Abstract
Background
Given the predominance of invasive fungal disease (IFD) amongst the non-immunocompromised adult critically ill population, the potential benefit of antifungal prophylaxis and the lack of generalisable tools to identify high risk patients, the aim of the current study was to describe the epidemiology of IFD in UK critical care units, and to develop and validate a clinical risk prediction tool to identify non-neutropenic, critically ill adult patients at high risk of IFD who would benefit from antifungal prophylaxis.
Methods
Data on risk factors for, and outcomes from, IFD were collected for consecutive admissions to adult, general critical care units in the UK participating in the Fungal Infection Risk Evaluation (FIRE) Study. Three risk prediction models were developed to model the risk of subsequent Candida IFD based on information available at three time points: admission to the critical care unit, at the end of 24 h and at the end of calendar day 3 of the critical care unit stay. The final model at each time point was evaluated in the three external validation samples.
Results
Between July 2009 and April 2011, 60,778 admissions from 96 critical care units were recruited. In total, 359 admissions (0.6 %) were admitted with, or developed, Candida IFD (66 % Candida albicans). At the rate of candidaemia of 3.3 per 1000 admissions, blood was the most common Candida IFD infection site. Of the initial 46 potential variables, the final admission model and the 24-h model both contained seven variables while the end of calendar day 3 model contained five variables. The end of calendar day 3 model performed the best with a c index of 0.709 in the full validation sample.
Conclusions
Incidence of Candida IFD in UK critical care units in this study was consistent with reports from other European epidemiological studies, but lower than that suggested by previous hospital-wide surveillance in the UK during the 1990s. Risk modeling using classical statistical methods produced relatively simple risk models, and associated clinical decision rules, that provided acceptable discrimination for identifying patients at ‘high risk’ of Candida IFD.
Trial registration
The FIRE Study was reviewed and approved by the Bolton NHS Research Ethics Committee (reference: 08/H1009/85), the Scotland A Research Ethics Committee (reference: 09/MRE00/76) and the National Information Governance Board (approval number: PIAG 2-10(f)/2005).
Similar content being viewed by others
Background
Once seen typically in immunocompromised patients, invasive fungal disease (IFD) amongst the critically ill is now predominantly found in the adult, general, non-immunocompromised population [1]. Over 5000 cases of IFD due to Candida species (Candida IFD) occur in the UK each year, with 40 % occurring in critical care units [2]. Epidemiological trends in IFD among the critically ill are changing, due in large part to the increased incidence of IFD risk factors and to new therapeutic strategies.
A number of randomised controlled trials have evaluated prophylactic and or empiric therapy with antifungals in non-neutropenic, critically ill patients. Despite patient heterogeneity, these trials have demonstrated a beneficial effect of antifungal prophylaxis on the risk of developing proven IFD and suggested a reduction in mortality [3]. Given that the effectiveness of antifungal prophylaxis has only been demonstrated in groups at high risk of IFD and that more widespread use of antifungal drugs may promote resistance and drive up costs, it is necessary to establish a method to identify high risk patient groups, who stand to benefit most from an antifungal prophylactic strategy. Several risk models and clinical decision rules have been proposed for identifying patients at high risk of IFD [4–8]. However, model generalisability has been limited owing to the restricted study population, being either post-surgical patients [7, 8], or patients already colonised with Candida [8].
Given the potential benefit of antifungal prophylaxis and lack of generalisable tools to identify high risk patients, the aim of the current study was to describe the epidemiology of IFD in UK critical care units and to develop and validate a clinical risk prediction tool to predict the risk of Candida IFD at three decision time points: at admission to the critical care unit, at 24 h following admission, and at the end of the third calendar day following admission, in order to identify non-neutropenic, critically ill adult patients at high risk of IFD who would benefit from antifungal prophylaxis.
Methods
Study design
Based on the results of a systematic review of the literature conducted by our research team [5] and on expert clinical opinion, data on risk factors for, and outcomes from, IFD were collected for consecutive admissions to adult, general critical care units in the UK participating in the Fungal Infection Risk Evaluation (FIRE) Study [9]. These data were linked with additional patient data collected for national clinical audits – the Case Mix Programme (in England, Wales and Northern Ireland) and the Scottish Intensive Care Society Audit Group (in Scotland) – to form the FIRE Study database. For all sources, data were collected prospectively and abstracted by trained data collectors and underwent extensive validation.
FIRE Study database
For all critical care unit admissions, data were extracted on age, sex, medical history, surgical status, acute severity of illness, primary reason for admission, therapies received, IFD, fungal colonisation, mortality and length of stay.
Severe comorbidities were defined using the APACHE II method [10] and must have been evident in the 6 months prior to critical care unit admission. Surgery within up to 7 days prior to admission to the critical care unit was classified as either emergency/urgent or scheduled/elective. Acute severity of illness was summarised using the APACHE II Score [10] and the ICNARC Physiology Score [11] assessed during the first 24 h following admission to the critical care unit.
Data on the following therapies were collected: total parenteral nutrition; systemic antimicrobials; immunosuppressive therapy; central venous catheters, organ support; and antifungal use. Corticosteroids were included as immunosuppressives. Organ support was recorded throughout the critical care unit stay, defined according to the UK Department of Health Critical Care Minimum Dataset [12].
Fungal colonisation was defined as the presence of yeasts in any sample reported on a microbiology system and was recorded as the date that a positive report was available – i.e. the point at which a treatment decision could be made based on this knowledge.
IFD was defined as a blood culture or sample from a normally sterile site (including, but not restricted to: cerebrospinal fluid; peritoneal fluid; pleural fluid; and pericardial fluid; and excluding bronchoalveolar lavage, urine and sputum) positive for yeast/mould cells in a microbiological or histopathological report. This definition was based on the Revised Definitions of Invasive Fungal Disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group [13]. This definition was chosen to best capture Candida IFD and was recognised to under-represent IFD due to other species. Timing of IFD was defined by the date on which the positive sample was collected. All patients reported to have IFD (pre critical care unit or during critical care unit stay), and a random sample of 2 % of those reported not to have IFD, were independently rechecked against hospital notes and microbiology records by the local investigator, blinded to the original data. For admissions that developed IFD during critical care unit stay, the timing of the first systemic antifungal was also reported relative to the timing of IFD.
Patients were followed up for mortality and length of stay until death or final discharge from acute hospital and those transferred to another acute hospital.
Descriptive epidemiology
Analyses were performed using Stata/SE Version 10.1 (StataCorp LP, College Station TX). For the purpose of summarising case mix, outcomes and antifungal use, the cohort was divided into groups of: admissions with IFD positive for Candida albicans (Candida albicans IFD); admissions with IFD positive for other Candida species (non-albicans Candida spp IFD); and admissions with no IFD either prior to or during the critical care unit stay (no IFD). Admissions with IFD positive for Candida of unknown species or non-Candida species were excluded due to small numbers. Admissions with IFD positive for both Candida albicans and non-albicans Candida species were included in the Candida albicans subgroup.
Development and validation of risk prediction models
Three risk prediction models were developed to model the risk of subsequent Candida IFD based on information available at three time points: admission to critical care unit, at the end of 24 h and at the end of calendar day 3 of the critical care unit stay.
The following exclusions were applied for the development and validation of the risk model at admission: age less than 18 years; second and subsequent admissions of the same patient; neutropenia (absolute neutrophil count less than 1 × 109 l−1); active haematological malignancy; admission following solid organ transplant; IFD identified up to 7 days prior to admission; and receipt of systemic antifungals up to 7 days prior to admission.
For the models at 24 h and at the end of calendar day 3, exclusions were as above plus any of the following occurring before the decision time point: death or discharge from the critical care unit; IFD; or receipt of systemic antifungals.
The dataset was divided into the following development and validation samples: development sample – all admissions to a random sample of participating critical care units in England, Wales and Northern Ireland, July 2009 to December 2010 (selected to include approximately two thirds of all admissions); random validation sample – all admissions to the remaining units in England, Wales and Northern Ireland; temporal validation sample – all admissions to units in the development sample, January to March 2011; and geographical validation sample – all admissions to units in Scotland.
The risk prediction models were derived in the development sample using logistic regression models with robust standard errors to allow for clustering within critical care units. All candidate variables were included in a ‘full’ multivariable model and the model was progressively simplified using backwards-stepwise selection with the least statistically significant being removed at each step.
Model discrimination was assessed with the c index [14], equivalent to the area under the receiver operating characteristic curve [15], calibration by graphical plots of observed against expected risk, and overall fit by Brier’s score [16], the mean squared error between outcome and prediction. Bootstrapping with 200 bootstrap samples was used on the development sample to internally validate the final selected model at each time point and to estimate optimism adjusted measures of the model discrimination and overall fit [17]. The final model at each time point was evaluated in the three external validation samples: a random sample, a geographic sample and a temporal sample, as described above. Each sample was chosen to test different aspects of future performance. The models were evaluated in each validation sample separately and then in all three samples combined using the same measures as in the development sample.
The sensitivity, specificity, positive predictive value and negative predictive value of the risk model at the end of calendar day 3 was compared with that of existing clinical decision rules identified from the systematic review of the literature using the full validation dataset. For comparison with the clinical decision rules, three alternative risk thresholds were applied to the risk predictions from the FIRE Study model corresponding to predicted risks of >0.5 % (F1), >1 % (F2) and >2 % (F3). The following existing clinical decision rules were included in the comparison: three rules presented in Ostrosky-Zeichner et al. [6] (OZ1-OZ3) and three rules presented in Paphitou et al. [7] (P1-P3).
Results
Between July 2009 and April 2011, a total of 96 critical care units with 60,778 admissions participated in the FIRE Study (see Fig. 1). The units were representative of all UK adult general critical care units in terms of geographical distribution, hospital teaching status and number of beds.
In total, 359 admissions (0.6 %) were admitted with, or developed, Candida IFD of which 66 % were Candida albicans (see Fig. 2). The most common non-albicans Candida species was Candida glabrata (17 %).
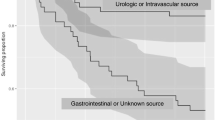
Admissions with non-albicans Candida spp IFD were compared to admissions with Candida albicans IFD as shown in Table 1. The critical care unit and acute hospital lengths of stay and mortality rates were comparable between the Candida albicans and non-albicans Candida spp. IFD subgroups but substantially higher when compared with admissions with no IFD. Overall crude critical care unit and acute hospital mortality for admissions with any Candida species IFD were 29.9 and 39.6 %, respectively.
The most common Candida IFD infection site was blood (57 %; Additional file 1: Table S1) corresponding to a rate of candidaemia of 3.3 per 1000 admissions. Of the 359 total admissions with IFD, approximately half had pre-critical care unit IFD (identified from a sample taken pre-admission or on calendar day 1 or 2) with the other half developing IFD during their critical care unit stay (identified from a sample taken from calendar day 3 onwards; Additional file 2: Figure S1). The median day for Candida IFD developed during the critical care unit stay was day 7 (IQR 4 to 12). The incidence of Candida IFD developed during the critical care unit stay was 3.1 cases per 1000 admissions.
Both Candida spp. IFD subgroups received similar organ support and systemic antifungals (Additional file 3: Table S2). In the admissions with Candida IFD that received systemic antifungals, 27 % received these before the first sample from which IFD was identified, 47 % within 3 days of the first positive sample and 27 % more than 3 days after the first positive sample.
Characteristics of the patients included in the development and validation samples are reported in the Additional file 4: Table S3. Of 46 potential variables, 19 candidate variables were selected for the admission model and, following backwards elimination, the final model contained seven variables with 11 parameters (c index 0.705; Table 2 and Additional file 5: Table S4). The 24-h model contained seven variables with 10 parameters (c index 0.824) and the end of calendar day 3 model contained five variables with seven parameters (c index 0.835). Validation of the models is reported in Table 3. The end of calendar day 3 model performed the best with a c index of 0.709 in the full validation sample.
The comparison of clinical decision rules based on the end of calendar day 3 FIRE Study model with existing clinical decision rules is reported in Table 4. The best performing of the existing rules was rule OZ3. This rule had better sensitivity, specificity, PPV and NPV than rules P1 and P3, while the remaining rules (OZ1, OZ2 and P2) gave higher sensitivity but at the cost of treating 43 to 83 % of admissions. Applying thresholds of >0.5 % (F1) or >1 % (F2) to risk predictions from the FIRE Study model gave similar performance to rule OZ3, with rule F1 giving slightly higher sensitivity but lower specificity and rule F2 giving slightly lower sensitivity but higher specificity (Additional file 6: Figure S2).
Discussion
The FIRE Study is the first multicentre study to report specifically on Candida IFD in UK critical care units and to develop and validate risk prediction tools to identify critically ill non-neutropenic adults at risk for IFD. Candida albicans accounted for two thirds of Candida IFD. Blood was the most common infection site, accounting for more than half of all Candida IFD. The risk prediction tools were developed at three decision time points: at admission to the critical care unit, at 24 h following admission, and at the end of calendar day 3 following admission. The final model at admission to the critical care unit had fair discrimination (c index ~0.7). When additional information from the first 24 h following admission was added, discrimination improved (c index ~0.8) and this level of discrimination was maintained at the end of calendar day 3. When clinical decision rules were defined based on cut-points of predicted risk at the end of calendar day 3, the performance of these rules was similar to the best performing rule from the literature.
The major strength of the FIRE Study is the huge sample size of admissions to a large number of critical care units across the UK, providing extremely representative and generalisable results. The definition of Candida IFD was chosen to be consistent with current international consensus definitions and was carefully validated. However, despite the huge sample size, the low rate of Candida IFD observed (although clearly good for patients) made robust statistical modelling challenging. The observed rate of Candida IFD was approximately half that anticipated from the literature and consequently the resulting models had a lower number of events per variable than ideal, which may have contributed to the drop in model performance when assessed in the validation samples. Model performance was worst when applied in the geographical validation sample, despite a similar rate of Candida IFD, suggesting that particular care should be taken in transferring the models to different geographical settings. A further potential limitation is the reliance on routinely available data – for example, the definition of Candida colonisation was based on samples sent for microbiological evaluation in normal clinical practice, and we did not specify any particular screening schedule. This may contribute to variation across critical care units, and therefore to how well the models validated in external data.
The rate of candidaemia in the present study, at 3.3 per 1000 admissions, is similar to other reports from European critical care units [18–20]. A previous hospital-wide surveillance from six sentinel hospitals in the UK identified that 45 % of candidaemia was reported from the critical care unit, corresponding to an incidence of 7.4 per 1000 admissions [21]. A prospective study in 24 French critical care units demonstrated a candidaemia incidence of 6.7 per 1000 admissions [22].
The distribution of Candida species in the present study is also similar to that of other critical care units in Western Europe [20]. A retrospective analysis of the EPIC II study, examining Candida bloodstream infections in 14,414 patients to 1265 critical care units in 76 countries, demonstrated varying proportions of Candida albicans and non-albicans Candida spp. infections [23]. Seventy two percent of the Candida infections in Western European units were due to Candida albicans, as compared to 66 % in the present study and 79 % in the previous UK sentinel hospital study [21]. Variations in proportions of non-albicans Candida spp infections may be due to differential use of fluconazole prophylaxis and subsequent emergence of resistant strains [1]. An analysis of Candida isolates reported to the Communicable Disease Surveillance Centre from England and Wales between 1990 and 1999 found that Candida albicans was responsible for 60 % of all clinically significant isolates [24]. Annual reporting between 1990 and 1999 showed increasing rates of reported Candida species. However, more recent data from the Health Protection Agency have shown a decline in both the total number of Candida bloodstream infections and the proportion of these due to Candida albicans (51 % in 2010) [25].
A recent systematic review of the literature, conducted as part of the FIRE Study [5], identified only one previous study with the explicit aim of developing a risk model for IFD, the “Candida score” [8]. The Candida score was developed in a cohort of 1699 patients admitted to 73 critical care units in Spain and has subsequently been externally validated among 1107 patients admitted to 36 critical care units in Spain, Argentina and France [26]. There are substantial differences between the rationale and approach of the Candida score and the FIRE Study. The Estudio de Prevalencia de Candidiasis (EPCAN) project, from which the Candida score was developed, recruited only patients staying at least 7 days in the critical care unit, and the Candida score was developed using only those diagnosed with colonisation by Candida species at any time during the critical care unit stay. The FIRE Study demonstrated that the risk factors, and the strength of their association with Candida IFD, varied between admission to the critical care unit and the end of calendar day 3. A model developed using data from later in the critical care unit stay may therefore not accurately reflect risk earlier in the stay, limiting the usefulness of the Candida score for making early decisions regarding antifungal prophylaxis. Similarly, the comparison of clinical decision rules based on the FIRE Study models with existing rules was limited to the decision time point at the end of calendar day 3 as all previous clinical decision rules have been based solely on data from patients staying at least 3 days [6, 7].
Conclusion
In summary, incidence of Candida IFD in UK critical care units in this study was consistent with reports from other European epidemiological studies, but lower than that suggested by previous hospital-wide surveillance in the UK during the 1990s. Risk modeling using classical statistical methods produced relatively simple risk models, and associated clinical decision rules, that provided acceptable discrimination for identifying patients at ‘high risk’ of Candida IFD.
Abbreviations
- APACHE II:
-
Acute physiology and chronic health evaluation II
- EORTC/MSG:
-
European organization for research and treatment of cancer/invasive fungal infections cooperative group and the national institute of allergy and infectious diseases mycoses study group
- EPCAN:
-
The estudio de prevalencia de candidiasis
- EPIC II:
-
Extended prevalence of infection in the ICU study
- FIRE:
-
Fungal infection risk evaluation study
- HTA:
-
Health technology assessment
- ICNARC:
-
Intensive care national audit & research centre
- IFD:
-
Invasive fungal disease
- NHS:
-
National health service
- NIHR:
-
National institute for health research
References
Eggimann P, Garbino J, Pittet D. Epidemiology of Candida species infections in critically ill non-immunosuppressed patients. Lancet Infect Dis. 2003;3:685–702.
Health Protection Agency. Fungal diseases in the UK-the current provision of support for diagnosis and treatment: assessment and proposed network solution. 2006.
Playford EG, Webster AC, Sorrell TC, Craig JC. Antifungal agents for preventing fungal infections in non-neutropenic critically ill and surgical patients: systematic review and meta-analysis of randomized clinical trials. J Antimicrob Chemother. 2006;57:628–38.
Blumberg HM, Jarvis WR, Soucie JM, Edwards JE, Patterson JE, Pfaller MA, et al. Risk factors for candidal bloodstream infections in surgical intensive care unit patients: the NEMIS prospective multicenter study. The National Epidemiology of Mycosis Survey. Clin Infect Dis. 2001;33:177–86.
Muskett H, Shahin J, Eyres G, Harvey S, Rowan K, Harrison D. Risk factors for invasive fungal disease in critically ill adult patients: a systematic review. Crit Care. 2011;15:R287.
Ostrosky-Zeichner L, Sable C, Sobel J, Alexander BD, Donowitz G, Kan V, et al. Multicenter retrospective development and validation of a clinical prediction rule for nosocomial invasive candidiasis in the intensive care setting. Eur J Clin Microbiol Infect Dis. 2007;26:271–6.
Paphitou NI, Ostrosky-Zeichner L, Rex JH. Rules for identifying patients at increased risk for candidal infections in the surgical intensive care unit: approach to developing practical criteria for systematic use in antifungal prophylaxis trials. Med Mycol. 2005;43:235–43.
Leon C, Ruiz-Santana S, Saavedra P, Almirante B, Nolla-Salas J, Alvarez-Lerma F. A bedside scoring system (“Candida score”) for early antifungal treatment in nonneutropenic critically ill patients with Candida colonization. Crit Care Med. 2006;34:730–7.
Harrison D, Muskett H, Harvey S, Grieve R, Shahin J, Patel K, Sadique Z, Allen E, Dybowski R, Jit M, Edgeworth J, Kibbler C, Barnes R, Soni N, Rowan K. Development and validation of a risk model for identification of non-neutropenic, critically ill adult patients at high risk of invasive Candida infection: the Fungal Infection Risk Evaluation (FIRE) Study. Health Technology Assessment. 2013;17(3).
Knaus WA, Draper EA, Wagner DP, Zimmerman JE. APACHE II: a severity of disease classification system. Crit Care Med. 1985;13:818–29.
Harrison DA, Parry GJ, Carpenter JR, Short A, Rowan K. A new risk prediction model for critical care: the Intensive Care National Audit & Research Centre (ICNARC) model. Crit Care Med. 2007;35:1091–8.
NHS. Critical care minimum data set. 2010
De Pauw B, Walsh TJ, Donnelly JP, Stevens DA, Edwards JE, Calandra T, et al. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis. 2008;46:1813–21.
Harrell FE, Califf RM, Pryor DB, Lee KL, Rosati RA. Evaluating the yield of medical tests. JAMA. 1982;247:2543–6.
Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36.
Brier GW. Verification of forecasts expressed in terms of probability. Mon Weather Rev. 1950;75:1–3. SRC - GoogleScholar.
Efron B. Estimating the error rate of a prediction rule: improvement on cross-validation. J Am Stat Assoc. 1983;78:316–31. SRC - GoogleScholar.
Blot SI, Vandewoude KH, Hoste EA, Colardyn FA. Effects of nosocomial candidemia on outcomes of critically ill patients. Am J Med. 2002;113:480–5.
Nolla-Salas J, Sitges-Serra A, León-Gil C, Martínez-González J, León-Regidor MA, Ibáñez-Lucía P, et al. Candidemia in non-neutropenic critically ill patients: analysis of prognostic factors and assessment of systemic antifungal therapy. Study Group of Fungal Infection in the ICU. Intensive Care Med. 1997;23:23–30.
Leleu G, Aegerter P, Guidet B, R. Collège des Utilisateurs de Base de Données en. Systemic candidiasis in intensive care units: a multicenter, matched-cohort study. J Crit Care. 2002;17:168–75.
Kibbler CC, Seaton S, Barnes RA, Gransden WR, Holliman RE, Johnson EM, et al. Management and outcome of bloodstream infections due to Candida species in England and Wales. J Hosp Infect. 2003;54:18–24.
Bougnoux M-E, Kac G, Aegerter P, d’Enfert C, Fagon J-Y, G. CandiRea Study. Candidemia and candiduria in critically ill patients admitted to intensive care units in France: incidence, molecular diversity, management and outcome. Intensive Care Med. 2008;34:292–9.
Kett DH, Azoulay E, Echeverria PM, Vincent J-L. Candida bloodstream infections in intensive care units: analysis of the extended prevalence of infection in intensive care unit study. Crit Care Med. 2011;39:665–70.
Lamagni TL, Evans BG, Shigematsu M, Johnson EM. Emerging trends in the epidemiology of invasive mycoses in England and Wales (1990–9). Epidemiol Infect. 2001;126:397–414.
Health Protection Agency. Candidaemia in England, Wales and Northern Ireland, Health Protection Report HCAL, vol. 5, 2010.
Leon C, Ruiz-Santana S, Saavedra P, Galvan B, Blanco A, Castro C. Usefulness of the “Candida score” for discriminating between Candida colonization and invasive candidiasis in non-neutropenic critically ill patients: a prospective multicenter study. Crit Care Med. 2009;37:1624–33.
Acknowledgements
The authors wish to thank the FIRE Study Steering Group (B Riley, R Barnes, J Edgeworth, R Grieve, D Harrison, M Jit, C Kibbler, R McMullan, K Rowan, N Soni, T Stambach), Professor Tim Walsh as Chair and the members of the UK CRN Critical Care Specialty Group, the FIRE Study Team in the ICNARC Clinical Trials Unit and the patients and staff in the participating critical care units.
Funding
The FIRE study project was funded by the National Institute for Health Research (NIHR) Health Technology Assessment (HTA) Programme (project number 07/29/01) and has been published in full in Health Technology Assessment. The views and opinions expressed therein are those of the authors and do not necessarily reflect those of the HTA Programme, NIHR, NHS or the Department of Health.
Availability of data and materials
Anonymised study data are available from the authors, subject to any necessary approvals.
Authors’ contributions
JS, EJA, KP, HM, SEH, SB, JE, MB, CCK, RAB, KMR, DAH all contributed to the study design, interpretation of data and manuscript preparation. JS, EJA, KP, DAH contributed to the analysis.
Competing interests
JE has received payment for lectures from MSD and for travel expenses from Novartis; his institution has received grant funding from Pfizer. CCK has received payment for consultancy and lectures from Astellas, Gilead, MSD and Pfizer. RAB has been a paid advisory board member or received payment for lectures and/or travel expenses from Astellas, Gilead, MSD and Pfizer; her institution has received grant funding from Gilead and Pfizer. The other authors report no conflicts of interest.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The FIRE Study was reviewed and approved by the Bolton NHS Research Ethics Committee (reference: 08/H1009/85) and had approval under Section 251 of the NHS Act 2006 (approval number: PIAG 2-10(f)/2005). In Scotland, the FIRE Study was reviewed by the Scotland A Research Ethics Committee and classified as an audit project not requiring ethics approval.
Author information
Authors and Affiliations
Consortia
Corresponding author
Additional files
Additional file 1: Table S1.
Site of Candida invasive fungal disease (N = 359). (DOC 31 kb)
Additional file 2: Figure S1.
Timing of Candida invasive fungal disease relative to critical care unit admission (N = 359). (DOC 27 kb)
Additional file 3: Table S2.
Therapies received, fungal colonisation, mortality and length of stay by invasive fungal disease subgroup. (DOC 61 kb)
Additional file 4: Table S3.
Description of patients included at each time point in each of the development and validation samples. (DOC 37 kb)
Additional file 5: Table S4.
Final risk models at admission, 24 h and end of calendar day 3. (DOC 44 kb)
Additional file 6: Figure S2.
Area under the operating curve for clinical decision rules. (DOC 110 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Shahin, J., Allen, E.J., Patel, K. et al. Predicting invasive fungal disease due to Candida species in non-neutropenic, critically ill, adult patients in United Kingdom critical care units. BMC Infect Dis 16, 480 (2016). https://doi.org/10.1186/s12879-016-1803-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-016-1803-9