Abstract
Background
Plant height (PH) and spike compactness (SC) are important agronomic traits that affect yield improvement in wheat crops. The identification of the loci or genes responsible for these traits is thus of great importance for marker-assisted selection in wheat breeding.
Results
In this study, we used a recombinant inbred line (RIL) population with 139 lines derived from a cross between the mutant Rht8-2 and the local wheat variety NongDa5181 (ND5181) to construct a high-density genetic linkage map by applying the Wheat 40 K Panel. We identified seven stable QTLs for PH (three) and SC (four) in two environments using the RIL population, and found that Rht8-B1 is the causal gene of qPH2B.1 by further genetic mapping, gene cloning and gene editing analyses. Our results also showed that two natural variants from GC to TT in the coding region of Rht8-B1 resulted in an amino acid change from G (ND5181) to V (Rht8-2) at the 175th position, reducing PH by 3.6%~6.2% in the RIL population. Moreover, gene editing analysis suggested that the height of T2 generation in Rht8-B1 edited plants was reduced by 5.6%, and that the impact of Rht8-B1 on PH was significantly lower than Rht8-D1. Additionally, analysis of the distribution of Rht8-B1 in various wheat resources suggested that the Rht8-B1b allele has not been widely utilized in modern wheat breeding.
Conclusions
The combination of Rht8-B1b with other favorable Rht genes might be an alternative approach for developing lodging-resistant crops. Our study provides important information for marker-assisted selection in wheat breeding.
Similar content being viewed by others
Background
The global human population is predicted to continually expand and reach 10 billion by 2050, significantly increasing the need for the safe and reliable food production. The improvement of crops using advanced technologies provides an effective strategy to meet food production demands in the future [1]. Wheat represents one of the most important staple crops worldwide, and the identification of quantitative trait loci (QTLs) important for agronomical traits, such as plant height (PH) and spikelet compactness (SC), offers critical information to ensure food security [2].
PH is tightly associated with lodging resistance and thus influences grain yield. To date, a total of 25 reduced height genes (Rht1–Rht25) have been documented in wheat [3], and these dwarf genes have been classified into gibberellic acid (GA)-sensitive or -insensitive based on response to GA treatment. The GA-insensitive Rht-B1b (Rht1) and Rht-D1b (Rht2) genes on chromosomes 4B and 4D, which encode truncated DELLA proteins, significantly increase the harvest index, and their use in breeding resulted in the well-known ‘Green Revolution’ [4, 5]. their allelic variations such as Rht-B1c (Rht3) [6], Rht-B1e (Rht11) [7], Rht-B1p (Rht17) [8], and Rht-D1c (Rht10) [9] have been identified. Moreover, several GA-sensitive dwarf genes including Rht8 [10, 11], Rht12 [12], Rht13 [13], Rht18 [14], and Rht24 [15] have been cloned or intensively studied in wheat. Rht12 was located on chromosome 5A and mutations in GA2oxA13 gene produced tall overgrowth phenotype in the Rht12 background [12]. A missense mutation of NB-LRR gene in Rht13 caused height reduction [13]. The dominant Rht18 gene was identified by isolating and sequencing chromosome 6A of overgrowth mutants, and the dwarf phenotype of this mutant was found to be caused by the increased expression of GA2oxA9 resulting in a reduction of active GA content [14]. Map-based cloning suggested that GA2oxA9 was the causal gene of Rht24, which affected GA homeostasis and led to plant height reduction [15].
Since the Rht8 gene does not influence coleoptile length, it well complements Rht-B1b and Rht-D1b weakness, and it has been widely used in wheat breeding for several decades [16, 17]. Rht8 was mapped on the short arm of chromosome 2D, and the SSR marker Xgwm261 was regarded as a perfect diagnostic marker for Rht8 previously [17, 18]. Using two wheat mutants Rht8-2 and Rht8-3 for construction of segregation populations, the Rht8 gene was cloned recently, and it was found to encode an RNase H-like protein that affects bioactive GA content and changes plant height [10]. Similar results were obtained simultaneously in a map-based cloning study using a wheat variety containing the Rht8 gene [11], and both studies also showed that the dwarf allele of Rht8 was positively selected during wheat breeding [10, 11].
Previous analyses of near isogenic lines (NILs) [19] and transgenic plants [11] have suggested that Rht8 not only reduces PH but also significantly decreases spike length (SL) and thus increases SC. The modification of SL or SC plays an important role in the improvement of yield potential in wheat [20, 21], and identification of associated QTLs is critical for wheat improvement. In hexaploid wheat, spike morphology is regulated by three major genes, namely Q, C (Compactum), and S (Sphaerococcum) located on chromosomes 5A, 2D, and 3D, respectively [22,23,24]. These genes exert pleiotropic effects on SC and SL, PH, and grain shape. To date, a large number of QTLs associated with spike morphology have been identified on nearly all wheat chromosomes [25]. In addition, the VRN, Ppd, and Eps genes have also been found to be involved in SL and development and to affect SC in wheat [26,27,28].
In the present study, we dissected the genetic control of PH and SC by performing QTL mapping using a recombinant inbred line (RIL) population derived from a cross between the wheat variety ND5181 and the mutant line Rht8-2 and identified natural variation in the homoeologous gene Rht8 that contributes to PH change for qPH2B.1. The linkage markers and genes influencing PH and SC identified here can be applied to molecular breeding and the promotion of wheat production.
Materials and methods
Plant materials and phenotypic evaluation
We established an F7 RIL population comprising 139 lines derived from a cross between wheat cultivar NongDa5181 (ND5181) and the mutant Rht8-2 using the single seed descent method. The RIL population and the parents were planted in the Zhongpuchang and Changping experimental fields at the Institute of Crop Sciences, Chinese Academy of Agricultural Sciences (39°97′N, 116°34′E; Beijing, China) during the 2021–2022 crop season. Each genotype was planted in one row (15 seeds per row), which was 1.5 m long with a row spacing of 0.3 m. PH and SC (calculated by dividing spikelet number per spike by the SL) were measured at maturity with eight replicates for each line [29].
Genotyping and QTL mapping
Genomic DNA of the RIL population and parent lines was extracted and assessed as previously described [29]. The DNA samples were hybridized to the GenoBaits Wheat 40 K Panel containing 202,971 markers. Genotyping was performed at the MOLBREEDING (Shijiazhuang) Biotech Co., Ltd. (http://www.molbreeding.com). A total of 15,258 homozygous SNPs were selected from ND5181 and Rht8-2 for follow-up analyses. The BIN and MAP functions of IciMapping 4.1 were used to remove redundant markers and construct the genetic map, respectively. The genetic linkage map contained 1847 Bin markers across 21 chromosomes. The threshold of the logarithm of odds (LOD) score was set to 2.5, and the Kosambi map function was used to calculate the map distance from recombination frequencies. Composite interval mapping (ICIM) on IciMapping 4.1 was selected to identify QTLs for PH and SC. The mean values of the phenotypic traits in each line were used for QTL analysis. QTLs detected in two environments were regarded as stable QTLs.
KASP marker development and QTL validation
We developed the KASP assay based on the SNPs identified within or around ND5181 and Rht8-2 from the 40 K SNP array genotyping data. The online primer design pipeline PolyMarker (http://polymarker.tgac.ac.uk/) was used to design specific primers. The KASP assays were performed as previously described [29]. End-point fluorescence data were screened using the microplate reader FLUOstar Omega SNP (BMG LABTECH, Germany) and analyzed by the Klustering Caller Software. The KASP markers were tested on the two parents, and then the developed polymorphic KASP markers were used for the identification of genotypes in the F7 RIL population. QTL analysis was conducted using IciMapping 4.1.
Cloning and sequencing of Rht8-B1
Gene-specific primers were designed to amplify the full-length sequence of Rht8-B1. The genomic DNA of ND5181 and Rht8-2 was extracted and used as a template in PCR. The components of each 20 µL reaction were as follows: 10 µL buffer, 4 µL dNTPs, 1 µL genomic DNA, 0.8 µL forward primer, 0.8 µL reverse primer, 0.4 µL KOD FX, and 3 µL ddH2O. The reaction conditions were 94 ℃ for 2 min, followed by 32 cycles of 98 ℃ for 10 s, annealing at 65 ℃ for 20 s, and 68 ℃ for 2 min, with a final extension of 68 ℃ for 5 min. The PCR products were sequenced at the Shanghai Sangon Biotech Co., Ltd. (https://www.sangon.com/).
Generating Rht8-B1 and Rht8-D1 mutants by gene editing
For CRISPR/Cas9-based gene editing, single guide RNA (sgRNA) target sequences were designed and plant transformations were performed as previously described [10]. The sgRNA sequence targeting Rht8 genes was 5’-GCCGCCGGAGAGCAGCTGCC-3’. We sequenced the Rht8 genes in T2 plants and validated the mutations produced by CRISPR/Cas9-based gene editing. Single mutants of Rht8-B1 and Rht8-D1 were successfully selected, and the heights of edited and wild-type (WT) plants recorded and compared.
Quantitative RT-PCR
Quantitative RT-PCR was performed according to our previous report [30]. Briefly, total RNA was isolated from the first internode below the spike using TRNzol-A+ Reagent (Tiangen Biotech), and then purified using an RNA purification kit (Tiangen). The first-strand cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad), and the SsoFast EvaGreen Supermix Kit (Bio-Rad) was used for quantitative RT-PCR. This experiment was conducted on a CFX 96 Real-Time System (Bio-Rad) following the manufacturer’s instructions. The actin gene was used as an internal control. The primers used for quantitative RT-PCR are listed in Supplementary Table 5.
Distribution analysis of Rht8-B1b in wheat accessions
A total of 305 worldwide accessions with genotypic information obtained from the Wheat Union Database (http://wheat.cau.edu.cn/WheatUnion/) were used for analysis of allelic variation in Rht8-B1 [31,32,33,34]. The frequency of Rht8-B1b in wheat accessions from different geographical regions was calculated according to the number of accessions carrying Rht8-B1b allele.
Accession numbers
Sequence data of Rht8-B1a and Rht8-B1b have been deposited in the GenBank data library under the accession numbers OQ512875 and OQ512876, respectively.
Statistical analysis
Statistical analyses, namely Student’s t-tests and correlation analyses, were performed using SPSS v21.0 software (IBM, USA).
Results
Phenotypic variation of two parent lines and the RIL population
We have previously identified a semi-dwarf wheat mutant line Rht8-2 with high yield potential [10]. To explore QTLs associated with important agronomic traits, we constructed a RIL population including 139 lines derived from a cross between the wheat variety ND5181 and the mutant line Rht8-2. The two parent lines showed significant differences in PH and SC in two environments (Zhongpuchang and Changping) (Fig. 1A). Specifically, compared with ND5181, the PH of Rht8-2 was 11 cm shorter, and SC was larger by 0.45 (Fig. 1; Table S1). In the RIL population, PH and SC displayed obvious transgressive segregation. PH and SC showed normal distributions in the two environments, suggesting they are controlled by multiple genes (Fig. 2). Pairwise correlation analysis between PH and SC showed correlation coefficients of -0.45 and -0.41 with statistical significance in the Zhongpuchang and Changping field experiments, indicating that a negative correlation between PH and SC in the RIL population.
The frequency distributions of plant height, and spikelet compactness in the RIL population. A-B. Frequency distribution of plant height in the Zhongpuchang (A) and Changping (B) environments. C-D. Frequency distribution of spikelet compactness in in the Zhongpuchang (C) and Changping (D) environments. Phenotypic values of the two parental lines are marked by vertical arrows
QTL mapping analysis
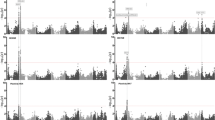
A total of 23 QTLs associated with PH and SC were detected in the two environments on chromosomes 1A, 1B, 2B, 2D, 3A, 3B, 4B, 5A, 5B, 6B, 6D, 7A, 7B, and 7D. Among these QTLs, qPH2B.1, qPH2D, qPH4B, qSC1B, qSC2B.1, qSC2D.1, and qSC7D, were detected in both environments (Fig. 3; Table 1).
Three stable QTLs associated with PH were identified on chromosomes 2B, 2D, and 4B in both environments. The major QTL qPH4B showed the highest LOD scores (30.1 in Zhongpuchang and 40.2 in Changping) and explained 29.7% and 41.8% of the phenotypic variation, respectively. qPH2D had LOD scores 22.7 (Zhongpuchang) and 23.6 (Changping) and accounted for 19.2% and 17.8% of the phenotypic variation, respectively, and qPH2B.1 had LOD scores of 11.5 (Zhongpuchang) and 4.0 (Changping) and explained 7.9% and 2.1% of the phenotypic variation, respectively. The qPH4B allele from ND5181 decreased PH, while the alleles of qPH2D and qPH2B.1 from Rht8-2 reduced PH (Table 1).
We identified four stable QTLs associated with SC on chromosomes 1B, 2B, 2D, and 7D in the two environments. qSC2B.1 had LOD scores 24.6 and 12.7 and explained 25.0% and 15.0% of the phenotypic variation in Zhongpuchang and Changping, respectively; qSC2D.1 had LOD scores of 13.6 and 19.4 and explained 13.0% and 27.6% of the phenotypic variation, respectively; qSC1B had LOD scores of 5.0 and 6.2 and explained 3.8% and 7.0% of the phenotypic variation, respectively; and qSC7D had LOD scores of 2.8 and 5.4 and explained 1.9% and 5.6% of the phenotypic variation, respectively. Among these QTLs in ND5181, the qSC1B and qSC7D alleles increased SC and the qSC2B.1 and qSC2D.1 alleles decreased SC (Table 1).
Rht8-B1 is the candidate gene of qPH2B.1
To validate the mapped region of qPH2B.1, we successfully developed nine KASP makers around the qPH2B.1 region based on the results of 660 K SNP array analysis between the ND5181 and Rht8-2 varieties. These markers validated this region and delimited it to a physical interval of 3.5 Mb between markers 2B-4 and 2B-5. This QTL showed a LOD score of 3.4 and explained 11.8% of the observed phenotypic variation (Fig. 4). Given that this region included Rht8-B1 (TraesCS2B02G073600), a homoeologous gene of Rht8, we sequenced the region and uncovered two genetic variants (39,418,567–39,418,568, GC in ND5181 and TT in Rht8-2) in the coding region of the gene, which resulted in an amino acid change from G (ND5181) to V (Rht8-2) at the 175th position. We then analyzed the effects of Rht8-B1 on PH and SC in the RIL population and found that TT allele reduced PH by 6.2% and 3.6%, shortened SL by 6.6% and 4.7%, and increased SC by 5.8% and 6.3% in the Zhongpuchang and Changping field experiments, respectively (Fig. 4).
Rht8-B1 exhibited a lower impact on PH reduction than Rht8-D1
We compared the effects of Rht8-B1 and the previously reported Rht8-D1 gene on PH by knocking out both genes in a Fielder background and evaluating the PH of the T2 lines. The results showed that the PH of edited plants was significantly lower than that of control plants. Specifically, edited Rht8-B1 and Rht8-D1 plants showed a PH reduction of 5.6% and 17.5%, respectively, compared with the WT control, suggesting that Rht8-B1 had a smaller effect on PH than Rht8-D1 (Fig. 5; Figure S1).
Comparison of the expression of Rht8-B1 and Rht8-D1
To examine the difference in expression patterns between Rht8-B1 and Rht8-D1, we analyzed their transcript levels using the Hexaploid Wheat Expression Dataset [35] and found that although both genes were highly expressed in the stem at the jointing stage, the expression of Rht8-D1 was significantly higher than that of Rht8-B1 (Fig. 6A). Our previous study suggested that a frameshift mutation in Rht8-D1 caused the dwarfism phenotype in Rht8-2 and that its expression was significantly lower in the mutant compared with that in WT [10]. To investigate the effects of mutation of Rht8-D1 on the B subgenome of Rht8, we analyzed the expression of Rht8-B1 in the first internode below the spike of the mutant Rht8-2 and WT. The results showed that the expression of Rht8-B1 was remarkably higher in Rht8-2 (Fig. 6B), indicating that the mutation of Rht8-D1 affected the transcript level of Rht8-B1.
Distribution of Rht8-B1 in wheat varieties worldwide
We used a total of 305 worldwide accessions from the Wheat Union Database (http://wheat.cau.edu.cn/WheatUnion/), namely 193 accessions from China and 112 accessions from other countries, for the analysis of Rht8-B1 allelic variation. We found that 68 Chinese accessions (35.2%) contained the Rht8-B1b TT allele, compared with only 6 accessions (5.4%) in other countries (Fig. 7). Of the Chinese accessions, 20.3% of the 118 modern varieties and 58.7% of the 75 landrace accessions had Rht8-B1b allele, (Table S2). These results suggest that Rht8-B1b allele was not widely used historically for wheat breeding.
Discussion
Semi-dwarf wheat usually possesses high lodging resistance with high yield stability [4]. The identification of novel genes regulating PH is pivotal for improving lodging resistance through wheat breeding. In this study, we identified Rht8-B1 as a novel regulator of PH by performing genetic mapping and gene editing analyses. By combining mapping using the Wheat40K array and molecular markers, qPH2B.1 was mapped within a physical interval of 36.5–40.0 Mb between markers 2B-4 and 2B-5 on chromosome 2B of the Chinese Spring reference genome v1.0 (Fig. 4A). The Rht8-B1 gene was located to position ~ 39.4 Mb on chromosome 2B, making it a candidate gene for qPH2B.1. Genotyping of Rht8-B1 in the RIL population showed that genetic variants in this gene were associated with PH (Fig. 4B). Further gene editing of Rht8-B1 validated its function in the regulation of PH (Fig. 5). These results suggest Rht8-B1 is the causal gene of qPH2B.1.
We recently identified Rht8 on chromosome 2D (Rht8-D1) by performing map-based cloning using two dwarf mutants, Rht8-2 and Rht8-3; this gene encodes a Ribonuclease H-like protein that modifies PH by regulating the bioactive GA content [10]. Here, we used Rht8-2 for the construction of a RIL population and found a major QTL – qPH2D – on chromosome 2D with a high LOD score, which probably represents Rht8-D1. In contrast to the Rht8-D1 (qPH2D) locus, qPH2B.1 had a minor effect on PH (Figs. 3 and 4 A), which is consistent with gene editing results (Fig. 5) and reports by Chai et al. [11]. The larger effect of Rht8-D1 on PH relative to Rht8-B1 is also in accordance with the higher expression of Rht8-D1 in the stem (Fig. 6A). Previous studies extensively investigated the function of the Rht8 orthologous gene TAC4/sg2 in rice [36, 37]. A stop-gain mutation in TAC4 was found to lead to a greater tiller angle and a dwarf phenotype that may result from changes in IAA content and distribution. In addition, an 8-bp deletion in sg2 was found to result in a smaller grain phenotype due to repressed cell expansion in spikelet hulls and a semi-dwarf phenotype. These observations indicate that orthologous genes to Rht8 affect multiple agronomic traits and play different roles, highlighting the need for further exploration of the effects of Rht8-B1 and Rht8-D1 on PH in the future.
The semidwarf Rht8-B1b allele has high potential for utilization in wheat breeding. By analyzing the genetic sequence of Rht8-B1 in the mutant Rht8-2 and WT variety Jing411, we found that the Rht8-B1b allele was derived from the WT, indicating that it is a natural variant. Importantly, the distribution of the Rht8-B1b allele in global wheat varieties suggests it has not been widely utilized historically in wheat breeding outside of China (Fig. 7). Similar to Rht8-D1b, the Rht8-B1b allele had no effects on thousand grain weight. Several studies have suggested that dwarf or semidwarf genes are associated with a decrease in thousand grain weight and ultimately affect wheat yield [38,39,40]. The combination of Rht8-B1b and other dwarf genes in wheat breeding can thus be an alternative approach for developing lodging-resistant wheat.
We found consistency between the stable QTLs associated with PH and SC detected in this study and previous reports. For example, Rht-B1b was located at ~ 30.8 Mb on chromosome 4B within the chromosomal region corresponding to qPH4B [5]; qSC2D.1 was identified between molecular markers A61578 and A61731 in the interval 20.8–30.3 Mb, and is closely linked to Rht8-D1, which was reported to significantly reduce SL [11]; qSC7D was located between molecular markers A202015 and A202077 in the interval 584.5–588.2 Mb, and is closely linked to WAPO1-7D, which regulates spikelet number per spike [41]. We also identified several putatively novel QTLs for PH and SC, including qPH7B.1 on chromosome 7BL, which had a LOD score 10.3 and explained 7.0% of the phenotypic variation in PH, and qSC1B located on the long arm of chromosome 1B between molecular markers A18513 and A19081, which is associated with SC.
Conclusion
We identified seven stable QTLs for PH and SC in two environments using a RIL population. Using mapping and gene editing analyses we found that Rht8-B1 is the causal gene of qPH2B.1 and affects PH variation. Rht8-B1 had less of an effect on PH than Rht8-D1 and has not been widely utilized in wheat breeding. This implies that combining Rht8-B1b with other favorable Rht genes has great potential for breeding lodging-resistant wheat varieties. Our study revealed novel markers and QTLs, providing important information for marker-assisted selection in wheat.
Data Availability
All data generated or analyzed during this study are included in the main text article and its supplementary files. The variant data for this study have been deposited in the European Variation Archive (EVA) at EMBL-EBI under accession number PRJEB60409 (http://www.ebi.ac.uk/eva/?eva-study=PRJEB60409).
References
Hickey LT, Hafeez AN, Robinson H, Jackson SA, Leal-Bertioli SCM, Gao MTester, Godwin CX, Hayes ID, Wulff BJ. Breeding crops to feed 10 billion. Nat Biotechnol. 2019;37(7):744–54.
Xiao J, Liu B, Yao YY, Guo ZF, Jia HY, Kong LR, Zhang AM, Ma WJ, Ni ZF, Xu SB, et al. Wheat genomic study for genetic improvement of traits in China. Sci China Life Sci. 2022;65(9):1718–75.
Mo YJ, Vanzetti LS, Hale I, Spagnolo EJ, Guidobaldi F, Al-Oboudi J, Odle N, Pearce S, Helguera M, Dubcovsky J. Identification and characterization of Rht25, a locus on chromosome arm 6AS affecting wheat plant height, heading time, and spike development. Theor Appl Genet. 2018;131(10):2021–35.
Hedden P. The genes of the Green Revolution. Trends Genet. 2003;19:5–9.
Peng JR, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Harberd NP. Green revolution’ genes encode mutant gibberellin response modulators. Nature. 1999;400:256–611.
Wen W, Deng QY, Jia HY, Wei LZ, Wei JB, Wan HS, Yang LM, Cao WJ, Ma ZQ. Sequence variations of the partially dominant DELLA gene Rht-B1c in wheat and their functional impacts. J Exp Bot. 2013;64(11):3299–312.
Divashuk MG, Vasilyev AV, Bespalova LA, Karlov GI. Identity of the Rht-11 and Rht-B1e reduced plant height genes. Russ J Genet. 2012;48(7):761–3.
Bazhenov MS, Divashuk MG, Amagai Y, Watanabe N, Karlov GI. Isolation of the dwarfing Rht-B1p (Rht17) gene from wheat and the development of an allele-specific PCR marker. Mol Breed. 2015;35(11). https://doi.org/10.1007/s11032-11015-10407-11031.
Li YY, Xiao JH, Wu JJ, Duan JL, Liu Y, Ye XG, Zhang X, Guo XP, Gu YQ, Zhang LC, et al. A tandem segmental duplication (TSD) in green revolution gene Rht-D1b region underlies plant height variation. New Phytol. 2012;196(1):282–91.
Xiong HC, Zhou CY, Fu MY, Guo HJ, Xie YD, Zhao LS, Gu JY, Zhao SR, Ding YP, Li YT, et al. Cloning and functional characterization of Rht8, a “Green Revolution” replacement gene in wheat. Mol Plant. 2022;15(3):373–6.
Chai LL, Xin MM, Dong CQ, Chen ZY, Zhai HJ, Zhuang JH, Cheng XJ, Wang NJ, Geng J, Wang XB, et al. A natural variation in ribonuclease H-like gene underlies Rht8 to confer “Green Revolution” trait in wheat. Mol Plant. 2022;15(3):377–80.
Buss W, Ford BA, Foo E, Schnippenkoetter W, Borrill P, Brooks B, Ashton AR, Chandler PM, Spielmeyer W. Overgrowth mutants determine the causal role of gibberellin GA2oxidaseA13 in Rht12 dwarfism of wheat. J Exp Bot. 2020;71(22):7171–8.
Borrill P, Mago R, Xu TY, Ford B, Williams SJ, Derkx A, Bovill WD, Hyles J, Bhatt D, Xia XD, et al. An autoactive NB-LRR gene causes Rht13 dwarfism in wheat. Proc Natl Acad Sci U S A. 2022;119(48):e2209875119.
Ford BA, Foo E, Sharwood R, Karafiatova M, Vrana J, MacMillan C, Nichols DS, Steuernagel B, Uauy C, Dolezel J, et al. Rht18 semidwarfism in wheat is due to increased GA 2-oxidaseA9 expression and reduced GA content. Plant Physiol. 2018;177(1):168–80.
Tian XL, Xia XC, Xu DA, Liu YQ, Xie L, Hassan MA, Song J, Li FJ, Wang DS, Zhang Y, et al. Rht24b, an ancient variation of TaGA2ox-A9, reduces plant height without yield penalty in wheat. New Phytol. 2021;233:738–50.
Botwright TL, Rebetzke GJ, Condon AG, Richards RA. Influence of the gibberellin-sensitive Rht8 dwarfing gene on leaf epidermal cell dimensions and early vigour in wheat (Triticum aestivum L). Ann Bot. 2005;95(4):631–9.
Zhang XK, Yang SJ, Zhou Y, He ZH, Xia XC. Distribution of the Rht-B1b, Rht-D1b and Rht8 reduced height genes in autumn-sown chinese wheats detected by molecular markers. Euphytica. 2006;152(1):109–16.
Korzun V, Roder M, W GM, Law JWA. Genetic analysis of the dwarfing gene (Rht8) in wheat. Part I. Molecular mapping of Rht8 on the short arm of chromosome 2D of bread wheat (Triticum aestivum L). Theor Appl Genet. 1998;96:1104–9.
Kowalski AM, Gooding M, Ferrante A, Slafer GA, Orford S, Gasperini D, Griffiths S. Agronomic assessment of the wheat semi-dwarfing gene Rht8 in contrasting nitrogen treatments and water regimes. Field Crops Res. 2016;191:150–60.
Li T, Deng GB, Su Y, Yang Z, Tang YY, Wang JH, Qiu XB, Pu X, Li J, Liu ZH, et al. Identification and validation of two major QTLs for spike compactness and length in bread wheat (Triticum aestivum L.) showing pleiotropic effects on yield-related traits. Theor Appl Genet. 2021;134(11):3625–41.
Wu XY, Cheng RR, Xue SL, Kong ZX, Wan HS, Li GQ, Huang YL, Jia HY, Jia JZ, Zhang LX, et al. Precise mapping of a quantitative trait locus interval for spike length and grain weight in bread wheat (Triticum aestivum L). Mol Breed. 2013;33(1):129–38.
Fan XL, Cui F, Ji J, Zhang W, Zhao XQ, Liu JJ, Meng DY, Tong YP, Wang T, Li JM. Dissection of pleiotropic QTL regions controlling wheat spike characteristics under different nitrogen treatments using traditional and conditional QTL mapping. Front Plant Sci. 2019;10:187.
Johnson EB, Nalam VJ, Zemetra RS, Riera-Lizarazu O. Mapping the compactum locus in wheat (Triticum aestivum L.) and its relationship to other spike morphology genes of the Triticeae. Euphytica. 2007;163(2):193–201.
Sourdille P, Tixier MH, Charmet G, Gay G, Cadalen T, Bernard S, Bernard M. Location of genes involved in ear compactness in wheat (Triticum aestivum) by means of molecular markers. Mol Breed. 2000;6:247–55.
Hu JM, Wang XQ, Zhang GX, Jiang P, Chen WY, Hao YC, Ma X, Xu SS, Jia JZ, Kong LR, et al. QTL mapping for yield-related traits in wheat based on four RIL populations. Theor Appl Genet. 2020;133(3):917–33.
Guedira M, Xiong M, Hao YF, Johnson J, Harrison S, Marshall D, Brown-Guedira G. Heading date QTL in winter wheat (Triticum aestivum L.) coincide with major developmental genes VERNALIZATION1 and PHOTOPERIOD1. PLoS ONE. 2016;11(5):e0154242.
Wang L, Niu JS, Li QY, Qin Z, Ni YJ, Xu HX. Allelic variance at the vernalization gene locus Vrn-D1 in a group of sister wheat (Triticum aestivum) lines and its effects on development. JAS. 2014;153(4):588–601.
Yan LL, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci U S A. 2006;103:19581–6.
Zhou CY, Xiong HC, Li YT, Guo HJ, Xie YD, Zhao LS, Gu JY, Zhao SR, Ding YP, Song XY, et al. Genetic analysis and QTL mapping of a novel reduced height gene in common wheat (Triticum aestivum L). J Integr Agric. 2020;19(7):1721–30.
Xiong H, Zhou C, Guo H, Xie Y, Zhao L, Gu J, Zhao S, Ding Y, Liu L. Transcriptome sequencing reveals hotspot mutation regions and dwarfing mechanisms in wheat mutants induced by gamma-ray irradiation and EMS. J Radiat Res. 2020;61(1):44–57.
Hao CY, Jiao CZ, Hou J, Li T, Liu HX, Wang YQ, Zheng J, Liu H, Bi ZH, Xu FF, et al. Resequencing of 145 landmark cultivars reveals asymmetric sub-genome selection and strong founder genotype effects on wheat breeding in China. Mol Plant. 2020;13(12):1733–51.
Walkowiak S, Gao L, Monat C, Haberer G, Kassa MT, Brinton J, Ramirez-Gonzalez RH, Kolodziej MC, Delorean E, Thambugala D, et al. Multiple wheat genomes reveal global variation in modern breeding. Nature. 2020;588(7837):277–83.
Yang ZZ, Wang ZH, Wang WX, Xie XM, Chai LL, Wang XB, Feng XB, Li JH, Peng HR, Su ZQ, et al. ggComp enables dissection of germplasm resources and construction of a multiscale germplasm network in wheat. Plant Physiol. 2022;188(4):1950–65.
Zhou Y, Zhao XB, Li YW, Xu J, Bi AY, Kang LP, Xu DX, Chen HF, Wang Y, Wang YG, et al. Triticum population sequencing provides insights into wheat adaptation. Nat Genet. 2020;52(12):1412–22.
International Wheat Genome Sequencing C. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Sci. 2014;345(6194):1251788.
Huang YS, Dong H, Mou CL, Wang P, Hao QX, Zhang M, Wu HM, Zhang FL, Ma TF, Miao R, et al. Ribonuclease H-like gene small Grain2 regulates grain size in rice through brassinosteroids signal pathway. J Integr Plant Biol. 2022;10:1883–900.
Li H, Sun HY, Jiang JH, Sun XY, Tan LB, Sun CQ. TAC4 controls tiller angle by regulating the endogenous auxin content and distribution in rice. Plant Biotechnol J. 2021;19(1):64–73.
Casebow R, Hadley C, Uppal R, Addisu M, Loddo S, Kowalski A, Griffiths S, Gooding M. Reduced height (rht) alleles affect wheat grain quality. PLoS ONE. 2016;11(5):e0156056.
Daoura BG, Chen L, Du YY, Hu YG. Genetic effects of dwarfing gene Rht-5 on agronomic traits in common wheat (Triticum aestivum L.) and QTL analysis on its linked traits. Field Crops Res. 2014;156:22–9.
Yang ZY, Zheng JC, Liu CY, Wang YS, Condon AG, Chen YF, Hu YG. Effects of the GA-responsive dwarfing gene Rht18 from tetraploid wheat on agronomic traits of common wheat. Field Crops Res. 2015;183:92–101.
Kuzay S, Xu YF, Zhang JL, Katz A, Pearce S, Su ZQ, Fraser M, Anderson JA, Brown-Guedira G, DeWitt N, et al. Identification of a candidate gene for a QTL for spikelet number per spike on wheat chromosome arm 7AL by high-resolution genetic mapping. Theor Appl Genet. 2019;132(9):2689–705.
Acknowledgements
We thank the anonymous reviewers for constructive suggestions that helped to improve the manuscript.
Funding
This work was financially supported by the National Key Research and Development Program of China (Grant No. 2022YFD1200700), the Crop Varietal Improvement and Insect Pests Control by Nuclear Radiation, the China Agriculture Research System of MOF and MARA (Grant No. CARS-03), and the Agricultural Science and Technology Innovation Program (Grant No. CAAS-ZDRW202109).
Author information
Authors and Affiliations
Contributions
L. L. and X. L. conceived the project and revised the manuscript. C. Z., H. X, and M. F. conducted most of the experiments and analyzed the data. H. G., L. Z., Y. X., J. G., S. Z., Y. D. Y. L. helped in RIL population planting and data analysis. C. Z., and H. X. wrote the first draft of the manuscript. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Experimental research and field studies on plants including the collection of plant material are compliant with relevant guidelines and regulations. The authors confirm that all the experimental research done on cultivated plants and carried out in accordance with relevant institutional, national, and international guidelines, standards, and legislation. The plants used were cultivated under controlled conditions and thus posed no threat to other plant species, and no specific permission was required to collect the required sample material used in this study. The plants were cultivated with seeds obtained from the Institute of Crop Sciences, Chinese Academy of Agricultural Sciences (ICS-CAAS), Beijing.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhou, C., Xiong, H., Fu, M. et al. Genetic mapping and identification of Rht8-B1 that regulates plant height in wheat. BMC Plant Biol 23, 333 (2023). https://doi.org/10.1186/s12870-023-04343-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-023-04343-3