Abstract
Background
Non-tuberculous mycobacteria (NTM), which are ubiquitous micro-organisms occurring in humans, animals and the environment, sometimes receive public health and veterinary attention as opportunistic disease-causing agents. In Tanzania, there is limited information regarding the diversity of NTM species, particularly at the human-livestock-wildlife interface such as the Serengeti ecosystem, where potential for cross species infection or transmission may exist.
Methods
Mycobacterial DNA was extracted from cultured isolates obtained from sputum samples of 472 suspect TB patients and 606 tissues from wildlife species and indigenous cattle. Multiplex PCR was used to differentiate NTM from Mycobacterium tuberculosis complex (MTBC) members. NTM were further identified to species level by nucleotide sequencing of the 16S rRNA gene.
Results
A total of fifty five (55) NTM isolates representing 16 mycobacterial species and 5 isolates belonging to the MTBC were detected. Overall, Mycobacterium intracellulare which was isolated from human, cattle and wildlife, was the most frequently isolated species (20 isolates, 36.4%) followed by M. lentiflavum (11 isolates, 20%), M. fortuitum (4 isolates, 7.3%) and M. chelonae-abscessus group (3 isolates, 5.5%). In terms of hosts, 36 isolates were from cattle and 12 from humans, the balance being found in various wildlife species.
Conclusion
This study reveals a diversity of NTM species in the Serengeti ecosystem, some of which have potential for causing disease in animals and humans. The isolation of NTM from tuberculosis-like lesions in the absence of MTBC calls for further research to elucidate their actual role in causing disease. We are also suggesting a one health approach in identifying risk factors for and possible transmission mechanisms of the NTM in the agro-pastoral communities in the Serengeti ecosystem.
Similar content being viewed by others
Background
Non-tuberculous mycobacteria (NTM) are saprophytic acid-fast bacilli, which are present in water, soil and biofilms [1]-[3]. Various names such as anonymous, atypical, environmental, opportunistic and mycobacteria other than tubercle bacilli (MOTT) have been used interchangeably referring to NTM [4]. In contrast to obligate parasites such as M. tuberculosis and M. leprae, NTM are present in the environment as saprophytes causing disease opportunistically [2],[3]. Similarities of NTM isolates have been found in humans and animals, soil and natural open water sources, implying a possible transmission via the environment [3].
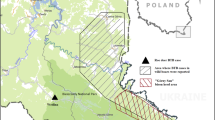
Map of the Serengeti ecosystem which shows study sites where tissues from animals and sputum samples from TB suspect patients were collected; A; Endulen Health centre, B; Bunda [Bunda district headquarter (HQ)], C; Mugumu (Serengeti district HQ), D; Waso-Loliondo (Ngorongoro district HQ), E; Serengeti National Park, F; Maswa Game Reserve.
NTM play a significant role as sources of human and animal infections [2],[5] with varied clinical manifestations [3] and should be a cause of concern in countries with high prevalence of human immunodeficiency virus (HIV) infection [6]. In Tanzania, NTM species including M. gordonae, M. smegmatis, M. fortuitum, M. phlei, M. flavescens and M. avium intracellulare have been detected in milk samples collected from the coast region [7]. Studies conducted in Arusha, northern Tanzania found NTM to be more common than M. tuberculosis in patients with TB adenitis [8]. Moreover, invasive NTM infections in Tanzania due to M. sherrisii and M. avium complex sequevar M. avium complex-D has been diagnosed in HIV-infected patients [9], also NTM infection due to M. intracellulare has been found to be associated with non-HIV infected patients with a history of tuberculosis [10].
In the Serengeti ecosystem a very high interaction of humans-environment-livestock- wildlife exists, posing a risk for animal-and human-based contamination of the natural water sources and the transmission of NTM to other animals or humans [11]. However, little information is available on NTM species circulating in humans, domestic and wild animals and even fewer reports are available in Africa, although Botha et al.[12] have reported that the following NTMs have been found in human and animal specimens: M. abscessus, M. asiaticum, M. avium, M. brasilienses, M. chelonae, M. elephantis, M. engbackii, M. farcinogenes, M. fortuitum, M. gilven, M. gordonae, M. heraklionense, M. hiberniae, M. intracellulare, M. interjectum, M. lentiflavum, M. marseillense, M. moriokaense, M. nonchromogenicum, M. palustre, M. palveris, M. paraffinicum, M. phlei, M. senegalense, M. simiae, M. sherrisii, M. sphagni, M. terrae and M. vulneris.
The use of molecular techniques that target the 16S rRNA gene has been useful for diagnosis of NTM. This is particularly important in TB endemic regions where NTMs are underdiagnosed in TB patients due to limited availability of laboratory facilities [13]. Most health centres in developing countries depend on Ziehl-Neelsen (ZN) for diagnosis of tuberculosis which cannot distinguish M. tuberculosis complex (MTBC) from NTM [14],[15]. Diagnosis of pathogens using molecular diagnostic techniques is more rapid and improve accuracy in identification as compared to conventional methods which are time consuming [16]. Sequencing of the 16S rRNA gene in NTM can be applied in the speciation of bacteria, even those which are dead or are uncultivable [15],[16]. Therefore, we used this technique to determine species diversity of NTM in biological samples collected from humans, wildlife and livestock at the human-animal interface in the Serengeti ecosystem, Tanzania.
Methods
Study site
This study was conducted in the Serengeti ecosystem and surrounding districts in Bunda (S 2° 0' 0, E33° 49' 60), Serengeti (S2° 0' 0, E 34° 49' 60) and Ngorongoro (E35° and 36°E and S 2° and 4°) (Figure 1). The Serengeti ecosystem as previously described by Katale et al. [17] is comprised of Serengeti National Park (SNP), Ngorongoro Conservation Area (NCA), Loliondo Game Controlled Area (LGCA), Maswa Game Reserve and Ikorongo-Grumeti Game Reserve (IGGR). Livestock and wildlife in the ecosystem interact during grazing and drinking water increasing possibility of interspecies disease transmission or contamination of water sources, which could potentially lead to a zoonotic epidemic [17].
Sampling of livestock and wildlife tissues
Livestock
Four hundred and nighty nine (499) tissues (mediastinal, retropharyngeal, pre-crural and pre-scapular lymphnodes, lungs and liver) with lesions suggestive of mycobacterial infection were collected from approximately 1200 slaughtered indigenous zebu cattle (Bos indicus) in the Serengeti (n = 420), Bunda (n = 600) and Ngorongoro (n = 180) abattoirs (Records from registry books, District Meat Inspection [2012-2013]). The slaughter houses were located in district headquarters where cattle were purchased from different local markets available in the districts and transported to district abattoirs for slaughter. Meat inspections were conducted with assistance of meat inspectors at the slaughter houses. To ensure that only cattle carcasses from the study site were sampled, the history of the origin of the cattle was taken from the owners. Tissues were selected based on presence of tuberculous lesions in organs and their associated lymph nodes. The anatomical sites of bTB suspected lesions for each carcass were recorded. With assistance from meat inspectors tuberculous organs and their associated lymph nodes were palpated, incised and preserved separately in sterile zip lock bags and packed in a cool box before processing for storage in liquid nitrogen containers.
Wildlife
One hundred and seven (107) wildlife tissues were sampled opportunistically from dead animals in the Serengeti ecosystem. Tissues sampled included road kill, where only intact organs were sampled (2 impala and 1 black-backed jackal), and animals that died from old age or disease. Forty three (43) tissues which included mediastinal, retropharyngeal, mesenteric lympnodes, lungs, kidney and liver were sampled during trophy and meat cropping in Maswa Game Reserve (MGR) as per the schedule of the Ministry of Natural Resources and Tourism, Tanzania. Tissue specimens were preserved in liquid nitrogen containers for further analysis. Ziehl-Neelsen (ZN) was performed on tissues that showed growth on LJ Media.
Tissues processing and culture
Tissues of approximately 10 g were chopped into small pieces using sterile scalpel blades and forceps and transferred into stomacher bags containing 10 ml of sterile distilled water. Samples were then homogenized for 2 min using a blender (Stomacher 80 laboratory blender, Seward Medical, London, UK). The homogenates were transferred into universal containers, followed by addition of an equal amount of 3% oxalic acid to the homogenate in the universal container. The suspensions were left for 45 minutes at room temperature with occasional shaking before centrifugation at 3000 rpm (Mistral 1000 MSE, UK) for 15 min. The sediments were neutralized with 3-4 drops of 2% sodium hydroxide solution to which 3-4 drops of 0.1% phenol red indicator were added. The sediments were mixed before inoculation on Lowenstein Jensen (LJ) Media (BDH Chemicals Ltd, Poole, UK) containing pyruvate or glycerol and incubated at 37°C. LJ slopes were examined weekly for indications of macroscopic growth for 12 weeks. When growth was visible, smears were prepared, air dried, fixed with heat, stained with ZN and examined microscopically for the presence of acid fast bacilli (AFB).
Sputum sample collection and preparation for culture
Sputum samples were collected from 472 hospitalized and out-patients who attended TB clinics in Bunda, Serengeti and Ngorongoro district hospitals. Sputum samples were collected in 50ml screw-cap Falcon tubes in the early morning. Both hospitalized and outpatients with symptoms suggestive of TB in the study area, new patients who had not yet been started on anti-tuberculosis treatment were considered for inclusion in the study. Digestion and decontamination of sputum samples were carried out by adding an equal volume of Cetyl Pyridinium Chloride (CPC) solution to the sputum samples. The specimens were left for 15 min at room temperature and concentrated by centrifuging at 4000rpm for 15 min. The supernatants were poured off into a splash proof container. Twenty (20) ml of distilled water was added to the sediments and the pellets were suspended by inverting the tubes several times before centrifugation at 3500 rpm for 15 min. The supernatant was poured off and the deposits were inoculated onto two slopes of LJ media containing glycerol or pyruvate by using sterile loops. Specimens were incubated and examined weekly for bacterial growth. Smears on microscope slides were prepared from the specimen sediments and thereafter stained by the Ziehl-Neelsen (ZN) stain.
Extraction of DNA
Mycobacterial DNA was isolated from colonies of each isolate that were scraped from the surface of the LJ medium containing glycerol or pyruvate. The isolates were suspended into labeled cryovials containing 100μl of sterile distilled water. The suspensions were heated in a water bath at 80°C for 1 hour to inactivate the bacteria and then centrifuged at 3000 rpm (Mistral 1000 MSE, UK) for 5 min. The supernatants were stored at −80°C until further analysis.
Mycobacterium genus typing
Multiplex PCR was performed using six primers as described by Wilton & Cousins et al. [18]. Briefly, six oligonucleotide primers namely MYCGEN-F 5’-AGA GTT TGA TCC TGG CTC AG-3’, MYCGEN-R 5’-TGC ACA CAG GCC ACA AGG GA-3’, which amplify a specific PCR product from the 16s rRNA gene of all know mycobacteria were used. MYCAV-R 5’-ACC AGA AGA CAT GCG TCT TG-3’ and MYCINT-F 5’-CCT TTA GGC GCA TGT CTT TA-3’ which amplify the hyper variable region of the 16S rRNA gene of M. intracellulare (MYCINT-F) and M. avium (MYCAV-R) respectively. Two primers (TB-F, TB-R) (TB-F 5’-GAA CAA TCC GGA GTT GAC AA-3’ and TB-R 5’-AGC ACG CTG TCA ATC ATG TA-3’ (TIB Molbiol® syntheselabor GmbH, Germany) which target for the MPB70 gene were used to specify M. tuberculosis complex from the mycobacteria. Briefly, one PCR reaction consisted of 2 μl DNA, 6.2 μl H20 Qiagen®, 10 μl of master mix and 0.3 μl for each of the primers. Initially, DNA was denatured at 95°C for 10 min, annealing involved 35 cycles for 1 min, 0.5 min and 1 min at 95°C, 61°C and 72°C, respectively. Extension was done at 72°C for 10 min and cooling at 4°C infinitely at 4°C. In all cases of DNA amplification water was included as negative controls and M. tuberculosis complex DNA as positive controls in each run. Electrophoresis was run at 100 V in 1.5% agarose (3.75 g in 250 ml of 1 × TAE buffer) (Sigma®) gel and visualized under UV light. During electrophoresis, Ethidium bromide was used as a staining reagent. The PCR products were calibrated using hundred base pair (100 bp) DNA ladder, blue loading dye (Promega Corporation, Madison, USA).
Amplification of DNA for Mycobacterial speciation of NTM
PCR amplification of mycobacterial DNA samples was done using 16S rRNA f- 5’ AGA GTT TGA TCC TGG CTC AG 3’ and 16S rRNA r- 5’ GCG ACA AAC CAC CTA CGA G 3’ primers. Each PCR reaction contained: 1 μl of DNA template, 2.5 μl of 10× buffer, 2 μl 25 mM MgCl2, 1 μl 10 mM dNTPs, 5 μl Q-buffer, 0.5 μl of each primer (50pmol/μl), 0.125 μl HotStarTaq DNA polymerase (Qiagen, Germany) in a 24.5 μl reaction with ddH2O. A negative control (no template) and a positive control (DNA template from M. tuberculosis H37Rv) were included to assay for contamination of the reagents and successful PCR amplification, respectively. Amplification was done by activating the Taq polymerase at 95’C for 15 min, followed by 45 cycles of 94°C for 30 s, 60°C for 1 min and 72°C for 30 s, followed by a final elongation step at 72°C for 10 min.
Nucleotide sequencing of non-tuberculous mycobacteria
To identify mycobacterial species, polymerase chain reactions (PCRs) were conducted followed by sequencing of the 16S rRNA gene [19] using 1.1pmol/μl16S rRNA forward primer (16S-27f (5'- GWA TTA CCGCGG CKG CTG -3'). Sequencing was done using an ABI sequencer (Applied Biosystem Inc.) at the Central Analytical Facility (CAF) of Stellenbosch University, South Africa. Accession numbers and obtained sequences were edited and analysed using the Ribosomal Differentiation of Microorganisms (RIDOM) project (http://www.ridom-rdna.de) and the National Center for Biotechnology Information (NCBI) Blast sequence alignment tool (http://blast.ncbi.nlm.nih.gov).
Ethical consideration
This study was approved by the Research Ethics committee of the Muhimbili University of Health and Allied Sciences (MUHAS)(Ref.MU/PGS/PhD/R/Vol.1), The National Institute for Medical Research (NIMR) (Ref. No. NIMR/HQ/R.8a/Vol. IX/1299), Tanzania, Tanzania Wildlife Research Institute (TAWIRI) and the Ministry of Natural Resources and Tourism, Tanzania (Ref. No. HA 403/563/01/74). Written informed consent was taken from human subjects and exclusion from the study is without any obligations on both sides.
Results
The 606 tissue specimens collected from livestock and wildlife species showed that 5 specimens (0.8%) were identified as MTBC members and 43 specimens (7.1%) were characterized as NTM species, which consisted mostly of M. intracellulare (20 of the 43 samples, 46.5%). Other NTM species identified were M. lentiflavum isolated from 6 cattle specimens, 3 buffalo specimens, 1 Thompson gazelle specimen and 1 isolate from a warthog. M. simiae, M. confluentis, M. neoaurum, M. nonchromogenicum, M. terrae and M. thermoresistibile were isolated from cattle only (see Table 1). No growth was observed for the other 558 (92.1%) tissue specimens cultured on LJ medium. From the 472 human sputum samples, only 12 specimens (2.9%) were NTM positive which were M. Chelonae-abscessus group (n = 3), M. genavense, M. gilvum, M. gordonae, M. intermedium, M. poriferae and M. spaghni (n = 2). No M. avium species were identified from human or animal specimens. Most of NTM species isolated in this study have been found in other countries such as Uganda, Kenya, Zambia and South Africa (Table 2). No report was found whether M. genavense, M. poriferae and M. spaghi have been isolated in these countries (Table 2). All members of the mycobacteria genus gave a PCR product of 1030 bp. M. intracellulare generated a PCR fragment of 850 bp in addition to the 1030 bp genus product. PCR fragments of 372 bp in addition to the 1030 bp product were generated by isolates from the M. tuberculosis complex.
Discussion
We found the overall prevalence of NTM in livestock and wildlife to be 7.1%, which is not far from results estimated based on comparative skin tests conducted in the same ecosystem in indigenous cattle. This study showed non-specific reaction (atypical mycobacterium) to avian and bovine tuberculin purified proteins derivatives (PPDs) to be 10.6% in indigenous zebu [17]. However, we were unable to find either NTM or MTBC in a high proportion (92.1%) of tissues with apparent tuberculous lesions, indicating that the lesions might be caused by other pathogens or there was a failure of egg based LJ medium to culture mycobacteria from these lesions. Liquid media is more efficient for recovery of mycobacteria than egg based solid medium [20]. A report by McCarthy et al. [21] found large differences in recovery of NTM between solid or liquid media, a study that cultured NTM from patients with HIV. However, in the present study liquid medium was not used for isolation of mycobacterium. Neither, was the HIV status of human TB suspected patients available to correlate the NTM infection with HIV status.
This study has shown that, 16 species of NTM were isolated from humans, indigenous cattle and wildlife, in the Serengeti ecosystem, of which the majority were M. intracellulare, M. lentiflavum and M. fortuitum. Other NTM species including M. cholenae-abscessus group, M. simiae, M. neoaurum, M. nonchromogenicum, M. spaghni, M. terrae, M. confluentis, M. genavense, M. gilvum, M. gordonae, M. intermedium, M. poriferae and M. thermoresistibile were isolated at a lower rate, ranging between 5.5% and 1.8%. Comparison of the NTM species isolated in this study indicate that most of them have previously been found in humans, animals, or the environment in Ethiopia, Kenya, Uganda, South Africa and Zambia (Table 2). In the present study, given the nature of sample acquisition for both cattle and wildlife specimens, isolation of M. gordonae and M. terrae complex as environmental contaminants [22] might occur. Thus there is unjustifiable potential for specimens’ environmental contamination which might obscure our findings. M. genavense, M. poriferae and M. spaghni are not common or have not previously been isolated in East Africa (Table 2). In contrast to our study where all NTM species were isolated from sputum samples and animals tissues. Previous studies found M .simiae, M. fortuitum, M. gordonae, M. intermedium, M. intracellulare, M. nonchromogenicum and M. terrae in soil and water (Table 2). The correlation of NTM between soil on one hand and animals on the other hand indicates that NTM are readily exchanged between animals and environments [23]. However, in our study no any environmental sample was taken to correlate between NTM species found in animals and environments.
M. intracellulare, which is a member of Mycobacterium avium complex (MAC) was the most frequently isolated species, consistent with a report from study of NTM in Uganda, South Africa and Australia [24],[25]. However, this finding differs from other studies in Kenya, South Africa and Uganda where M. mucogenicum, M. terrae and M. fortuitum were the most frequently isolated species respectively [14],[16]. The high prevalence of M. intracellulare is of concern in this setting, given a high HIV prevalence and the ability of M. intracellulare to cause pulmonary and extrapulmonary TB in such individuals [26],[27] such as has been reported in Kenya [28]. In the current study, M. intracellulare species were isolated from human, cattle and wildlife.
We also found M. fortuitum, M. chelonae-abscessus group, M. genavense, M. gilvum, M. gordonae, M. intermedium, M. poriferae and M. spaghni in TB suspect patients who had clinical signs suggestive of tuberculosis. However the role of these NTM in TB disease causation as well as possible influence in the diagnostic tests for tuberculosis is not known in our cases. This finding is consistent with previous studies where these species were isolated from respiratory samples from patients with chronic bronchitis, pulmonary TB, sub-acute pneumonia and healed pulmonary TB [29]-[32]. In Uganda, M. fortuitum was the most prevalent NTM species isolated from respiratory secretions in infants and adolescence [13].
We isolated from tuberculous lesions of indigenous cattle a number of NTM species which include M. simiae, M. confluentis, M. neoaurum, M. nonchromogenicum, M. terrae and M. thermoresistibile. Apparently these animals had neither M. bovis nor other MTBC and it may be important to examine the role of these NTM in disease causation. It is also potentially important to test the effect of exposure of animals to NTM on the specificity and/or sensitivity of bovine tuberculosis (bTB) diagnostic test [33] such as skin test and interferon gamma assay due to potential cross-reactivity.
Lastly, we detected a low prevalence of M. nonchromogenicum in cattle and M. lentiflavum from indigenous cattle and wildlife. M. nonchromogenicum, part of the M. terrae complex, has also been isolated from nasal mucus from cattle in a herd infected with bTB [34] and in immunosuppressed individuals with pulmonary disease [35]. Infections due to M. terrae complex are rare [36]. However, M. nonchromogenicum has been associated with bacteremia in AIDS patients [25], lung infection in humans and has been detected from patients with chronic skin ulcers unresponsive to antibiotic treatment, arthritis [37], tenosynovitis [38], pulmonary infection [37],[39] and meningitis [40], signifying its public health and veterinary importance. Together with M, terrae, M. nonchromogenicum is widely distributed in Africa although proportions differ in different countries [23].
The ubiquitous nature of NTM suggests value for investigation of their clinical relevance as well as effects on interpreting other positive TB test results, for example, we do not know what the influence of NTM on immunity and immunological based TB diagnostic tests, given the fact that mycobacteria share many common antigens.
Conclusion
In conclusion, there is a very wide range of NTM species from humans and animals in the Serengeti ecosystem and understanding their transmission dynamics will require a one health approach involving sampling and sequencing of samples from livestock, wildlife, humans and their environment. Furthermore, there is a need to investigate the influence of NTM in disease causation and efficacy on screening and diagnostic tests.
References
Hatherill M, Hawkridge T, Whitelaw A, Tameris M, Mahomed H, Moyo S, Hanekom W, Hussey G: Isolation of Non-tuberculous mycobacteria in children investigated for pulmonary tuberculosis. 1(1). PLoS One. 2006, 1 (1): e21-10.1371/journal.pone.0000021. doi:10.1371/journal.pone.0000021.
Fine PE, Floyd S, Stanford JL, Nkhosa P, Kasunga A, Chaguluka S, Warndorff DK, Jenkins PA, Yates M, Ponnighaus JM: Environmental mycobacteria in northern Malawi: implications for the epidemiology of tuberculosis and leprosy. Epidemiol Infect. 2001, 126: 379-387. 10.1017/S0950268801005532.
Katoch VM: Infection due to non-tuberculous mycobacteria (NTM). Indian J Med Res. 2004, 120: 290-304.
Garcia-Jimenez WL, Benıtez-Medina JM, Martınez R, Carranza J, Cerrato R, Garcıa-Sanchez A, Risco D, Moreno JC, Sequeda M, Gomez L, Fernández-Llario P, Hermoso-de-Mendoza J: Non-tuberculous Mycobacteria in Wild Boar (Sus scrofa) from Southern Spain: Epidemiological, Clinical and Diagnostic Concerns.Transbound Emerg Dis 2012. doi:10.1111/tbed.12083.,
van Ingen J, Boeree MJ, Dekhuijzen PN, van Soolingen D: Environmental sources of rapid growing nontuberculous mycobacteria causing disease in humans. Clin Microbiol Infect. 2009, 15: 888-893. 10.1111/j.1469-0691.2009.03013.x.
Falkinham JO: Epidemiology of infection by nontuberculous mycobacteria. Clin Microbiol Rev. 1996, 9: 177-215.
Mdegela RH, Kusiluka LJM, Kapaga AM, Karimuribo ED, Turuka FM, Bundala A, Kivaria F, Kabula B, Manjurano A, Loken T, Kambarage DM: Prevalence and determinants of mastitis and milk-borne zoonoses in smallholder dairy farming sector in kibaha and morogoro districts in eastern Tanzania. J Veterinary Med Ser B. 2004, 51: 123-128. 10.1111/j.1439-0450.2004.00735.x.
Mfinanga SGM, Morkve O, Kazwala RR, Cleaveland S, Sharp MJ, Kunda J, Nilse R: Mycobacterial adenitis, role of Mycobacterium bovis, non tuberculous mycobacteria, HIV infection, and risk factors in Arusha, Tanzania. East Afr Med J. 2004, 81: 171-178. 10.4314/eamj.v81i4.9150.
Crump JA, van Ingen J, Morrissey AB, Boeree MJ, Mavura DR, Swai B, Thielman NM, John A, Bartlett Grossman H, Maro VP, van Soolingen D: Invasive disease caused by nontuberculous mycobacteria, Tanzania. Emerg Infect Dis. 2009, 15: 53-55. 10.3201/eid1501.081093. doi:10.3201/eid1501.081093
Haraka F, Rutaihwa LK, Battegay M, Reither K: Mycobacterium intracellulare infection in non-HIV infected patient in a region with a high burden of tuberculosis.BMJ Case Reports 2012, 1–3. doi:10.1136/bcr.01.2012.5713.,
Cleaveland S, Mlengeya T, Kazwala RR, Michel A, Kaare MT, Jones SL, Eblate E, Shirima GM, Packer C: Tuberculosis in Tanzanian Wildlife. J Wildl Dis. 2005, 41: 446-453. 10.7589/0090-3558-41.2.446.
Botha L, van Pittius NCG, van Helden PD: Mycobacteria and disease in southern Africa. Transbound Emerg Dis. 2013, 60: 147-156. 10.1111/tbed.12159.
Gopinath K, Singh S: Non-tuberculous mycobacterium in TB endemic countries: are we neglecting the danger?. PLoS Negl Trop Dis. 2010, 4: e615-10.1371/journal.pntd.0000615.
Asiimwe B, Bagyenzi G, Ssengooba W, Mumbowa F, Mboowa G, Wajja A, Mayanja-Kiiza H, Musoke P, Wobudeya E, Kallenius G, Joloba M: Species and genotypic diversity of non-tuberculous mycobacteria isolated from children investigated for pulmonary tuberculosis in rural Uganda. BMC Infect Dis. 2013, 13: 88-10.1186/1471-2334-13-88.
Dang NA, Kuijper S, Walters E, Claassens M, van Soolingen D, Vivo-Truyols G, Janssen H, Kolk AHJ: Validation of biomarkers for distinguishing mycobacterium tuberculosis from Non-tuberculous mycobacteria using Gas chromatography–mass spectrometry and chemometrics. PLoS One. 2013, 8: e76263-10.1371/journal.pone.0076263. doi:76210.71371/journal.pone.00762663
Covert TC, Rodgers MR, Reyes AL, Stema GN: Occurrence of nontuberculous mycobacteria in environmental sample. Appl Environ Microbiol. 1999, 65: 2492-2496.
Katale BZ, Mbugi EV, Michel AL, Keyyu JD, Kendal S, Kibiki GS, Godfrey-Faussett P, Karimuribo ED, Kazwala RR, van Helden P, Matee MI: Prevalence and risk factors for infection of bovine tuberculosis in indigenous cattle in the Serengeti ecosystem, Tanzania. BMC Vet Res. 2013, 9: 267-10.1186/1746-6148-9-267.
Wilton S, Cousins D: Detection and identification of multiple mycobacterial pathogens by DNA amplification in a single tube. PCR Methods Appl. 1992, 1: 269-273. 10.1101/gr.1.4.269.
Harmsen D, Dostal S, Roth A, Niemann S, Rothgänger J, Sammeth M, Albert J, Frosch M, Richter E: RIDOM: Comprehensive and public sequence database for identification of Mycobacterium species. BMC Infect Dis. 2003, 3: 26-10.1186/1471-2334-3-26.
Abe C, Hosojima S, Fukasawa Y, Kazumu Y, Takahashi M, Hirano K, Mori T: Comparison of MB-Check, BACTEC, and egg-based media for recovery of mycobacteria. J Clin Microbiol. 1992, 30: 878-881.
McCarthy KD, Cain KP, Winthrop KL, Udomsantisuk N, Lan NT, Sar B, Kimerling ME, Kanara N, Lynen L, Monkongdee P, Tasaneeyapan T, Varma JK: Nontuberculous mycobacterial disease in patients with HIV in Southeast Asia. Am J Respir Crit Care Med. 2012, 185: 981-988. 10.1164/rccm.201107-1327OC.
Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Gwen H, Iademarco MI, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ, Winthrop K: An Official ATS/IDSA Statement: Diagnosis, treatment, and prevention of non tuberculous mycobacterial Diseases. Am J Respir Crit Care Med. 2007, 175: 367-416. 10.1164/rccm.200604-571ST.
Gcebe N, Rutten V, van Pittius NC, Michel A: Prevalence and distribution of non-tuberculous mycobacteria (NTM) in cattle, African buffaloes (Syncerus caffer) and their environments in South Africa. Transbound Emerg Dis. 2013, 1: 74-84. 10.1111/tbed.12133.
Kankya C, Muwonge A, Djonne B, Munyeme M, Opuda-Asibo J, Skjerve E, Oloya J, Edvardsen V, Johansen TB: Isolation of non-tuberculous mycobacteria from pastoral ecosystems of Uganda: public health significance. BMC Publ Health. 2011, 11: 320-10.1186/1471-2458-11-320.
Hoefsloot W, van Ingen J, Andrejak C, Angeby K, Bauriaud R, Bemer P, Beylis N, Boeree MJ, Cacho J, Chihota V: The geographic diversity of nontuberculous mycobacteria isolated from pulmonary samples: a NTM-NET collaborative study. Eur Respir J. 2013, 42: 1604-1613. 10.1183/09031936.00149212.
Nishiuchi Y, Maekura R, Kitada S, Tamaru A, Taguri T, Kira Y, Hiraga T, Hirotani A, Yoshimura K, Miki M, Ito M: The Recovery of Mycobacterium avium-intracellulare Complex (MAC) from the Residential Bathrooms of Patients with Pulmonary MAC. Clin Infect Dis. 2007, 45: 347-351. 10.1086/519383.
Han XY, Tarrand JJ, Infante R, Jacobson KL, Truong M: Clinical significance and epidemiologic analyses of mycobacterium avium and mycobacterium intracellulare among patients without AIDS. J Clin Microbiol. 2005, 43: 4407-4412. 10.1128/JCM.43.9.4407-4412.2005.
Nyamogoba HDN, Mbuthia G, Mining S, Kikuvi G, Biegon R, Mpoke S, Menya D, Waiyaki PG: HIV co-infection with tuberculous and non-tuberculous mycobacteria in western Kenya: challenges in the diagnosis and management. Afr Health Sci. 2012, 12: 305-311. http://dx.doi.org/10.4314/ahs.v12i3.9
de Lima CAM, Gomes HM, Oelemann MAC, Ramos JP, Caldas PC, Campos CED, Pereira MAS, Montes FFO, de Oliveira MSC, Suffys PN, Moura MM: Nontuberculous mycobacteria in respiratory samples from patients with pulmonary tuberculosis in the state of Rondônia, Brazil. Mem Inst Oswaldo Cruz. 2013, 108: 457-462. 10.1590/S0074-0276108042013010.
Rammaert B, Couderc L-J, Rivaud E, Honderlick P, Zucman D, Mamzer M-F, Cahen P, Bille E, Lecuit M, Lortholary O, Catherinot E: Mycobacterium genavense as a cause of subacute pneumonia in patients with severe cellular immunodeficiency. BMC Infect Dis. 2011, 11: 311-10.1186/1471-2334-11-311.
Ito A, Kishi F, Saito N, Kazumi Y, Mitarai S: Pulmonary mycobacterium intermedium disease in an elderly Man with healed pulmonary tuberculosis. J Clin Microbiol. 2005, 43: 1473-1474. 10.1128/JCM.43.3.1473-1474.2005.
Ballester F, Pujol I, Alcaide F, Pizarro I, Simó JM, Joven J, Camps J: First human isolate of mycobacterium poriferae in the sputum of a patient with chronic bronchitis. J Clin Microbiol. 2011, 49: 3107-3108. 10.1128/JCM.00436-11.
Thacker TC, Robbe-Austerman S, Harris B, Van Palmer M, Waters WR: Isolation of mycobacteria from clinical samples collected in the United States from 2004 to 2011. BMC Vet Res. 2013, 9: 100-10.1186/1746-6148-9-100.
McCorry TP, McCormick CM, Hughes MS, Pollock JM, NeillS D: Mycobacterium nonchromogenicum in nasal mucus from cattle in a herd infected with bovine tuberculosis. Vet Microbiol. 2004, 99: 281-285. 10.1016/j.vetmic.2003.12.006.
Molteni C, Gazzola L, Cesari M, Lombardi A, Salerno F, Tortoli E, Codecasa L, Penati V, Franzetti F, Gori A: Mycobacterium lentiflavum Infection in Immunocompetent Patient. Emerg Infect Dis. 2005, 11: 119-122. 10.3201/eid1101.040523.
Eskesen AN, Skråmm I, Steinbakk M: Infectious tenosynovitis and osteomyelitis caused by Mycobacterium nonchromogenicum. Scand J Infect Dis. 2007, 39: 179-180. 10.1080/00365540600798817.
Chen H-W, Lai C-C, Tan C-K: Arthritis caused by Mycobacterium terrae in a patient with rheumatoid arthritis. Int J Infect Dis. 2009, 13: e145-e147. 10.1016/j.ijid.2008.09.002.
Ridderhof JC, Wallace RJ, Kilburn JO, Butler WR, Warren NG, Tsukamura M, Steele LC, Wong ES: Chronic tenosynovitis of the hand due to Mycobacterium nonchromogenicum: use of high performance liquid chromatography for identification of isolates. Rev Infect Dis. 1991, 13: 857-864. 10.1093/clinids/13.5.857.
Sawai T, Inoue Y, Doi S, Izumikawa K, Ohno H, Yanagihara K, Higashiyama Y, Miyazaki Y, Hirakata Y, Tashiro T, Kohno S: A case of Mycobacterium nonchromogenicum pulmonary infection showing multiple nodular shadows in an immunocompetent patient. Diagn Microbiol Infect Dis. 2006, 54: 311-314. 10.1016/j.diagmicrobio.2005.11.005.
Lai C-C, Chen H-W, Liu W-L, Ding L-W, Lin C-L, Lu G-D, Hsueh P-R: Fatal meningitis caused by mycobacterium nonchromogenicum in a patient with nasopharyngeal Carcinoma. Clin Infect Dis. 2008, 46: 325-326. 10.1086/524897.
Acknowledgement
This study was funded by the Welcome Trust Grant [WT087546MA] to the Southern African Centre for Infectious Diseases and Surveillance (SACIDS). The Muhimbili University of Health and Allied Sciences (MUHAS) is acknowledged for supporting the research through its SIDA-Sarec small grant [000/3177]. The Tanzania Wildlife Research Institute (TAWIRI) as well as the Medical and Veterinary Authorities in Bunda, Serengeti and Ngorongoro districts for logistic support during sample collection.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
BZK designed the study, conducted field work, analysed the results and drafted the manuscript; EVM designed the study, participated in field work and reviewed the manuscript; LB analysed the results and reviewed the manuscript; JDK, SK, HD, ALM, RRK, MMR, and PVH critically reviewed the manuscript; MIM coordinated field work and critically reviewed the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Katale, B.Z., Mbugi, E.V., Botha, L. et al. Species diversity of non-tuberculous mycobacteria isolated from humans, livestock and wildlife in the Serengeti ecosystem, Tanzania. BMC Infect Dis 14, 616 (2014). https://doi.org/10.1186/s12879-014-0616-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-014-0616-y