Abstract
Background
The definition of sepsis is regularly updated; however, there is no standard diagnostic test. To improve diagnosis and prognostic prediction, the aim of this study was to determine the predictive value of circulating plasma mitochondrial DNA (mtDNA) levels in patients admitted to the emergency department (ED) with sepsis.
Methods
A total of 107 patients hospitalized from June 2018 to January 2019 were divided into the sepsis (n = 72) and septic shock (n = 35) groups based on the sepsis-3 definition. Clinical and laboratory data were measured within 24 h of admission. The mtDNA concentrations in clarified plasma were estimated by quantitative polymerase chain reaction. Binary logistic regression analysis and the receiver operating characteristic (ROC) curve were used to determine predictive value of mtDNA and other markers for sepsis outcome (28-day mortality).
Results
The median plasma mtDNA levels on admission were significantly higher in the septic shock patients than in the sepsis patients (134,252(IQR 70215–203,184) vs. 59,945(IQR 13274–95,319) copies/μL, P < 0.01), and were also higher in non-survivors than in survivors within 28 days (165,291(IQR 89919–272,228)vs. 63,025(IQR 17031–98,401)copies/μL, P < 0.01). Binary logistic regression showed that plasma lactate and mtDNA levels were independent risk factors for 28-day mortality [odds ratio (OR) 1.341, 95% confidence interval (CI) 1.035–1.736, P = 0.026 and OR 13.299, 95%CI 2.765–63.956, P = 0.001, respectively). The area under the ROC curve values for plasma mtDNA levels, lactate concentration, and their combined were 0.781 (p < 0.001, 95%CI 0.671–0.891), 0.733 (p < 0.001, 95%CI 0.635–0.832), and 0.799 (p < 0.001, 95%CI 0.698–0.901), respectively. The calibration test for the combined variable showed X2 of 2.559 and P = 0.923.
Conclusion
A higher plasma mtDNA level was associated with a poor prognosis of sepsis in the emergency room, and could serve as a predictor of sepsis for 28-day mortality.
Similar content being viewed by others
Background
The updated definition of sepsis is a life-threatening organ dysfunction caused by a dysregulated host response to infection [1], and remains a common and lethal syndrome. Although outcomes have improved, sepsis-related mortality is still higher than 25–30%, and reaches up to 40–50% in cases of septic shock as a subset of sepsis with particularly profound circulatory, cellular, and metabolic abnormalities [1, 2]. Sepsis is a major public health concern, accounting for more than $20 billion (5.2%) of total US hospital costs in 2011 [3]., Moreover, the reported incidence of sepsis is increasing [4, 5], likely reflecting the aging population with more comorbidities, greater recognition of the condition [6], and, in some countries, reimbursement-favorable coding [7]. There is currently no gold-standard diagnostic test of sepsis or septic shock. Although monitoring of lactate levels is used as a prognostic guide or indicator of illness severity, the value of this marker is highly controversial. Some suggested that elevated lactate levels represent an important marker of “cryptic shock” in the absence of hypotension. Others voiced concern about its specificity and that the non-availability of lactate measurement in resource-poor settings would preclude a diagnosis of septic shock [1]. Lactate level is a sensitive, albeit nonspecific, stand-alone indicator of cellular or metabolic stress rather than “shock” [8] .Therefore, there is an urgent need to find a specific factor that can accurately predict the severity and mortality risk of sepsis.
Mitochondrial DNA (mtDNA) has many similarities with bacterial DNA because of their shared common ancestry. Accordingly, increasing evidence points to mtDNA as a danger signal that is recognized by the innate immune system to directly modulate the inflammatory response [9]; thus, extracellular mtDNA can activate signaling pathways and promulgate inflammation [10]. This raises the possibility that mtDNA may serve as a surrogate of disease severity or even a predictor of mortality in critically ill patients. In a seminal series of studies, Zhang et al. [11,12,13] demonstrated that patients admitted with trauma showed significant elevations of mtDNA concentrations in the plasma and injured tissues. These results were validated in a rat model of trauma/hemorrhagic shock in which the plasma mtDNA levels were elevated for 7 days after injury [11]. Nakahira et al. [14] further demonstrated that an elevated mtDNA level was associated with intensive care unit (ICU) mortality among multiple cohorts, and could improve risk prediction in the field of critical care illness.
Thus, both basic research and clinical trials have now provided clear evidence that circulating mtDNA should be considered a new subtype of a damage-associated molecular pattern (DAMP) that is elevated in critically ill diseases such as sepsis, trauma, or hemorrhagic shock. However, data regarding the role of plasma mtDNA in adult sepsis is conflicting, and the clinical ramifications of this finding remain elusive. Moreover, clinical trials provide conflicting evidence about the association of mtDNA with mortality in critically ill patients [15], and the consensus definition and clinical criteria for sepsis change every few years. Since some recent studies have been performed on the basis of the newest criteria sepsis-3, we used this consensus definition in the present study to determine the clinical value of plasma mtDNA levels as a biomarker for predicting the mortality of sepsis in patients at the emergency department (ED) of a Chinese hospital.
Methods
Study design, setting, and population
This observational study was conducted in the ED of Shanghai South Campus, Renji Hospital, which is an urban university tertiary-care hospital with approximately120,000 ED visitors per year. From June 2018 to January 2019, 116 consecutive non-traumatic patients who fulfilled the sepsis-3 criteria defined by the American College of Chest Physicians/Society of Critical Care Medicine (ACCP/SCCM) were enrolled in the study. The exclusion criteria were as follows: younger than 18 years old, terminal stage of disease (malignant cancer of any type), end-stage renal disease, and patients who declined to participate in the study themselves or via their relatives. Ultimately, 107 consecutive patients were enrolled, and nine patients were excluded. All procedures performed in studies involving human participants were in accordance with the ethical standards of the Shanghai Jiaotong University School of Medicine, Renji Hospital Ethics Committee (NO.2016-109 k) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. All of the patients provided written informed consent.
Definitions and grouping
The patients were divided into a sepsis group and a septic shock group according to the sepsis-3 criteria [1]. That is, sepsis was clinically identified as an infection with a SOFA score ≥ 2, and patients with septic shock were clinically identified according to vasopressor requirement to maintain a mean arterial pressure of 65 mmHg or greater and a serum lactate level > 2 mmol/L (> 18 mg/dL) in the absence of hypovolemia.
Septic cardiomyopathy (SCM) was defined as an acute syndrome of cardiac dysfunction that is unrelated to cardiac ischemia in patients with sepsis [16], B-type natriuretic peptide (BNP) and troponin elevations appear to reflect SCM.
Preparation and quantification of plasma DNA
Blood samples were drawn from patients in the two groups within 24 h after admission, transferred into ethylenediaminetetraacetic acid-coated blood collection tubes, and processed within 2 h after venipuncture [14]. The samples were left to rest for 30 min and then centrifuged immediately at 1914×g for 10 min to separate the plasma from the cellular components. The plasma samples were stored at − 80 °C for batch analysis.
DNA was isolated from the plasma using the QIAamp Blood Mini Kit (#51106; Qiagen GmbH, Hilden, Germany) according to the manufacturer’s manual. All samples were thawed on ice, and the level of the mtDNA gene cytochrome b was measured by a SYBR Green dye-based quantitative real-time polymerase chain reaction (qPCR) assay (TaKaRa, Japan) using an ABI Prism7900HT detection system with the following primers: forward, ATGACCCCAATACGCAAAAT; reverse, CGAAGTTTCATCATGCGGAG. The PCR mixture was set up in a reaction volume of 10 μL using 5 μL of 2× SYBR green Master Mix (2×), 0.5 μL forward primer (1 μM), 0.5 μL reverse primer (1 μM), 3 μL of nuclease-free H2O, and 1 μL of plasma extract. The following thermocycler conditions were used: 3-min incubation at 95 °C, followed by 40 cycles of initial denaturation at 95 °C for 30 s, annealing at 54 °C for 45 s, and elongation at 68 °C for 1 min. The concentration of mtDNA was determined using a standard curve generated by qPCR with construct plasmids (pMD® 18-T Vector; Sangon Biotech, Shanghai) containing the human mitochondrial cytochrome B gene sequences described above. Calibrators were prepared by serial 10-times dilution of the stock solution and contained 102 to 108 mtDNA copies/μL.
Statistical analysis
Clinical and laboratory data are expressed as number (percent) or median [interquartile range (IQR), i.e., 25th–75th percentile], as appropriate. Statistical calculations were performed with IBM SPSS software version 26.0 (SPSS Inc., Chicago, IL, USA). Bivariate comparisons were conducted with the Mann-Whitney U test for continuous variables, and with the chi-squared test or Fisher’s exact tests for categorical variables. Bonferroni adjustment was adopted to select variables (treatment process and sepsis indused organ injury werec excluded). The influence of clinical and baseline laboratory data, including respiratory tract infection (RSTI),coronary heart disease (CHD), in addition to the log mtDNA concentration and lactate levels (which adjusted P <0.002), on 28-day mortality was evaluated in bivariate logistic regression models to determine independent predictors. Receiver operating characteristic (ROC) curve analyses were used to assess the predictive value of mtDNA on mortality by Sigma Plot 14.0, and cut-off values were calculated according to Youden’s index. The sensitivities and specificities were further used to calculate the positive and negative likelihood ratios. The Hosmer-Lemeshow goodness of fit test was used for verifying model calibration. All statistical tests were two-tailed, and P < 0.05 was considered to indicate a statistically significant difference.
Results
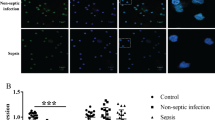
The baseline characteristics of the sepsis (n = 72) and septic shock group (n = 35) on admission to the ED are presented in Table 1. The median age for the septic shock patients was 73 (64–83) years, and 60% of the patients were male. Similarly, the median age for the septic patients was 68 (56.25–80) years, and 59.72% were male. There was no significant difference in underlying chronic medical problems between the two groups at the time of enrollment except for CHD (P < 0.01). There was a significant difference in the primary site of infection, including respiratory tract infection (P < 0.001) and urinary tract infection (P = 0.014) between the two groups. Clinical parameters such as CRRT (P = 0.002), requirement of mechanical ventilation (P < 0.001), vasopressor use (P < 0.001), and SOFA score (P < 0.001) were all significantly higher in the septic shock group. There was no significant difference in peripheral blood risk factors such as white blood cell count, hemoglobin, platelet count, C-reactive protein (CRP), and procalcitonin (PCT) between the two groups. The median mtDNA level and median lactate concentration were significantly higher in the septic shock patients than those of the sepsis patients on admission (P = 0.001 and P < 0.0001, respectively). The plasma mtDNA concentration did not correlate to the commonly used acute-phase markers CRP and PCT (γ = − 0.001, P = 0.994 and γ = − 0.145, P = 0.141, respectively), but was correlated with the SOFA score (γ = 0.344, P < 0.001).
In addition, the overall 28-d (P < 0.001) and 90-d (P < 0.001) mortality rates were significantly higher in the septic shock patients. To determine the predictive value of plasma mtDNA and other variables following sepsis, we compared their levels between survivors and non-survivors. As shown in Table 1, the median plasma mtDNA and lactate levels in non-survivors were significantly higher than those of the survivors (both P = 0.001). Box plots of plasma levels of mtDNA in the sepsis/septic-shock group and survivors/non-survivors group are shown in Fig. 1. Univariate analysis (Table 2) indicated a difference in clinical and baseline characteristics between the survivors and non-survivors, including respiratory tract infection (RSTI)(P<0.001) CHD (P < 0.001)Lac(P = 0.019) and Log mtDNA (P<0.001). These significant variables were then included in the bivariate logistic regression model, demonstrating that lactate and log mtDNA levels were independent predictors of survival (P = 0.017 and P = 0.003, respectively; Table 2).
Since there was maximum overlap in the combined variable (log mtDNA + lactate) in septic patients and non-survivors, the ROC curve was analyzed (Table 3 and Fig. 1). Both plasma log mtDNA and lactate concentrations could effectively distinguish survivors and non-survivors in the ED (Table 3 and Fig. 2); however, the combined variable was the strongest predictor of mortality with area under the curve (AUC) values of 0.635–0.832 for lactate and 0.671–0.891 for log mtDNA, and moderate to high sensitivities and specificities (Table 3). The cut-off value for lactate was 2.29 (Youden’s index = 0.413) and was 5.01 (Youden’s index = 0.535) for log mtDNA (Table 3). For comparison, the AUC value for the combined variable was 0.698–0.901, with higher sensitivity and specificity, and the cut-off value was 5.46 (Table 3). The calibration test for the combined variable showed an X2 value of 2.559 and P = 0.923 (Fig. 3).
Discussion
We investigated plasma mtDNA levels in ED patients admitted with sepsis or septic shock, which were significantly higher in the septic shock group and correlated with the SOFA score and sepsis 28-d mortality. Moreover, the mtDNA levels showed superior prognostic prediction value than that of lactate levels.
Based on the sepsis 3.0 definition, a poor prognosis of sepsis is associated with organ injury, which is in line with the positive correlation of plasma mtDNA levels and severity of illness or SOFA score, even after controlling for potential confounders. We chose to focus our study on the level of free mtDNA in the plasma based on the observations by Zhang et al. [11] suggesting that mtDNA directly induces inflammation due to homology with pathogen-associated molecular patterns, thus acting as a DAMP. Our results are also consistent with those of Nakahira et al. [14] who conducted one of the first clinical trials on this topic in 2013, demonstrating significantly higher mtDNA levels in patients who died within 28 days of ICU admission than those of the survivors.
Moreover, mtDNA has been shown to improve risk prediction compared to that of commonly measured biomarkers such as lactate and PCT. In 2015, Bhagirath et al. [17] published a translational study aimed at elucidating the role of nuclear DNA (nucDNA), mtDNA, and bacterial DNA in sepsis. They showed that the levels of plasma nucDNA and mtDNA were 200- and 50-fold greater in the patients with sepsis than those of healthy controls, respectively. Timmermans et al. [18] conducted a prospective observational study by collecting samples from 121 septic shock patients in the ICU on day 1, 3, 5, 6, 9, 14, 21, and 28, demonstrating that the levels of nucDNA and mtDNA were significantly elevated and remained elevated at all time points relative to those of healthy controls. Nevertheless nucDNA, but not mtDNA, levels were associated with mortality from septic shock, which is inconsistent with our findings. We speculate that these differences are due to variations in the study population and diagnostic criteria between studies. Kung et al. [19] demonstrated that both plasma nucDNA and mtDNA concentrations on admission were significantly higher in non-survivors than in survivors, and both levels increased shortly after severe infection and then gradually decreased after antimicrobial therapy. Moreover, the nucDNA levels were significantly higher than the mtDNA levels for the same group. In a recent study, Yang et al. [20] indicated that the relative mononuclear cell mtDNA copy number in non-survivors was significantly lower than that of survivors, which was also an important predictor of clinical outcome, as patients with low copy numbers had higher 28-d mortality rates. This suggests that the nucDNA was first elevated in the initial stages of sepsis, accompanied by a depletion of mtDNA copy numbers, followed by the release of mtDNA in the plasma becoming DNA fragments, or mtDNA DAMPs, which are associated with the susceptibility and pathogenesis of sepsis-associated organ injury [21, 22]. The potential contribution of mtDNA DAMPs to organ injury is also supported by persuasive evidence from cell culture and animal models in which administration of exogenous mtDNA fragments or prevention of their accumulation had concordant effects on cytotoxicity, cellular dysfunction, and tissue inflammation [11, 23,24,25,26,27].
Innocenti et al. [28] demonstrated that higher lactate levels and decreased clearance were associated with increased short-term and intermediate-term mortality in patients with sepsis, regardless of the presence of shock. Timmermans et al. [18] also suggested that the plasma mtDNA level on admission was a more powerful predictor than lactate concentration, which is more commonly used for outcome prediction in clinical practice. This conclusion is consistent with our present findings. Therefore, we hypothesized that the plasma mtDNA might become elevated at an earlier stage than serum lactate; however, further prospective study is needed to verify this possibility.
Limitations
Several limitations of the study deserve consideration. First, we only measured plasma levels on admission in the ED and did not measure levels serially, and therefore could not assess the variation of these levels in the ICU or over time. Second, large-scale prospective studies are warranted to evaluate the prognostic contribution of plasma DNA on clinical outcomes. Third, we did not measure intracellular mtDNA levels, and thus cannot extrapolate these findings to the variation in cellular mtDNA content. Finally, we did not examine the mtDNA levels of healthy controls for comparison owing to limitations of funding and ethical considerations.
Conclusions
Plasma mtDNA levels were associated with the SOFA score and a poor prognosis of patients with sepsis in the emergency room. Plasma mtDNA could serve as a predictor of 28-d mortality of patients with sepsis in the ED.
Availability of data and materials
The datasets used during the current study are available from the corresponding author on reasonable request.
Abbreviations
- mtDNA:
-
Mitochondrial DNA
- ICU:
-
Intensive care unit
- DAMP:
-
Damage-associated molecular pattern
- ED:
-
Emergency department
- SOFA:
-
Sequential Organ Failure Assessment
- SCM:
-
Septic cardiomyopathy
- BNP:
-
B-type natriuretic peptide
- qPCR:
-
Quantitative real-time polymerase chain reaction
- RSTI:
-
Respiratory tract infection
- CHD:
-
Coronary heart disease
- ROC:
-
Receiver operating characteristic
- CRP:
-
C-reactive protein
- PCT:
-
Procalcitonin
- AUC:
-
Area under the curve
- nucDNA:
-
Nuclear DNA
References
Seymour CW, Liu VX, Iwashyna TJ, Brunkhorst FM, Rea TD, Scherag A, Rubenfeld G, Kahn JM, Shankar-Hari M, Singer M, Deutschman CS, Escobar GJ, Angus DC. Assessment of clinical criteria for Sepsis: for the third international consensus definitions for Sepsis and septic shock (Sepsis-3). JAMA. 2016;315(8):762–74. https://doi.org/10.1001/jama.2016.0288.
Vincent JL, Marshall JC, Namendys-Silva SA, Francois B, Martin-Loeches I, Lipman J, Reinhart K, Antonelli M, Pickkers P, Njimi H, Jimenez E, Sakr Y. Assessment of the worldwide burden of critical illness: the intensive care over nations (ICON) audit. Lancet Respir Med. 2014;2(5):380–6. https://doi.org/10.1016/s2213-2600(14)70061-x.
Torio CM, Andrews RM. National Inpatient Hospital Costs: the Most expensive conditions by payer, 2011: statistical brief #160. In: Healthcare cost and utilization project (HCUP) statistical briefs. Rockville (MD): Agency for Healthcare Research and Quality (US); 2006.
Iwashyna TJ, Cooke CR, Wunsch H, Kahn JM. Population burden of long-term survivorship after severe sepsis in older Americans. J Am Geriatr Soc. 2012;60(6):1070–7. https://doi.org/10.1111/j.1532-5415.2012.03989.x.
Gaieski DF, Edwards JM, Kallan MJ, Carr BG. Benchmarking the incidence and mortality of severe sepsis in the United States. Crit Care Med. 2013;41(5):1167–74. https://doi.org/10.1097/CCM.0b013e31827c09f8.
Dellinger RP, Levy MM, Rhodes A, Annane D, Gerlach H, Opal SM, Sevransky JE, Sprung CL, Douglas IS, Jaeschke R, Osborn TM, Nunnally ME, Townsend SR, Reinhart K, Kleinpell RM, Angus DC, Deutschman CS, Machado FR, Rubenfeld GD, Webb SA, Beale RJ, Vincent JL, Moreno R. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2012. Crit Care Med. 2013;41(2):580–637. https://doi.org/10.1097/CCM.0b013e31827e83af.
Rhee C, Gohil S, Klompas M. Regulatory mandates for sepsis care--reasons for caution. N Engl J Med. 2014;370(18):1673–6. https://doi.org/10.1056/NEJMp1400276.
Kraut JA, Madias NE. Lactic acidosis. N Engl J Med. 2014;371(24):2309–19. https://doi.org/10.1056/NEJMra1309483.
Boyapati RK, Tamborska A, Dorward DA, Ho GT. Advances in the understanding of mitochondrial DNA as a pathogenic factor in inflammatory diseases. F1000Research 6:169; 2017. https://doi.org/10.12688/f1000research.10397.1.
Di Caro V, Walko TD 3rd, Bola RA, Hong JD, Pang D, Hsue V, Au AK, Halstead ES, Carcillo JA, Clark RS, Aneja RK. Plasma mitochondrial DNA--a novel DAMP in pediatric Sepsis. Shock. 2016;45(5):506–11. https://doi.org/10.1097/SHK.0000000000000539.
Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, Hauser CJ. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464(7285):104–7. https://doi.org/10.1038/nature08780.
West AP, Shadel GS, Ghosh S. Mitochondria in innate immune responses. Nat Rev Immunol. 2011;11(6):389–402. https://doi.org/10.1038/nri2975.
Zhang Q, Itagaki K, Hauser CJ. Mitochondrial DNA is released by shock and activates neutrophils via p38 map kinase. Shock. 2010;34(1):55–9. https://doi.org/10.1097/SHK.0b013e3181cd8c08.
Nakahira K, Kyung SY, Rogers AJ, Gazourian L, Youn S, Massaro AF, Quintana C, Osorio JC, Wang Z, Zhao Y, Lawler LA, Christie JD, Meyer NJ, Mc Causland FR, Waikar SS, Waxman AB, Chung RT, Bueno R, Rosas IO, Fredenburgh LE, Baron RM, Christiani DC, Hunninghake GM, Choi AM. Circulating mitochondrial DNA in patients in the ICU as a marker of mortality: derivation and validation. PLoS Med. 2013;10(12):e1001577; discussion e1001577. https://doi.org/10.1371/journal.pmed.1001577.
Harrington JS, Choi AMK, Nakahira K. Mitochondrial DNA in Sepsis. Curr Opin Crit Care. 2017;23(4):284–90. https://doi.org/10.1097/MCC.0000000000000427.
Beesley SJ, Weber G, Sarge T, Nikravan S, Grissom CK, Lanspa MJ, Shahul S, Brown SM. Septic cardiomyopathy. Crit Care Med. 2018;46(4):625–34. https://doi.org/10.1097/ccm.0000000000002851.
Bhagirath VC, Dwivedi DJ, Liaw PC. Comparison of the Proinflammatory and Procoagulant properties of nuclear, mitochondrial, and bacterial DNA. Shock. 2015;44(3):265–71. https://doi.org/10.1097/shk.0000000000000397.
Timmermans K, Kox M, Scheffer GJ, Pickkers P. Plasma nuclear and mitochondrial DNA levels, and markers of inflammation, shock, and organ damage in patients with septic shock. Shock. 2016;45(6):607–12. https://doi.org/10.1097/shk.0000000000000549.
Kung CT, Hsiao SY, Tsai TC, Su CM, Chang WN, Huang CR, Wang HC, Lin WC, Chang HW, Lin YJ, Cheng BC, Su BY, Tsai NW, Lu CH. Plasma nuclear and mitochondrial DNA levels as predictors of outcome in severe sepsis patients in the emergency room. J Transl Med. 2012;10:130. https://doi.org/10.1186/1479-5876-10-130.
Yang Y, Yang J, Yu B, Li L, Luo L, Wu F, Wu B. Association between circulating mononuclear cell mitochondrial DNA copy number and in-hospital mortality in septic patients: a prospective observational study based on the Sepsis-3 definition. PLoS One. 2019;14(2):e0212808. https://doi.org/10.1371/journal.pone.0212808.
Parikh SM, Yang Y, He L, Tang C, Zhan M, Dong Z. Mitochondrial function and disturbances in the septic kidney. Semin Nephrol. 2015;35(1):108–19. https://doi.org/10.1016/j.semnephrol.2015.01.011.
Shen X, Han G, Li S, Song Y, Shen H, Zhai Y, Wang Y, Zhang F, Dong N, Li T, Yao Y, Zhu H. Association between the T6459C point mutation of the mitochondrial MT-CO1 gene and susceptibility to sepsis among Chinese Han people. J Cell Mol Med. 2018;22(11):5257–64. https://doi.org/10.1111/jcmm.13746.
Diebel LN, Liberati DM, Ledgerwood AM, Lucas CE. Changes in lymph proteome induced by hemorrhagic shock: the appearance of damage-associated molecular patterns. J Trauma Acute Care Surg. 2012;73(1):41–50; discussion 51. https://doi.org/10.1097/TA.0b013e31825e8b32.
El Kebir D, Jozsef L, Pan W, Wang L, Filep JG. Bacterial DNA activates endothelial cells and promotes neutrophil adherence through TLR9 signaling. J Immunol. 2009;182(7):4386–94. https://doi.org/10.4049/jimmunol.0803044.
Schneberger D, Caldwell S, Kanthan R, Singh B. Expression of toll-like receptor 9 in mouse and human lungs. J Anat. 2013;222(5):495–503. https://doi.org/10.1111/joa.12039.
Sun S, Sursal T, Adibnia Y, Zhao C, Zheng Y, Li H, Otterbein LE, Hauser CJ, Itagaki K. Mitochondrial DAMPs increase endothelial permeability through neutrophil dependent and independent pathways. PLoS One. 2013;8(3):e59989. https://doi.org/10.1371/journal.pone.0059989.
Yao X, Wigginton JG, Maass DL, Ma L, Carlson D, Wolf SE, Minei JP, Zang QS. Estrogen-provided cardiac protection following burn trauma is mediated through a reduction in mitochondria-derived DAMPs. Am J Phys Heart Circ Phys. 2014;306(6):H882–94. https://doi.org/10.1152/ajpheart.00475.2013.
Innocenti F, Meo F, Giacomelli I, Tozzi C, Ralli ML, Donnini C, Tassinari I, Caldi F, Zanobetti M, Pini R. Prognostic value of serial lactate levels in septic patients with and without shock. Intern Emerg Med. 2019. https://doi.org/10.1007/s11739-019-02196-z.
Acknowledgements
We wish to thank our colleagues at Department of Rheumatology, Ren ji Hospital for their help and advice, Dr. Shuang Ye and Dr. Haiting Wang for his expert advice assistance in this study.
Funding
This study was partly supported by grants from the foundation of Health and Family Planning Commission of Shanghai Province (CN) (NO.201640142).
Author information
Authors and Affiliations
Contributions
LFW: acquisition of the data, drafting of the manuscript. WZ: analysis and interpretation of the data. KWW: statistical expertise and figure drawing. SJH: critical revision of the manuscript for important intellectual content. YC: study concept and design. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Shanghai Jiaotong University School of Medicine, Renji Hospital Ethics Committee (NO. 2016-109 k) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. All of the patients provided written informed consent.
Consent for publication
Not applicable.
Competing interests
All of the authors report no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, L., Zhou, W., Wang, K. et al. Predictive value of circulating plasma mitochondrial DNA for Sepsis in the emergency department: observational study based on the Sepsis-3 definition. BMC Emerg Med 20, 25 (2020). https://doi.org/10.1186/s12873-020-00320-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12873-020-00320-3