Abstract
Background
Streptococcus suis is a zoonotic pathogen that causes invasive infections in humans and pigs. It has been reported that S. suis infection in humans is mostly caused by serotype 2. However, human cases caused by other serotypes have rarely been reported. This is the first report of a human case of infection with S. suis serotype 31 in Thailand.
Case presentation
A 55-year-old male alcohol misuser with liver cirrhosis was admitted with sepsis to a hospital in the Central Region of Thailand. He had consumed a homemade, raw pork product prior to the onset of illness. He was alive after treatment with ceftriaxone and no complication occurred. An isolate from blood culture at the hospital was suspected as viridans group Streptococcus. It was confirmed at a reference laboratory as S. suis serotype 31 by biochemical tests, 16S rDNA sequencing, and multiplex polymerase chain reaction for serotyping, but it was untypable by the co-agglutination test with antisera against recognized S. suis serotypes, suggesting loss of capsular material. The absence of a capsule was confirmed by transmission electron microscopy. The isolate was confirmed to be sequence type 221, with 13 putative virulence genes that are usually found in serotype 2 strains.
Conclusion
We should be aware of the emergence of S. suis infections caused by uncommon serotypes in patients with predisposing conditions. Laboratory capacity to identify S. suis in the hospital is needed in developing countries, which can contribute to enhanced surveillance, epidemiological control, and prevention strategies in the prevalent area.
Similar content being viewed by others
Background
Streptococcus suis is a zoonotic pathogen that causes invasive infections in humans who have been in close contact with infected pigs or contaminated pork-derived products, and this disease has lately received increasing attention worldwide [1]. Of the 29 serotypes that have been described, most of the clinical isolates from human cases are serotype 2 strains [1]. However, human cases caused by serotypes 1, 4, 5, 14, 16, 21 and 24, as well as an untypable strain, have been reported; in particular, serotypes 5, 14 and 24 and an uncapsulated strain have been found in Thailand [1–3]. Serotypes other than serotype 2 cause different clinical manifestations such as septic arthritis and peritonitis caused by serotype 5, meningitis by serotype 4 and 21, sepsis by serotype 24, and peritonitis by serotype 16 [1]. The diversity of infections caused by serotypes other than serotype 2 is indicative of the awareness of diagnosticians of S. suis infections [1].
Phenotypically, S. suis resembles the viridans streptococcal species Streptococcus gordonii, Streptococcus sanguinis and Streptococcus parasanguinis, and therefore may be misidentified in many human diagnostic laboratories [4]. Therefore, molecular techniques as well as antiserum serotyping are needed for confirmation of S. suis as well as its serotype. We report here a case of sepsis caused by S. suis serotype 31 in a patient in Thailand, using a polyphasic approach to identify this bacterium, as well as the results of genetic characterization of putative virulence gene profile, capsule gene (cps) locus, and genetic relatedness of S. suis serotype 31. The information from this study may increase awareness of the emergence of S. suis infections caused by uncommon strains, which will be important in the development of surveillance systems and epidemiological control and prevention strategies.
Case presentation
A 55-year-old man was admitted to a hospital in Suphanburi Province, a central region in Thailand on December 8, 2012. He had an underlying condition with liver cirrhosis from a history of hepatitis B infection and alcohol misuse. Before admission, he had a history of raw pork product consumption (4 days prior to onset). Physical examination revealed a temperature of 38.6 °C, pulse rate of 102 beats/min, respiratory rate of 23 breaths/min, and blood pressure of 110/65 mmHg. No nuchal stiffness, ecchymosis, or hearing loss was found. Initial diagnosis in this case was sepsis based on systemic inflammatory response syndrome [5]. His white blood cell count was 13,600 cells/μL (70 % neutrophils) and platelet count was 129,000/mm3. A comprehensive metabolic panel revealed elevated creatinine level (1.95 mg/dL), serum albumin level of 3.4 mg/dL, blood urea nitrogen (BUN) level of 18 mg/dL, serum aspartate aminotransferase (AST) level of 660 IU/L, serum alanine aminotransferase (ALT) level of 130 IU/L, and creatine phosphokinase (CPK) level of 740 U/L. A bacterial isolate (no. 43640) was separated from the blood of the patient and identified as viridans group Streptococcus at the hospital laboratory. The isolate was sent to a reference laboratory for confirmation of its species. It was identified as S. suis by biochemical tests, multiplex polymerase chain reaction (PCR), and 16S rRNA gene sequencing (99 % identity) [6–8]. Based on these results, this case was finally diagnosed as sepsis. The patient was treated with ceftriaxone and discharged on December 14, 2012.
Multiplex PCR demonstrated that this isolate belonged to serotype 31; however, antiserum serotyping revealed that this strain did not agglutinate with the antisera of any recognized S. suis serotypes [7, 9]. Further analysis of this strain by transmission electron microscopy confirmed almost complete absence of capsular material around the bacterial cells (Fig. 1) [10], showing the isolate to be unencapsulated. The isolate was susceptible to penicillin (MIC ≤0.12 μg/mL), ceftriaxone (MIC ≤1 μg/mL), erythromycin (MIC ≤0.25 μg/mL), levofloxacin (MIC ≤2.0 μg/mL), clindamycin (MIC ≤0.25 μg/mL), and vancomycin (MIC ≤1.0 μg/mL), and was resistant to tetracycline (MIC ≥8.0 μg/mL). Since breakpoints for S. suis are not defined in the 2014 Clinical and Laboratory Standards Institute guidelines, breakpoints for the viridans group streptococci were used instead.
Multilocus sequence typing (MLST) analysis of this isolate showed that it belonged to sequence type (ST) 221 of clonal complex (CC) 221/234 (Fig. 2) [11]. PCR and sequencing were used to study the presence of 29 genes previously associated with S. suis serotype 2 virulence. These genes include the muramidase-released protein gene (mrp), the extracellular factor gene (epf), the suilysin gene (sly), the arginine deaminase gene (arcA) [12], the factor H-binding surface protein gene (fhb) [13], the fibronectin-binding protein gene (fbps) [14], an infection-related factor gene (trag) [15], the serum opacity factor gene (ofs) [16], the S-ribosylhomocysteinase gene (luxS) [17], the hyaluronate lyase gene (hyl) [18], the glutamine synthetase gene (glnA) [19], an amylopullulanase gene (apuA) [20], an enolase gene (eno) [21], an IgA protease gene [22], the subtilisin-like protease gene (sspA) [23], the sortase A gene (srtA) [24], the sortase BCD gene (srtBCD), the sortase E gene (srtE), the sortase F gene (srtF), the sortase G gene (srtG) [25], the zinc uptake regulator gene (zur) [26], the transcriptional regulator gene (rgg) [27], orphan response regulator genes including covR and revS [28, 29], the two-component regulatory system gene (ciaRH) [30], the divalent-cation-related ABC transporter genes (adcR and fur) [31], an iron-transporter gene (feoB) [32], and the virulence-related gene (virA) [33]. Our analysis of this isolate revealed the presence of 13 genes found in virulent serotype 2 strains. The IgA protease gene, arcA, luxS, glnA, apuA, eno, sspA, srtA, covR, zur, fur, adcR and feoB were present in this isolate; however, the well-recognized virulence markers (epf/mrp/sly) were absent.
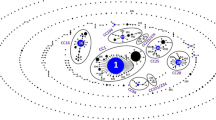
An eBURST analysis of the entire S. suis MLST database (accessed on May 15, 2015). Clonal complexes relevant to human infection in Thailand are circled and labeled. S. suis serotype 31 (ST221) in this study belonged to CC221/234 (bold circle). Clonal complexes and the predicted founders STs are indicated by blue dots. The size of the dots is relative to the number of isolates with the respective ST present in the database
The entire cps locus of the unencapsulated serotype 31 strain 43640 (accession number KM576773) as well as cpsA and cpsB of five encapsulated S. suis serotype 31 strains (p523, p346, p369, p567 and p559; accession numbers KM884773–KM884777) isolated from slaughtered pigs were amplified and sequenced [see Additional file 1: Table S1 and Additional file 2: Table S2]. Our isolate showed an intact cpsA-cpsO (Fig. 3) as described for serotype 31 reference strain 92–4172 (accession number AB737835). No insertions, deletions, or frameshifts were found in the genes of the cps locus. In addition, the nucleotide sequence of cpsA-cpsO in this isolate showed a high identity (97 %) with the reference serotype 31. Comparison of the CpsA–CpsO protein sequences of both strains also revealed high identities (97.75–100 %), except for the CpsA and CpsB proteins, which had lower identities (93.11 and 84.71 %, respectively) with proteins of the reference strain (Fig. 3).
Following the analysis of cpsA and cpsB, the CpsA protein of the unencapsulated strain 43640 was found to be composed of 479 amino acids with the highest identity to serotype 9-CpsA (96.66 %), followed by serotype 10-CpsA (96.45 %) and serotype 24-CpsA (95.62 %) (Fig. 4). The CpsB protein sequence comprised 229 amino acids that were identical to the reference serotype 31 as well as other serotypes. The sequence presented a greater identity to CpsB of the reference serotype 13 (91.27 %) and 24 (91.27 %) strains than to the reference serotype 31 (84.71 %) strain (Fig. 4). As shown in Fig. 4, the CpsA and CpsB sequences of five additional encapsulated S. suis serotype 31 strains isolated from pigs clustered with the reference serotype 31 strain with a high percentage identity (99–100 %), even though these sequences in unencapsulated strain no. 43640 were distant.
Discussion
This is believed to be the first report of an unencapsulated S. suis serotype 31 infection in humans. In general, identification of S. suis as well as its serotypes can be achieved with a polyphasic approach using biochemical testing, antisera serotyping, and/or PCR assays [1]. As mentioned above, human laboratories are less able to identify this organism because of limited resources/technology, knowledge of S. suis, or phenotypic resemblance between S. suis and the viridans streptococcal species or others. Our study demonstrated that the hospital laboratory misidentified S. suis as viridians group Streptococcus. Even though Thailand has a reference laboratory to confirm unidentified bacteria from hospitals using a polyphasic approach, laboratory capacity should be increased, especially in hospital laboratories, to increase their knowledge of S. suis identification.
The strain isolated from this case belonged to ST221 in CC221/234. To the best of our knowledge, only two other unencapsulated strains have been recovered from human cases [3, 34], although unencapsulated S. suis has frequently been isolated from pigs [1, 35]. It is particularly interesting that CC221/234 did not contain any serotype 2 strains, but rather only serotype 24 strains (ST221 and ST234) (http://ssuis.mlst.net/sql/burstspadvanced.asp). This CC is not related to other CCs, especially the CCs that are associated with human and pig infections, such as CC1, CC16, CC20, CC25, CC27 and CC104 (Fig. 2) [1, 11, 36]. We have shown that CC221/234 S. suis serotype 24 can cause sepsis, similar to the isolate in this study [2]. This suggests that CC221/234 is a newly emerging, human infectious clone that does not include serotype 2 strains.
Our isolate revealed the presence of 13 putative virulence genes in virulent serotype 2 strains, as described above. The presence of one or more of these genes may have contributed to the virulence of this strain. Published evidence indicates that mutations in one or more of these putative virulence genes are sufficient to decrease the virulence of S. suis. For example, ApuA promotes adhesion to porcine epithelium and mucus in vitro [20]. The subtilisin-like protease (sspA) and the IgA protease are able to degrade natural proteins in host cells, for example gelatins, fibrinogens, and IgA antibody [22, 23]. Enolase plays an important role in adhesion to and invasion of brain microvascular endothelial cells [21]. Aranda et al. [32] demonstrated that full virulence of S. suis requires the FeoB transporter. Inactivation of glnA in S. suis resulted in reduced mortality and morbidity in murine infection models [19]. In addition, inactivation of zur, covR and luxS interferes with the expression of virulence factors, significantly contributing to attenuation of the virulence of S. suis [17, 26, 28].
The present isolate caused sepsis with signs and symptoms that did not differ from those of infection with S. suis serotype 2 strains. However, it is particularly interesting that patients with infections caused by bacteria other than serotype 2 also have predisposing conditions such as cirrhosis, asplenia and splenectomy: a likely explanation is that these infections are the result of immunocompromise [1–3, 34, 37]. The fact that the strain analyzed in this study was unencapsulated makes it even more atypical. A previous study reported that loss of the capsule may increase bacterial adhesion to host cells, increase biofilm formation that may allow bacteria to become persistent colonizers and to resist clearance by the host immune system and antibiotics, as well as induce the secretion of cytokines such as tumor necrosis factor-α, interleukin (IL-1β, IL-6, and IL-8) [10, 38–40]. Loss of the capsule may exacerbate the immunological response induced by the exposed cell wall components, leading to uncontrolled inflammatory reactions that may promote the development of a severe disease outcome. Moreover, alcohol abuse and liver cirrhosis may have contributed to the impaired neutrophil function in this patient [41, 42]. These observations may explain why the unencapsulated strain could survive in the blood circulation and cause disease.
Three hypotheses have been established to explain the unencapsulated phenotype. (1) Mutation in the promoter region of the cps locus may contribute to unencapsulation. Indeed, it has been shown that Staphylococcus aureus strains with mutations in the promoter region upstream of cap5(8)A fail to transcribe the capsule genes [43]. (2) Some amino acid substitutions that are not due to nonsense mutations and seem to result in an intact protein may also contribute to unencapsulation [44]. That study demonstrated that a single amino acid substitution each in Cps2E and Cps2F was the main cause of the capsule loss in S. suis serotype 2 or 1/2 isolates [44]. In the comparison between our isolate and reference serotype 31 strains, amino acid substitutions were found in the CpsA–CpsO protein sequences; only CpsG and CpsJ were found to have 100 % identity in their amino acid sequences. CpsE and CpsF of this unencapsulated strain revealed seven and one amino acid substitutions, respectively. Other protein sequences had substitutions in one position of CpsD, CpsH, CpsI, CpsN and CpsO; two positions of CpsC; three positions of CpsI; five positions of CpsK and CpsM; and nine positions of CpsL (data not shown). (3) S. suis capsules are believed to be synthesized via the Wzx/Wzy-dependent pathway, with a mature capsule translocated to the cell wall by the Wzd/Wze protein complex encoded by cpsB and cpsC [45]. In our unencapsulated strain, the cpsA and cpsB genes that encoded the CpsA and CpsB proteins had low similarity to those of the reference serotype 31 strain (Figs. 3 and 4). We hypothesize that dissimilar CpsB proteins may affect the compatibility of the protein–protein interactions for capsule processing in this unencapsulated strain. While CpsA is a capsule expression regulator, it appears to modulate the transcription of the capsule mRNA through specific binding to the capsule gene promoter [46, 47]. The dissimilarity of this protein in our isolate and in reference serotype 31 may also result in reduced capsule expression. Incompatibility of the protein–protein or protein–DNA interaction cannot be ruled out.
Similar to our study, Liu et al. [48] reported that the cps genes of S. suis strain YS54 were similar to reference serotype 29 strain 92-1191, except for cpsH and cpsI. The cpsH and cpsI of YS54 were similar to those of reference serotype 21 strain 14A, while the cpsH and cpsI of strain 14A shared no similarity with strain 91-1191 [48]. We speculated that horizontal gene transfer (HGT) may have been involved in cpsA-cpsB replacement/switching in our case; however, we do not know the source of the genes. HGT, via transformation, transduction or conjugation, can lead to the acquisition of entirely new sequences, as well as sequences that are homologous to existing DNA. The transfer of DNA via homologous recombination leads to the replacement of a region of the genome of a recipient cell by the corresponding region from the donor cell [49]. HGT events are frequently observed in Streptococcus such as Streptococcus pyogenes, Streptococcus agalactiae and Streptococcus pneumoniae; for example, genetic studies of S. pneumoniae serotype 6A and 6C capsule gene loci showed that substitution of wciN6C for wciN6A through homologous recombination resulted in a serotype switch from 6A to 6C [50].
Conclusion
Although the isolation rate for this bacterial strain is still low, the emergence of S. suis infections caused by uncommon serotypes as well as unencapsulated strains should raise awareness in patients with predisposing conditions. Increased laboratory capacity to identify S. suis in hospitals is necessary in Thailand as well as in developing countries to contribute towards enhanced surveillance, epidemiological control, and prevention strategies for public health. Further investigations, such as complementation assay, should be considered to demonstrate clearly the mechanisms of capsular loss in this particular isolate [44].
Consent
Written informed consent was obtained from the patient for publication of this Case report. A copy of the written consent is available for review by the Editor of this journal.
References
Goyette-Desjardins G, Auger JP, Xu J, Segura M, Gottschalk M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg Microbe Infect. 2014;3, e45.
Kerdsin A, Dejsirilert S, Sawanpanyalert P, Boonnark A, Noithachang W, Sriyakum S, et al. Sepsis and spontaneous bacterial peritonitis in Thailand. Lancet. 2011;378:960.
Kerdsin A, Takeuchi D, Gottschalk M, Hamada S, Akeda Y, Oishi K. A human case of Streptococcus suis infection caused by an unencapsulated strain. JMM Case Rep. 2014. doi:10.1099/jmmcr.0.002329.
Facklam R. What happened to the Streptococci: overview of taxonomy and nomenclature changes. Clin Microbiol Rev. 2002;15:613–30.
Muchart DJ, Bhaganjee S. American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference definitions of the systemic inflammatory response syndrome and allied disorders in relation to critically injured patients. Crit Care Med. 1997;25:1765–95.
Hommez J, Devriese LA, Henrichsen J, Castryck F. Identification and characterization of Streptococcus suis. Vet Microbiol. 1986;11:349–55.
Kerdsin A, Akeda Y, Hatrongjit R, Detchawna U, Sekizaki T, Hamada S, et al. Streptococcus suis serotyping by a new multiplex PCR. J Med Microbiol. 2014;63:824–30.
Chatellier S, Harel J, Zhang Y, Gottschalk M, Higgins R, Devriese LA, et al. Phylogenetic diversity of Streptococcus suis strains of various serotypes as revealed by 16S rRNA gene sequence comparison. Int J Syst Bacteriol. 1998;48:581–9.
Gottschalk M, Higgins R, Boudreau M. Use of polyvalent coagglutination reagents for serotyping of Streptococcus suis. J Clin Microbiol. 1993;31:2192–4.
Bonifait L, Gottschalk M, Grenier D. Cell surface characteristics of non-typeable isolates of Streptococcus suis. FEMS Microbiol Lett. 2010;311:160–6.
King SJ, Leigh JA, Heath PJ, Luque I, Tarradas C, Dowson CG, et al. Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype exchange. J Clin Microbiol. 2002;40:3671–80.
Silva LM, Baum CG, Rehm T, Wisselink HJ, Goethe R, Valentin-Weigand P. Virulence-associated gene profiling of Streptococcus suis isolates by PCR. Vet Microbiol. 2006;115:117–27.
Pian Y, Gan S, Wang S, Guo J, Wang P, Zheng Y, et al. Fhb, a novel factor H-binding surface protein, contributes to the antiphagocytic ability and virulence of Streptococcus suis. Infect Immun. 2012;80:2402–13.
De Greeff A, Buys H, Verhaar R, Dijkstra J, van Alphen L, Smith HE. Contribution of fibronectin-binding protein to pathogenesis of Streptococcus suis serotype 2. Infect Immun. 2002;70:1319–25.
Zhang H, Fan H, Lu C. Identification of a novel virulence-related gene in Streptococcus suis type 2 strains. Curr Microbiol. 2010;61:494–9.
Baums CG, Kaim U, Fulde M, Ramachandran G, Goethe R, Valentin-Weigand P. Identification of a novel virulence determinant with serum opacification activity in Streptococcus suis. Infect Immun. 2006;74:6154–62.
Cao M, Feng Y, Wang C, Zheng F, Li M, Liao H, et al. Functional definition of LuxS, an autoinducer-2 (AI-2) synthase and its role in full virulence of Streptococcus suis serotype 2. J Microbiol. 2011;49:1000–11.
King SJ, Allen AG, Maskell DJ, Dowson CG, Whatmore AM. Distribution, genetic diversity, and variable expression of the gene encoding hyaluronate lyase within the Streptococcus suis population. J Bacteriol. 2004;186:4740–7.
Si Y, Yuan F, Chang H, Liu X, Li H, Cai K, et al. Contribution of glutamine synthetase to the virulence of Streptococcus suis serotype 2. Vet Microbiol. 2009;139:80–8.
Ferrando ML, Fuentes S, de Greeff A, Smith H, Wells JM. ApuA, a multifunctional α-glucan-degrading enzyme of Streptococcus suis, mediates adhesion to porcine epithelium and mucus. Microbiol. 2010;156:2818–28.
Esgleas M, Li Y, Hancock MA, Harel J, Dubreuil JD, Gottschalk M. Isolation and characterization of α-enolase, a novel fibronectin-binding protein from Streptococcus suis. Microbiol. 2008;154:2668–79.
Zhang A, Mu X, Chen B, Liu C, Han L, Chen H, et al. Identification and characterization of IgA1 protease from Streptococcus suis. Vet Microbiol. 2010;140:171–5.
Bonifait L, Dominguez-Punaro MC, Vaillancourt K, Bart C, Slater J, Frenette M, et al. The cell envelope subtilisin-like proteinase is a virulence determinant for Streptococcus suis. BMC Microbiol. 2010;10:42.
Wang C, Li M, Feng Y, Zheng F, Dong Y, Pan X, et al. The involvement of sortase A in high virulence of STSS-causing Streptococcus suis serotype 2. Arch Microbiol. 2009;191:23–33.
Takamatsu D, Noshino H, Ishiji T, Ishii J, Osaki M, Fittipaldi N, et al. Genetic organization and preferential distribution of putative pilus gene clusters in Streptococcus suis. Vet Microbiol. 2009;138:132–9.
Feng Y, Li M, Zhang H, Zheng B, Han H, Wang C, et al. Functional definition and global regulation of Zur, a zinc uptake regulator in a Streptococcus suis serotype 2 strain causing streptococcal toxic shock syndrome. J Bacteriol. 2008;190:7567–78.
Zheng F, Ji H, Cao M, Wang C, Feng Y, Li M, et al. Contribution of the Rgg transcription regulator to metabolism and virulence of Streptococcus suis serotype 2. Infect Immun. 2011;79:1319–28.
Pan X, Ge J, Li M, Wu B, Wang C, Wang J, et al. The orphan response regulator CovR: a globally negative modulator of virulence in Streptococcus suis serotype 2. J Bacteriol. 2009;191:2601–12.
De Greeff A, Buys H, van Alphen L, Smith HE. Response regulator important in pathogenesis of Streptococcus suis serotype 2. Microb Pathog. 2002;33:185–92.
Li J, Tan C, Zhou Y, Fu S, Hu L, Hu J, et al. The two-component regulator system CiaRH contributes to the virulence of Streptococcus suis 2. Vet Microbiol. 2011;148:99–104.
Aranda J, Garrido ME, Cortes P, Llagostera M, Barbe J. Analysis of the protective capacity of three Streptococcus suis proteins induced under divalent-cation-limited conditions. Infect Immunol. 2008;76:1590–8.
Aranda J, Cortes P, Garrido ME, Fittipaldi N, Llagostera M, Gottschalk M, et al. Contribution of the FeoB transporter to Streptococcus suis virulence. Int Microbiol. 2009;12:137–43.
Li P, Liu J, Zhu L, Qi C, Bei W, Cai X, et al. VirA: a virulence-related gene of Streptococcus suis serotype 2. Microb Pathog. 2010;49:305–10.
Gomez E, Kennendy CC, Gottschalk M, Cunningham SA, Patel R, Virk A. Streptococcus suis-related prosthetic joint infection and streptococcal toxic shock-like syndrome in a pig farmer in the United States. J Clin Microbiol. 2014;52:2254–8.
Lakkitjaroen N, Takamatsu D, Okura M, Sato M, Osaki M, Sekizaki T. Loss of capsule among Streptococcus suis isolates from porcine endocarditis and its biological significance. J Med Microbiol. 2011;60:1669–76.
Schultsz C, Jansen E, Keijzers W, Rothkamp A, Duim B, Wagenaar JA, et al. Differences in the population structure of invasive Streptococcus suis strains isolated from pigs and from humans in the Netherlands. PLoS One. 2012;7, e33854.
Kopic J, Paradzik MT, Pandak N. Streptococcus suis infection as a cause of severe illness: 2 cases from Croatia. Scan J Infect Dis. 2002;34:683–4.
Benga L, Goethe R, Rohde M, Valentin-Weigand P. Non-encapsulated strains reveal novel insights in invasion and survival of Streptococcus suis in epithelial cells. Cell Microbiol. 2004;6:867–81.
Tanabe S, Bonifait L, Fittipaldi N, Grignon L, Gottschalk M, Grenier D. Pleiotropic effects of polysaccharide capsule loss on selected biological properties of Streptococcus suis. Can J Vet Res. 2010;74:65–70.
Segura M, Vanier G, Al-Numani D, Lacouture S, Olivier M, Gottschalk M. Proinflammatory cytokine and chemokine modulation by Streptococcus suis in a whole-blood culture system. FEMS Immunol Med Microbiol. 2006;47:92–106.
Shawcross DL, Wright GAK, Stadbauer V, Hodges SJ, Davies NA, Wheeler-Jones C, et al. Ammonia impairs neutrophil phagocytic function in liver disease. Hepatol. 2008;48:1202–12.
Tritto G, Bechlis Z, Sadblauer V, Davis N, Francis R, Shah N, et al. Evidence of neutrophil functional defect despite inflammation in stable cirrhosis. J Hepatol. 2011;55:574–81.
Cocchiaro JL, Gomez MI, Risley A, Solinga R, Sordell DO, Lee JC. Molecular characterization of the capsule locus from non-typable Staphylococcus aureus. Mol Microbiol. 2006;59:948–60.
Lakkitjaroen N, Takamatsu D, Okura M, Sato M, Osaki M, Sekizaki T. Capsule loss or death: The position of mutations among capsule genes sways the destiny of Streptococcus suis. FEMS Microbiol Lett. 2014;354:46–54.
Okura M, Takamatsu D, Maruyama F, Nozawa T, Nakagawa I, Osaki M, et al. Genetic analysis of capsular polysaccharide synthesis gene clusters from all serotypes of Streptococcus suis: potential mechanisms for generation of capsular variation. Appl Environ Microbiol. 2013;79:2796–806.
Hanson BR, Lowe BA, Neely MN. Membrane topology and DNA-binding ability of Streptococcal CpsA protein. J Bacteriol. 2011;193:411–20.
Hanson BR, Runft DL, Streeter C, Kumar A, Carion TW, Neely MN. Functional analysis of the CpsA protein of Streptococcus agalactiae. J Bacteriol. 2012;194:1668–78.
Liu Z, Zheng H, Gottschalk M, Bai X, Lan R, Ji S, et al. Development of multiplex PCR assays for the identification of the 33 serotypes of Streptococcus suis. PLoS One. 2013;8, e72070.
Smith JM, Dowson CG, Spratt BG. Localized sex in bacteria. Nature. 1991;349:29–31.
Park IH, Park S, Hollingshead SK, Nahm MH. Genetic basis for the new pneumococcal serotype, 6C. Infect Immun. 2007;75:4482–9.
Acknowledgments
This work was supported by research grants from Division of Research and Academic Services, Kasetsart University Chalermphrakiat Sakon Nakhon Province Campus, the Ministry of Health, Labor and Welfare of Japan, and The Japan Initiative for Global Research Network on Infectious Diseases launched by the Ministry of Education, Science, and Culture, Japan and the Natural Sciences and Engineering Research Council of Canada (NSERC) grant #154280.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RH, AK, YA, KO and SH participated in the design of the study; RH carried out microbiological analysis including PCR and sequencing; AK performed susceptibility testing; MG carried out co-agglutination and electron microscopy; RH, AK, YA and DT collected clinical data and analyzed the data; RH, YA and MG contributed reagents/materials/analysis tools; RH, AK, YA, MG and KO drafted the manuscript. All authors read and approved the final manuscript.
Authors’ information
Not applicable.
Availability of data and materials
Not applicable.
Additional files
Additional file 1: Table S1.
Primer sets used for PCR and sequencing of cps locus of the unencapsulated S. suis serotype 31 strain 43640. (DOCX 25 kb)
Additional file 2: Table S2.
Primer sets used for PCR and sequencing of cpsA and cpsB of the encapsulated S. suis serotype 31 isolated from pigs in this study. (DOCX 18 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Hatrongjit, R., Kerdsin, A., Gottschalk, M. et al. First human case report of sepsis due to infection with Streptococcus suis serotype 31 in Thailand. BMC Infect Dis 15, 392 (2015). https://doi.org/10.1186/s12879-015-1136-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-015-1136-0