. The current study was conducted with the objectives of identifying the rate of vaginal colonization with multidrug-resistant (MDR) Klebsiella spp. and E. coli among patients being admitted for term vaginal delivery, identifying the rate of vaginal colonization with Group B streptococci and enterococci among same patients, identifying the rate of peri-rectal colonization with multidrug-resistant Klebsiella spp. and E. coli among infants born to the same mothers and identifying the rates of potential transfer between mother and baby.
Methods
This was a prospective observational study which included data gathered using a questionnaire and a laboratory analysis. Subject recruitment and sample collection was conducted at the Professorial Obstetrics Unit, Teaching Hospital, Peradeniya, Sri Lanka and laboratory analysis was carried out in the Antimicrobial Laboratory, Department of Microbiology, Faculty of Medicine, University of Peradeniya, Sri Lanka.
Ethical approval was obtained from the institutional ethical review committee, Faculty of Medicine, University of Peradeniya, Sri Lanka. Informed written consent was obtained from the subjects prior to recruitment to the study.
Mothers being admitted for term vaginal delivery between 01/10/2015 to 06/01/2016 were included in the study while the admission of the baby to special baby care unit, heavy bleeding per-vagina and emergency lower segment Caesarian section (LSCS) were considered as exclusion criteria. Following data were collected from the participants, age of the mother, occupation, number of family members at home, parity, gestation, hospitalizations within the preceding 3 months, history of taking antibiotics within the preceding 3 months, mode of delivery, date of delivery, weight of the baby and date of discharge.
Subjects were selected according to convenience sampling method. Each subject was given a study number and details of the participant, her baby and samples were collected under the given number. Under each subject number, three swabs were collected as,
-
a.
A low-vaginal swab from mother collected at the time of admission to antenatal ward,
-
b.
A low-vaginal swab from mother collected at the time of discharge from postnatal ward,
-
c.
A peri-rectal swab from the baby of the mother collected on discharge.
All swabs were transported to the laboratory in Amie’s transport medium within 2 h of collection. Swabs were stored at − 20 °C till further testing.
All three swabs were inoculated on to quality controlled Blood agar and MacConkey agar plates and low vaginal swabs were then enriched in quality controlled Todd Hewitt broth. All plates and enrichment broth were incubated at 37 °C for 18–24 h. Lactose fermenting colonies of Gram negative bacilli or non-lactose fermenting, oxidase negative colonies of Gram negative bacilli were presumptively selected further identification. Up to ten morphologically distinct colonies were picked from per plate. These were purified and stored at − 80 °C before further identification and sensitivity testing.
Preliminary identification of Gram negative isolates was carried out using the oxidase test, oxidative/fermentative test (O/F Test) and motility test. Identification up to species level was done according to the Cowan and Steel’s Manuel for the Identification of Medical Bacteria including sugar fermentation tests and other biochemical tests. The analysis was restricted mainly to E. coli isolates and Klebiella species.
Preliminary identification of the Gram positives was carried out with the catalase test, bile esculin test and 6.5% NaCl salt tolerance test. Lancefield grouping of the selected isolates were confirmed by rapid latex agglutination method (Streptococcal Grouping kit, Thermo-Scientific, USA).
Antibiotic Sensitivity Testing (ABST) with disc diffusion method was done on Enterobacteriaceae isolates identified up to species level according to clinical and laboratory standards institute (CLSI) guidelines for cefotaxime, ceftriaxone, ceftazidime, imipenem, ertapenem, gentamicin and ciprofloxacin [14, 15].
Enterococcus isolates were sub cultured on vancomycin resistant enterococci (VRE) agar medium (Thermoscientific, 2016) to screen for vancomycin resistance. Minimum Inhibitory Concentration (MIC) for vancomycin was done on isolates identified as potential VRE using agar dilution method [14, 16].
Zone diameters for cefotaxime and ceftazidime for E. coli and Klebsiella species were used to identify potential ESBL producers according to the CLSI 2015 guidelines [14]. Those identified as potential ESBL producers were further tested with the combined disk test. All isolates demonstrating non-susceptibility for ertapenem and/or imipenem were tested for carbapenemase production by the Modified Hodge Test (MHT).
BlaCTX-M, blaSHV and blaTEM were looked for as genetic determinants of ESBL production by a previously described multiplex PCR [17]. In instances where the same Enterobacteriaceae with the same antibiograme were isolated from the mother and the neonate, these isolates were selected for typing with random amplification of polymorphic DNA (RAPD). RAPD was performed using previously described primers and PCR conditions [18]. Dendrogrammes were drawn using the JelJ software using the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) method [19]. Isolates that had a similarity of > 75% in the Dice index at a tolerance of 3.5 were considered as possibly having the same origin indicating potential transmission between the mother and baby. Enterobacteriaceae isolates that were non-susceptible for ≥3 antimicrobial categories as defined by Magiorakos et al. were identified as being MDR isolates [20].
Results
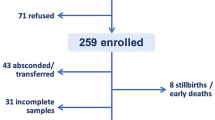
During the study period, 250 term pregnant women who were admitted to the study unit, were recruited for the study. On admission low-vaginal swabs (swab a) were collected from all the participants (250). Pre-delivery colonization rates were calculated including all 250 participants recruited.
Post-delivery swabs (swab b) were collected from 130 of the 250 participants. The 120 participants in whom a post-delivery swab was not available belonged to one of the following four categories; delivered via emergency LSCS (n = 56), discharged without delivery (n = 19), admission of the baby to special care baby units (n = 8) or discharged prior to research team collecting the swab (n = 37).
A peri-rectal swab was collected from 159 babies (swab c). All three swabs were collected from 120 participant mother-baby combinations while a maternal and a neonatal swab (a and/or b + c) was available in 159 participant pairs (Fig. 1). Differences in pre and post-delivery colonization rates were calculated only for the 130 participants where both swabs (a and b) were available. Potential mother to child transmissions were assessed in instance where at least one maternal swab and the peri-anal swab from the baby were available (n = 159).
Maternal ages ranged from 17 to 41 years with a mean age of 28.3 (SD 5.3 years). Majority of the mothers were in their first pregnancy (110/250, 44%). Hundred and eighty nine (75.6%) of the participants were housewives while the others were employed. Twenty five (10%) participants had one or more co-morbid conditions that were pre-existing or pregnancy associated. Fifteen of the participants had either gestational or non-gestational diabetes mellitus, 8 participants had gestational or essential hypertension, 2 participants were on treatment for hypothyroidism and 1 participant was diagnosed to have anaemia. The mean duration of hospitalization was 3.9 days (SD 2.1), the mean stay till delivery was 1.8 days (SD 1.9) and the mean duration of hospital stay for the baby was 2.2 days (SD 1.2).
Pre-delivery bacterial colonization (N = 250)
S. agalactiae colonization rate was 14.8% (n = 37) while the colonization rate for Enterococcus spp. was 24.8% (n = 62). All enterococcus isolates had vancomycin of MICs < 2 μg/ml.
The colonization rate for at least one Enterobacteriaceae isolate was 18.8% (n = 47). The genus specific colonization rate for Klebsiella spp. was 12.4% (n = 31). Two Klebsiella spp. isolates (2/31,6.45%) were confirmed to be ESBL producers harbouring all three genetic determinants tested (blaCTX-M, blaSHV and blaTEM). All Klebsiella spp. were sensitive to imipenem, meropenem and ertapenem as well as ciprofloxacin and levofloxacin. Only one isolate could be defined as a MDR organism.
The species specific colonization rate for E. coli was 5.6% (n = 14). Two E. coli isolates were confirmed to be ESBL producers (2/14, 14.3%) and harboured blaCTX-M only. All E. coli isolates were sensitive to imipenem, meropenem and ertapenem. All E. coli isolates were also sensitive to ciprofloxacin and levofloxacin. None of the E. coli were MDR organisms.
Pre-delivery colonization rate for ESBL producing Klebsiella spp. or E.coli was 1.6% (4/250). Carbapenem resistance rate for Klebsiella spp. or E.coli was 0%. The pre-delivery colonization rate for MDR Klebsiella spp. or E.coli was 0.4% (1/250).
Post-delivery colonization rates (N = 130)
Hundred and thirty participants were sampled in the post delivery period. S. agalactiae colonization rate was 10.8% (n = 14) while the colonization rate for Enterococcus spp. was 17.7% (n = 23). All enterococcus isolates had vancomycin of MICs < 2 μg/ml.
Fifty four patients (41.5%) were colonized with at least one Enterobacteriaceae. Thirty three (25.4%) patients carried Klebsiella spp. Three isolates (3/33, 9.1%) were confirmed to be ESBL producers. One of the ESBL producers carried all three determinants of ESBL, namely blaCTX-M, blaSHV and blaTEM, while one isolate each carried blaCTX-M, and blaTEM.. One isolate was found to be resistant to ertapenem and imipenem while being sensitive to meropenem. This isolate was found to be a carbapanamase producer by the MHT. Two isolates were resistant to ciprofloxacin and levofloxacin (2/33,6.1%). Only one isolate could be defined as a MDR organism for being resistant to more than three classes of antibiotics.
The species specific colonization rate for E. coli was 10.8% (n = 14). One isolate was confirmed to be an ESBL producer harbouring both blaCTX-M and blaTEM. All isolates were sensitive to meropenem, imipenem, ertapenem, ciprofloxacin and levofloxacin. None of the E. coli were MDRs.
Therefore, the post-delivery colonization rate with ESBL producing Klebsiella spp. or E. coli was 3.1% (4/130). Colonization rate for carbapenem resistant Klebsiella spp. or E. coli was 0.8% (1/130). The colonization rate for MDR Klebsiella spp. or E. coli among the post-delivery cohort was also 0.8% (1/130).
Comparison of pre and post-delivery colonization rates
This analysis was restricted to the 130 instance where a pre and a post-delivery swab were taken from the same patient. There was a significant increase in the colonization with any Enterobacteriaceae and Klebsiella spp. (Table 1). The reduction in the colonization rates for Enterococcus species was also significant. Changes seen in S. agalactiae and E. coli were not statistically significant.
Considering antibiotic resistance, the MDR Klebsiella spp. or E. coli rate in the 130 patients where a both swabs were available was 0.8% (1/130) in the pre-delivery period as well as the post delivery period. The colonization rates for ESBL producing Klebsiella spp. or E. coli were 2.3% (3/130) and 3.1% (4/130) during the pre and post delivery periods. The difference was not statistically significant.
Neonatal colonization (N = 159)
S. agalactiae colonization rate was 5.7% (n = 9) while the colonization rate for Enterococcus spp. was 11.9% (n = 19). MIC of one enterococcus for vancomycin was 4 μg/ml while it was < 2 μg/ml in the other isolates. Enterobacteriaceae were found in 34.0% (n = 54) of neonates while Klebsiella spp. were found among 21.4% (n = 34) and E. coli was found in 10.1% (n = 16) of the babies. Two E. coli isolates were confirmed to be ESBL producers while none of the Klebsiella species were found to be ESBL producers. The two ESBL producers harboured both blaCTX-M, and blaTEM. None of the Klebsiella spp. or E. coli isolates were resistant to carbapenems, ciprofloxacin or levofloxacin. None of the isolates were MDRs. Therefore, the colonization rate among the neonates with ESBL producing Klebsiella spp. or E. coli was 1.2% (2/159) and the rates were 0% for carbapenem resistant or MDR Klebsiella spp. or E. coli.
Potential transfer of organisms from mother to baby (N = 159)
This was assessed using the 159 pairs of maternal-neonatal pairss where either pre and/or post delivery swab from the mother and the neonatal peri-rectal swab was available. S. agalactiae was found in both the mother and the baby in four instances (2.5%). While Enterococcus species was found in mother –baby pairs in 13 instances (8.2%).
There were 23 (14.5%) instances where both the mother and the baby were colonized with the same Enterobacteriaceae with the same antibiogramme, where 4 (2.5%) maternal-neonatal paires were colonized with E. coli, 18 (11.3%) were with Klebsiella spp. (K. oxytoca − 3, K. pneumoniae – 15) and one was with Enterobacter cloacae (n = 1).
Analysis of RAPD results revealed that two of the four E. coli found in maternal neonatal pairs showed similarity of more than 75% in at a tolerance of 3.5 (Fig. 2). Of the 18 pairs of Klebsiella isolates, eight pairs showed similarity of more than 75% in at a tolerance of 3.5 (Fig. 3). The Enterobacter cloacae found in the maternal-neonatal pair showed a similarity of > 75% (Not shown in figure). Therefore, considering only the isolates with > 75% similarity in Dice index, eleven pairs isolates (11/159, 6.9%) could have been from the same origin therefore, potentially transferred between the mother and the baby. Only one (1/159, 0.6%) of these 11 was an ESBL producer and while none were MDR organisms.
Dendrogramme to demonstrate potential transfer from mother to baby – Escherichia coli isolates. Numbers along the axis indicate the percentage similarity in Dice index. Isolates demonstrating similarity > 75%: 190b – 190c and 149b – 149 c. a: pre-delivery low vaginal swab, b: post-delivery low vaginal swab, c: peri-rectal swab from the baby. The number denotes the study number allocated to a given mother-baby pair
Dendrogramme to demonstrate potential transfer from mother to baby – Klebsiella isolates. Numbers along the axis indicate the percentage similarity in Dice index. Isolates demonstrating similarity > 75%: 1b-1c, 72b-72c, 74b-74c, 82b-82c, 113b-113c, 139b-139c, 142a-142b-142c, 173b-173c. a: pre-delivery low vaginal swab, b: post-delivery low vaginal swab, c: peri-rectal swab from the baby. The number denotes the study number allocated to a given mother-baby pair. *We could not compare all Klebsiella isolates in a single gel due to inconsistencies of the marker used. Therefore, the three gels used were analysed separately as (a,b and c). However, isolates from a single mother-neonate pair were in the same gel
Discussion
We identified 47 expectant mothers to be colonized with Enterobacteriaceae giving a percentage of 18.8% while the genus specific colonization rate for Klebsiella spp. was 12.4% (n = 31) and the species specific colonization rate for E. coli was 5.6% (n = 14).
In a study conducted in Pakistan in 2008, the E. coli colonization among pre-delivery mothers has been reported as 13.7% (n = 100), while the colonization rate for Klebsiella pneumoniae was 10.5% (n = 77) and for β haemolytic streptococci it was 8.8% (n = 64) among 1923 subjects [21]. A study conducted in 2012 in Argentina revealed an 18.55% (n = 48) colonization rate for Enterobacteriaceae among 259 pregnant women, and the species specific colonization rates were 14.3% (n = 37) for E.coli and 1.2% (n = 3) Klebsiella pneumoniae [22]. E. coli colonization percentage in pregnant women in Assam, India during 2014 was reported as 16.26% among 246-screened samples [23]. Our overall colonization rate for Enterobacteriaecae was similar to the Argentinean study. Our colonization rate for E .coli at 5.6% was lower than that reported from the neighboring India and Pakistan. Our colonization rate for Klebsiella spp. at 12.4% was similar to that reported from Pakistan. These variations might reflect differences in sample collecting, processing or hygienic and environmental conditions in the given locales.
In the current study, we found a pre-delivery colonization rate of S. agalactieae was 14.8% (n = 37) while the colonization rate for Enterococcus spp. was 24.8% (n = 62). Colonization rate for S. agalactieae reported by Dissanayake et al. in the same study unit was 24% [24]. The difference may be due to the direct enrichment employed Dissanayake et al. whereas in this study, enrichment for GBS was done following initial plating of the swabs. A temporal shift could also explain the given difference as there has been a 5 year lapse between the studies.
We have identified a significant increase in the colonization rate with Enterobacteriaceae during the post-delivery period. This has been the case in previous studies for over decades, and the presence of altered blood and dead tissue in the vaginal tract during this period with the associated changes in the environment has been cited as the possible reasons [25].
The colonization rates for ESBL producing Klebsiella spp. or E .coli were 1.6, 3.1 and 1.2% among the pre-delivery, post-delivery and the neonatal periods. Considering the 130 participants where both pre and post delivery samples were available, the rates did not differ significantly. The rates identified are much lower than that reported from Madagascar and other areas for the pre-delivery colonization rates [7]. In Sri Lanka, hospital isolates have been reported to be much higher [26, 27]. However, the previous Sri Lankan studies were conducted considering patients who had clinical illnesses, such as urinary tract infections. Even among patients with community onset infections, the exposure to previous antibiotics has been high [27]. However, our study cohort was pregnant females, who have minimal health care exposure in hospital settings. Only 10% of the participants had any co-morbid conditions, this is also favourable for a lower rate of colonization with resistant organisms [28]. It is encouraging to see that despite high resistance rates among hospital isolates, the rates remain lower normal flora of healthy groups. Furthermore, the length of stay in hospital among the study cohort was relatively short (Mean, 3.9 days), further contributing to the lower ESBL colonization rate, even among the post-delivery period.
The colonization rates for MDR Klebsiella spp. or E. coli were 0.4, 0.8 and 0% among the pre-delivery, post-delivery and the neonatal periods. Considering the 130 participants where both pre and post delivery samples were available, the rates did not differ significantly. The explanations given for the lower ESBL colonization rates are valid for the lower rates of colonization with multi-drug resistant organisms as well.
Among the ESBL producing Enterobacteriaceae, the commonest genetic determinant of resistance was found to be blaCTX-M, keeping up with previous studies including more community onset samples and unlike those originating from health care associated samples [26, 29]..
The potential maternal-neonatal transfer rate identified in the study was 6.9% for Enterobacteriaceae and it was 0.6% for ESBL producers. While the rates are not too high, keeping a vigilant eye on changing epidemiology is needed.
As the transfer events for MDR organisms was low, we did not perform a risk factor analysis.
The potential for the different colonizers identified in this study to cause EOS differs and mortality associated with EOS caused by different organisms also differ [30]. Furthermore, there is scarcity of data on organisms causing EOS in Sri Lanka. Therefore, what we have identified here, as colonization rates among mothers and neonates who are healthy, can only be used as a surrogate to the pool of organisms that may emerge as causing EOS. Furthermore, we excluded the babies who were admitted to the special care baby units from the analysis due to logistic reasons. A follow-up study where infants are monitored to see if they develop subsequent neonatal sepsis and if so correlate the causative organisms for neonatal sepsis with the colonizers identified from the mother and baby would be useful to establish the actual risk of antibiotic resistant maternal colonizers causing sepsis.
There are a few limitations to this study. We collected a low vaginal swab from the mother and a peri-rectal swab from the child while a peri-rectal swab from the mother may have helped to identify more women colonized with resistant organisms [31]. Samples were stored in – 20 °C prior to testing, which is not the ideal but the only feasible method given the circumstances. We identified the resistance rates only among Klebsiella spp. and E. coli. Not all Enterobacteriaceae or Gram negatives were considered. We also did not perform sensitivity testing on the Group B streptococci. The sample size included was also relatively lower. We gathered patient details including co morbidities by direct questioning of the participant than by going through hospital records, which may not be the most accurate method to gather the data. As we had to change the molecular marker used for agarose gels in during the study period, we could not compare the potential transfer of all Klebsiella isolates in a single dendrogramme. However, as what we were interested in is to compare the nature of isolates obtained from a single maternal-neonate pair than the whole cohort, this did not affect the results.
Despite the limitations, our study is one of the very first in Sri Lanka which looks at the potential colonization with resistant organisms among a healthy population.
Conclusions
The lower rates of resistance identified among vaginal colonizers from pregnant females being admitted for delivery including ESBL producers, MDR organisms and carbapenem resistant isolates is encouraging.
However, steps need to be taken to periodically monitor this group of individuals as an increase resistance rates among vaginal colonizers in term pregnant women would have implications on the treatment options used for puerperal sepsis as well as early onset neonatal sepsis.
Abbreviations
- CLSI:
-
Clinical and Laboratory Standards Institute
- EOS:
-
Early onset neonatal sepsis
- ESBL:
-
Extended spectrum βlactamase
- GBS:
-
Group B streptococci
- MDR:
-
Multi drug resistant
- MHT:
-
Modified Hodge test
- MIC:
-
Minimum inhibitory concentration
- PCR:
-
Polymerase chain reaction
- RAPD:
-
Random amplification of polymorphic DNA
- SD:
-
Standard deviation
- UPMGA:
-
Unweighted Pair Group Method with Arithmetic Mean
- VRE:
-
Vancomycin resistant enterococci
References
Bilinski J, Robak K, Peric Z, Marchel H, Karakulska-Prystupiuk E, Halaburda K, Rusicka P, Swoboda-Kopec E, Wroblewska M, Wiktor-Jedrzejczak W, Basak GW. Impact of gut colonization by antibiotic-resistant bacteria on the outcomes of allogeneic hematopoietic stem cell transplantation: a retrospective, Single-Center Study. Biol Blood Marrow Transplant. 2016;22(6):1087–93. https://doi.org/10.1016/j.bbmt.2016.02.009. Epub 2016 Feb 18. PubMed PMID: 26900084
Woerther PL, Andremont A, Kantele A. Travel-acquired ESBL-producing Enterobacteriaceae: impact of colonization at individual and community level. J Travel Med. 2017;24(suppl_1):S29–34. https://doi.org/10.1093/jtm/taw101. Review.PubMed PMID: 28520999; PubMed Central PMCID: PMC5441303
Bonfanti P, Bellù R, Principe L, Caramma I, Condò M, Giani T, Rossolini GM, Luzzaro F. Mother-to-child transmission of KPC Carbapenemase-producing Klebsiella pneumoniae at birth. Pediatr Infect Dis J. 2017;36(2):228–9. https://doi.org/10.1097/INF.0000000000001403. PubMed PMID: 27846056
Viswanathan R, Singh AK, Basu S, Chatterjee S, Sardar S, Isaacs D. Multi-drug resistant gram negative bacilli causing early neonatal sepsis in India. Arch Dis Child Fetal Neonatal Ed. 2012;97(3):F182–7. https://doi.org/10.1136/archdischild-2011-300097. Epub 2011 Dec 7. PubMed PMID: 22155619
Polin RA. Committee on fetus and newborn. Management of neonates with suspected or proven early-onset bacterial sepsis. Pediatrics. 2012;129(5):1006–15. https://doi.org/10.1542/peds.2012-0541. Epub 2012 Apr 30. PubMed PMID: 22547779
Simonsen KA, Anderson-Berry AL, Delair SF, Davies HD. Early-onset neonatal sepsis. Clin Microbiol Rev. 2014;27(1):21–47. https://doi.org/10.1128/CMR.00031-13. Review. PubMed PMID: 24396135; PubMed Central PMCID: PMC3910904
Chereau F, Herindrainy P, Garin B, Huynh BT, Randrianirina F, Padget M, Piola P, Guillemot D, Delarocque-Astagneau E. Colonization of extended-spectrum-β-lactamase- and NDM-1-producing Enterobacteriaceae among pregnant women in the community in a low-income country: a potential reservoir for transmission of multiresistant Enterobacteriaceae to neonates. Antimicrob Agents Chemother. 2015;59(6):3652–5. https://doi.org/10.1128/AAC.00029-15. Epub 2015 Apr 6. PubMed PMID: 25845871; PubMed Central PMCID: PMC4432137
Edwards RK, Clark P, Sistrom CL, Duff P. Intrapartum antibiotic prophylaxis 1: relative effects of recommended antibiotics on gram-negative pathogens. Obstet Gynecol. 2002;100(3):534–9. PubMed PMID: 12220774
Lebon A, Moll HA, Tavakol M, van Wamel WJ, Jaddoe VW, Hofman A, Verbrugh HA, van Belkum A. Correlation of bacterial colonization status between mother and child: the Generation R Study. J Clin Microbiol. 2009;48(3):960–2. https://doi.org/10.1128/JCM.01799-09. Epub 2009 Nov 25. PubMed PMID: 19940045; PubMed CentralPMCID: PMC2832455
Denkel LA, Schwab F, Kola A, Leistner R, Garten L, von Weizsäcker K, Geffers C, Gastmeier P, Piening B. The mother as most important risk factor for colonization of very low birth weight (VLBW) infants with extended-spectrum β-lactamase-producing Enterobacteriaceae (ESBL-E). J Antimicrob Chemother. 2014;69(8):2230–7. https://doi.org/10.1093/jac/dku097. Epub 2014 Apr 11. PubMed PMID: 24729603
Li X, Xu X, Yang X, Luo M, Liu P, Su K, Qing Y, Chen S, Qiu J, Li Y. Risk factors for infection and/or colonisation with extended-spectrum β-lactamase-producing bacteria in the neonatal intensive care unit: a meta-analysis. Int J Antimicrob Agents. 2017;50(5):622–8. https://doi.org/10.1016/j.ijantimicag.2017.06.027. Epub 2017 Jul 18. Review. PubMed PMID: 28733213
Nordberg V, Jonsson K, Giske CG, Iversen A, Aspevall O, Jonsson B, Camporeale A, Norman M, Navér L. Neonatal intestinal colonization with extended-spectrum β-lactamase-producing Enterobacteriaceae-a 5-year follow-up study. Clin Microbiol Infect. 2018; pii: S1198-743X(18)30045–4. https://doi.org/10.1016/j.cmi.2017.12.028. [Epub ahead of print] PubMed PMID: 29326011
Seale J, Millar M. Perinatal vertical transmission of antibiotic-resistantbacteria: a systematic review and proposed research strategy. BJOG. 2014;121(8):923–8. https://doi.org/10.1111/1471-0528.12746. Epub 2014 Mar 27. Review. PubMed PMID: 24674346
Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: 23rd informational supplement M100-S25. Wayne, PA: Clinical and laboratory standards institute; 2015.
Clinical and Laboratory Standards Institute. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—9th ed. CLSI document M07-A9. Wayne, PA: Clinical and laboratory standards institute; 2012.
Clinical and Laboratory Standards Institute. Performance standards for antimicrobial disk susceptibility tests; approved standard—11th ed CLSI document M02-A11. Wayne, PA: Clinical and laboratory standards institute; 2012.
Monstein HJ, Ostholm-Balkhed A, Nilsson MV, Nilsson M, Dornbusch K, Nilsson LE. Multiplex PCR amplification assay for the detection of blaSHV, blaTEM and blaCTX-M genes in Enterobacteriaceae. APMIS. 2007;115(12):1400–8.
Barbarini D, Russello G, Brovarone F, Capatti C, Colla R, Perilli M, Moro ML, Carretto E. Evaluation of carbapenem-resistant Enterobacteriaceae in an Italian setting: report from the trench. Infect Genet Evol 2015 Mar;30:8–14. doi: https://doi.org/10.1016/j.meegid.2014.11.025. PubMed PMID: 25444940.
Heras J, Domínguez C, Mata E, Pascual V, Lozano C, Torres C, Zarazaga M. GelJ--a tool for analyzing DNA fingerprint gel images. BMC Bioinf. 2015 Aug 26;16:270. https://doi.org/10.1186/s12859-015-0703-0. PubMed PMID: 26307353; PubMedCentral PMCID: PMC4549892.
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012 Mar;18(3):268-281. https://doi.org/10.1111/j.1469-0691.2011.03570.x. Epub 2011 Jul 27. PubMed PMID: 21793988.
Mumtaz S, Ahmad M, Aftab I, Aktar N, Haassan MN, Hamid A. Aerobic vaginal pathogens and their sensitivity pattern. J Ayub Med Coll Abbottabad. 2008;20(1):113–7.
Villar HE, Aubert V, Baserni MN, Jugo MB. Maternal carriage of extended-spectrum beta-lactamase-producing Escherichia coli isolates in Argentina. J Chemother. 2013;25(6):324–7. https://doi.org/10.1179/1973947813Y.0000000081. Epub 2013 Dec 6. PubMed PMID: 24091027
Devi U, Berman N, Barua P, Malik V, Das JK, Baruah P, Mahanta J. Vaginal carriage of antibiotic resistant Escherichia coli by pregnant women; a concern for the neonates. Clin Microbiol. 2014;3(4):153–6.
Dissanayake BN, Herath GC, Gamage TM. Group B β hemolytic Streptococcus colonization in Pregnancy. Sri Lanka J Infect Dis. 2015;5(1), 13–18. DOI: http://doi.org/10.4038/sljid.v5i1.7918.
Goplerud CP, Ohm MJ, Galask RP. Aerobic and anaerobic flora of the cervix during pregnancy and the puerperium. Am J Obstet Gynecol. 1976;126(7):858–68. PubMed PMID: 793391
Patabendige CGUA, Chandrasiri NS, Karunanayake LI, Karunaratne GKD, Somaratne P, Elwitigala JP, Chandrasiri P. Antimicrobial resistance in resource-poor settings – Sri Lanka experience. Reg Health Forum. 2011;15:18–26.
Fernando MM, Luke WA, Miththinda JK, Wickramasinghe RD, Sebastiampillai BS, Gunathilake MP, Silva FH, Premaratna R. Extended spectrum beta lactamase producing organisms causing urinary tract infections in Sri Lanka and their antibiotic susceptibility pattern -a hospital based cross sectional study. BMC Infect Dis. 2017;17(1):138. https://doi.org/10.1186/s12879-017-2250-y. PubMed PMID: 28187754; PubMed Central PMCID: PMC5303299
Schoevaerdts D, Bogaerts P, Grimmelprez A, de Saint-Hubert M, Delaere B, Jamart J, Swine C, Glupczynski Y. Clinical profiles of patients colonized or infected with extended-spectrum beta-lactamase producing Enterobacteriaceae isolates: a 20 month retrospective study at a Belgian University Hospital. BMC Infect Dis. 2011;11(12) https://doi.org/10.1186/1471-2334-11-12. PubMed PMID: 21226923; PubMed Central PMCID: PMC3023698
Hall JM, Corea E, Sanjeewani HD, Inglis TJ. Molecular mechanisms of β-lactam resistance in carbapenemase-producing Klebsiella pneumoniae from Sri Lanka. J Med Microbiol. 2014;63(Pt 8):1087–92. https://doi.org/10.1099/jmm.0.076760-0. Epub 2014 May 22. PubMed PMID: 24855071
Shane AL, Sánchez PJ, Stoll BJ. Neonatal sepsis. Lancet 2017 Oct 14;390(10104):1770–1780. https://doi.org/10.1016/S0140-6736(17)31002-4. Epub 2017 Apr 20. Review. PubMed PMID: 28434651.
Zamfir M, Adler AC, Kolb S, Dammeyer A, Nasri L, Schomacher L, Karlin B, Franitza M, Hörmansdorfer S, Tuschak C, Valenza G, Ochmann U, Herr C. Evaluation of sampling locations in pregnant women and newborns for the detection of colonisation with antibiotic-resistant bacteria. Eur J Clin Microbiol Infect Dis 2017 Oct;36(10):1819–1826. https://doi.org/10.1007/s10096-017-2997-5. Epub 2017 May 4. PubMed PMID: 28474179.
Funding
Faculty of Medicine, University of Peradeniya is acknowledged for financial assistance through grant RG/EF/2015/04.
Availability of data and materials
Dataset generated and/or analyzed during the current study are not publicly available as consent for data sharing has not been obtained and ethical clearance for data sharing has not been taken but data can be obtained from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
DN – Data collection, sample collection, sample analysis, data analysis, drafting of the paper and approval of final paper. VL – Conceptualization of the study, designing of the study, data analysis, drafting of the paper and approval of final paper. CK – Designing of study, troubleshooting data collection, drafting and final approval of paper. CG - Designing of study, troubleshooting data collection, drafting and final approval of paper.AE – Sample analysis, Microbiological testing and troubleshooting of microbiological assays, final approval of paper. DA – Molecular biological testing, data analysis, drafting and final approval of the paper.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethical clearance was obtained from the institutional ethical review committee, Faculty of Medicine, University of Peradeniya and informed written consent was obtained from study participants.
Consent for publications
Not applicable.
Competing interests
Authors declare that they have no conflicts of interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Nanayakkara, D., Liyanapathirana, V., Kandauda, C. et al. Maternal vaginal colonization with selected potential pathogens of neonatal sepsis in the era of antimicrobial resistance, a single center experience from Sri Lanka. BMC Infect Dis 18, 351 (2018). https://doi.org/10.1186/s12879-018-3262-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-018-3262-y