Abstract
Background
As the king of all herbs, the medicinal value of ginseng is self-evident. The perennial nature of ginseng causes its quality to be influenced by various factors, one of which is the soil environment. During plant growth and development, MYB transcription factors play an important role in responding to abiotic stresses and regulating the synthesis of secondary metabolites. However, there are relatively few reports on the MYB transcription factor family in Panax ginseng.
Results
This study identified 420 PgMYB transcripts under 117 genes ID in the Jilin ginseng transcriptome database. Phylogenetic analysis showed that PgMYB transcripts in Jilin ginseng were classified into 19 functional subclasses. The GO annotation result indicated that the functional differentiation of PgMYB transcripts was annotated to 11 functional nodes at GO Level 2 in ginseng. Expression pattern analysis of PgMYB transcripts based on the expression data (TPM) that PgMYB transcripts were revealed spatiotemporally specific in expression patterns. We performed a weighted network co-expression network analysis on the expression of PgMYB transcripts from different samples. The co-expression network containing 51 PgMYB transcripts was formed under a soft threshold of 0.85, revealing the reciprocal relationship of PgMYB in ginseng. Treatment of adventitious roots of ginseng with different concentrations of NaCl revealed four up-regulated expression of PgMYB transcripts that can candidate genes for salt resistance studies in ginseng.
Conclusions
The present findings provide data resources for the subsequent study of the functions of MYB transcription factor family members in ginseng, and provide an experimental basis for the anti-salt functions of MYB transcription factors in Panax ginseng.
Similar content being viewed by others
Introduction
Panax ginseng, a member of the perennial herb genus Ginseng in the family of Araliaceae, has been used medicinally for millennia. As a traditional and valuable Chinese medicine, ginseng is mainly used to treat weakness and fatigue, improve mental function, exercise capacity, immune function, and diabetes-related diseases [1]. As we all know, the dried root of ginseng is the main medicinal part of ginseng, and ginsenoside is the main active ingredient of ginseng. Ginsenosides are found in different tissues such as roots, leaves, stems, flower buds, and berries [2]. Ginsenoside content is affected by the age of growth and is also affected by various conditions such as species and cultivation area [3, 4]. Meanwhile, abiotic stresses can seriously affect the quality of ginseng during the years of growth. Important regulators of transcription factors in plant growth and development and secondary metabolism. According to previous studies, transcription factors such as WRKY [5], MYB [6], NAC [7], and ERF [8] play important roles in plant response to abiotic stresses.
MYB transcription factors, one of the largest family transcription factors in plants [9], were first reported in maize [10]. The MYB structural domain usually consists of one to three incomplete repeats of approximately 52 amino acid residues in a helix-turn-helix conformation inserted into the main DNA groove [11]. The MYB transcription factor family has been divided into subgroups based on the number of DNA-binding domains they contain, such as 1R-MYB, R2R3-MYB, 3R-MYB and 4R-MYB. Interestingly, in recent years, research on R2R3-MYB has been hotly pursued. MYB research has progressed hotly, while 4R-MYB has been less reported. So far, MYB transcription factors have been reported in Arabidopsis [12], rice [13], soybean [14], Hedychium coronarium [15], watermelon [16] and other plants have been identified. In rice, OsMYB2 was associated with salt, cold and dehydration tolerance [17] GmMYB12B2 affects the expression level of key enzyme genes for flavonoid biosynthesis in transgenic Arabidopsis [18]. Activating R2R3-myb genes induce repressive R2R3-myb genes to balance anthocyanin and proanthocyanidin accumulation [19] MYB transcription factors play an important role in regulating secondary metabolite biosynthesis, turgor development and stress response in tea tree [20]. In ginseng, the expression of PgMYB1 was up-regulated by abscisic acid, salicylic acid, NaCl, and cold (chilling), and down-regulated by methyl jasmonate. These results suggest that PgMYB1 might be involved in responding to environmental stresses and hormones [21]. PgMYB2 protein can bind to the promoter of PgDDS, which in turn regulates the expression of ginsenosides [22]. Although there are reports of MYB family members in ginseng, only two ginseng MYB genes have been cloned, and they were both cloned using homologous sequences from heterologous species as candidate genes. This study provides a data resource for subsequent studies of the MYB transcriptional family in ginseng and provides candidate genes for ginseng research in salt resistance.
In this study, we identified and analyzed the MYB transcription factor family members in ginseng and obtained 420 PgMYB transcripts under 117 gene IDs. We identified the evolutionary relationships and conserved motifs present in PgMYB transcripts and uncovered the potential functions of these transcripts. Weighted network co-expression revealed co-expression relationships among PgMYB transcripts, suggesting a synergistic effect when they exercise their functions. After treatment of adventitious roots of ginseng with different concentrations of NaCl, the fluorescence quantification results showed that the gene expression of PgMYB01, PgMYB71–01, PgMYB71–03 and PgMYB71–05 were increased to different degrees.
Materials and methods
Identification of the MYB genes in Jilin ginseng
This study is based on the Jilin ginseng transcriptome database. In order to ensure the integrity of the MYB transcription factor family in Jilin ginseng as much as possible, we took three different approaches in our methodology [23]. First, the seed file of the hidden Markov model containing the MYB_DNA-binding (PF00249) structural domain was downloaded from the PFAM (http://pfam.xfam.org/) protein database, and the protein sequences containing MYB structural domain were retrieved from the Jilin Ginseng Protein Database using native HMMER software (http://hmmer.org/Download.html), and the nucleic acid sequences of Jilin ginseng were compared by tBlastn at 1.0E-06. Second, we downloaded the CDS and protein sequences associated with MYB transcription factors from the Korean Ginseng Genome website (http://ginsengdb.snu.ac.kr/pathway.php), respectively Jilin ginseng transcriptome database to obtain the relevant transcripts. Third, we chose to download 10 sequences of MYB proteins that have been verified to function from the NCBI database, which were from medicago [24], tobacco [25, 26], Arabidopsis [18, 27], strawberry [28], poplar [29], petunia [30], wheat [31], citrus [32]. Similarly, these 10 protein sequences were compared to the Jilin ginseng transcriptome database to obtain the corresponding transcripts. Subsequently, the nucleic acid sequence results obtained from the three comparisons were intersected to remove the duplicate information. Subsequently, the above results were uploaded to iTAK (http://itak.feilab.net/cgi-bin/itak/index.cgi), and finally, the CD Search in NCBI (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) for further identification of their structural domains and further validation of the structural domains of PgMYB proteins by SMART (http://smart.embl-heidelberg.de/). Finally, they were named PgMYB + numbers (Table S1).
Structural characteristics and phylogeny of the PgMYB transcripts
Through the SMART online tool analysis, we obtained 257 PgMYB transcripts containing the MYB structural domain (Table S2), which were analyzed by the MEGA X [33]. Phylogenetic analysis of these transcripts was performed using the Neighbor-Joining (NJ) method, bootstrap repeats 1000, other settings as default parameters, and finally, they were embellished on iTOL (https://itol.embl.de/). To understand the structure of these transcripts, we predicted their conserved motifs by the online tool MEME (https://meme-suite.org/meme/tools/meme). Also, the contained structural domains of PgMYB transcription factors were analyzed by NCBI CDD tool (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi). Finally, they were visualized by TBtools [34] to visualize them.
PgMYB gene duplication and chromosome localization
The 420 PgMYB genes from Jilin ginseng were compared locally with ginseng genomic data [35]. The transcript data with 100% match to the chromosome were selected for analysis. The positions of PgMYB transcripts on the chromosomes were then visualized by the online tool MG2C V2.1 (http://mg2c.iask.in/mg2c_v2.1/index.html). Subsequently, sequences with ≥99% similarity in the matched regions and transcripts ≥300 bp in length were selected for intra-species covariance analysis, and the circles of PgMYB transcripts on chromosomes with duplicated gene pairs were mapped by R language.
Functional differentiation of the PgMYB gene family
The resulting PgMYB transcripts were submitted to Blast2GO V6.0.3 [36] for gene ontology annotation. The annotation results were used to classify these transcripts into functions and count the number of transcripts annotated to different functions (Biological Process, Molecular Function, Cellular Component). The functional subclasses of PgMYB transcripts annotated under Level 2 were visualized by R language (Table S3).
Expressions and network analysis of the PgMYB transcripts
During variable splicing of genes, different transcripts are formed, and these transcripts form proteins with different potential functions through the process of translation. Therefore, we retrieved the expression data of PgMYB transcripts from the expression database of Jilin ginseng transcripts (from 42 farmer’s cultivar roots, 14 different tissues, and four different-year-old ginseng roots) and explored the expression patterns of these PgMYB transcripts (Table S4).
Weight correlation network analysis of the PgMYB gene transcripts
To explore the co-expression of PgMYB transcripts in Jilin ginseng, we chose to analyze the expression characteristics of these PgMYB gene expression data (TPM) of 60 different samples in R language using the WGCNA (Weight Correlation Network Analysis) program package. The soft threshold for constructing the pro joining matrix was set to 0.85, the minimum number of transcripts under the module (a collection of highly inner-connected genes) was set to 30, and the threshold for merging was chosen to be 0.25. Subsequently, the modules’ transcripts were formed into a visual network in Cytoscape V3.9.0 based on their concatenation relationships (Table S5). Finally, we plotted the transcripts in the module in an expression heat map in the R language.
Response of the PgMYB genes to salt stress
The adventitious root materials of Jilin ginseng were treated with different salt concentrations of NaCl (CK, 70 mM, 80 mM, and 90 mM) added to the B5 medium required for their growth. These adventitious root materials were treated with salt stress at 22 °C in the dark for 30 days. Subsequently, all treated adventitious root materials were fast frozen by liquid nitrogen and finally stored in a refrigerator at − 80 °C.
We extracted RNA from salt stress-treated ginseng adventitious roots using the TRIzol method (Biotake, Beijing, China). The cDNA was obtained by reverse transcription using the extracted RNA as a template according to the experimental procedure for reverse transcription provided by HiFiScript gDNA Removal cDNA Synthesis Kit (CWBIO, Beijing, China). cDNA was obtained by reverse transcription according to the experimental protocol provided by UltraSYBR One Step RT-qPCR Kit (Low ROX) (CWBIO, Beijing, China) to perform fluorescent quantitative PCR in ABI 7500 Fast Real-Time PCR System. Actin 1 gene is an internal reference gene [37]. The 10 μL reaction system consisted of 5 μL UltraSYBR Mixture (Low ROX), 0.2 μL each of right and antisense primers, 1 μL of cDNA template obtained by reverse transcription, and 3.6 μL of ddH2O. After three technical and biological replicates, the internal reference gene was normalized, and the expression of the target gene was calculated by the 2−△△Ct method. Finally, bar graphs were plotted by GraphPad prism V8.3.0 and significant differences were calculated between the treated and control groups.
Results
Identification of the MYB transcripts in Jilin ginseng
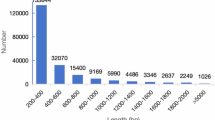
A total of 420 transcripts containing the MYB structural domain were identified in the Jilin ginseng transcriptome database, and these 420 transcripts were generated by alternate splicing of 117 genes. These transcripts were named PgMYB01 - PgMYB117. Table S1 contains the specific information of these transcripts. These include Transcript ID, Gene ID, Transcript Length (bp), DNA sequences (5′ - 3′), ORF (bp/aa), Conserve domain. These nucleic acid sequences range in length from 218 bp–4161 bp, with the shortest fragment length being PgMYB111, the longest fragment length was PgMYB109–01, and their amino acid lengths ranged from 33 aa – 1267 aa.
Phylogenetic analysis and sequence conservation of the PgMYB gene family
To understand the evolutionary relationships of PgMYB transcripts, we performed a phylogenetic analysis of 257 protein sequences containing MYB structural domains by the NJ method (Fig. 1). These protein sequences were mainly distributed in 19 subfamilies (a - s), among which subfamilies m and k contained 45 and 47 PgMYB transcripts, respectively, and subfamilies n, o and d contained two PgMYB transcripts each. We predicted 20 conserved motifs among these 257 transcripts by MEME analysis. Motif 1, motif 2, motif 6, and motif 8 were identified as conserved motifs of MYB. The structural domain prediction results of these PgMYB protein sequences by NCBI CDD revealed a total of 19 structural domain information, including 7 structural domain information of MYB, in addition to 12 other structural domains (Fig. 2). The different PgMYB transcripts contain different motifs and structural domains in different subclasses, which confers the diversity of PgMYB gene functions in ginseng.
Phylogenetic tree of the PgMYB proteins in Panax ginseng. The phylogenetic tree of the PgMYB gene family was constructed using 257 PgMYB gene. The subfamilies of the PgMYB gene family are indicated 19 subfamilies (a to s). The phylogenetic tree was constructed using the Neighbor-Joining (NJ) method with 1000 bootstrap replications
Functional differentiation of the PgMYB gene family
To understand the functions of individual PgMYB transcripts, 420 PgMYB transcripts were analyzed for GO function annotation (Table S3, Fig. 3). 397 PgMYB transcripts were annotated to any of the three major categories in the GO database (Biological Process (BP), Molecular Function (MF) and Cellular Component (CC)). The remaining 23 PgMYB transcripts did not receive annotation information. Of the 397 PgMYB transcripts annotated to function, 246 PgMYB transcripts were annotated to Biological Process, 181 PgMYB transcripts were annotated to Cellular Component, and 383 PgMYB transcripts were annotated to Molecular Function. 155 PgMYB at the level 2 level, these transcripts were annotated to different nodes under the three major categories, which include MF (binding (315), transcription regulator activity (48), catalytic activity (76)), BP (metabolic process (191), biological regulation (147), cellular process (209), regulation of biological process (144), response to stimulus (68), positive regulation of biological process (36)), CC (cellular, anatomical entity (170), protein entity (170), protein-containing complex (80)). Thus, it can be demonstrated that PgMYB transcripts play multiple functions in ginseng.
Functional categorization of the PgMYB transcripts by gene ontology (GO). A Venn diagram of the PgMYB transcripts among the biological process (BP), molecular function (MF) and cellular component (CC) functional categories. B The PgMYB transcripts are classified into 11 functional categories at Level 2, including two CC functional categories (Green), three MF functional category (Blue), and six BP functional categories (Red)
PgMYB gene duplication and chromosome localization
After the local Blast, a total of 138 gene transcripts could be fully matched to the chromosomes of ginseng, and these PgMYB transcripts were localized to 20 of the 24 chromosomes. It can be seen from Fig. 4 that chr2, chr13, chr22, and chr24 were not matched to any of the PgMYB transcripts. Interestingly, we found that PgMYB transcripts were also segmentally duplicated in ginseng by Synteny Block within ginseng. This just proves that the event of whole genome duplication (WGD) is also present in ginseng.
Chromosomal localization and synteny block of the PgMYB gene family in Panax ginseng. A Chromosomal localization of the MYB gene family in Jilin ginseng. B Synteny block of PgMYB transcriptional gene family members within the ginseng genome. Black arcs indicate synteny between genes, Chr: Chromosome, extrachromosomal scale represents the length of chromosome (Mb)
Expression characteristics of the PgMYB transcripts
To further characterize PgMYB transcripts in 14 tissue parts of 4-year-old ginseng (stem, fiber root, fruit peduncle, main root epiderm, fruit pedicel, rhizome, leaf peduncle, arm root, leaflet pedicel, leg root, leaf blade, fruit flesh, main root cortex, and seed), four different age stages of ginseng roots (5, 12, 18, and 25 years), and 42 farm cultivars (S1 - S42), we analyzed the expression of these transcripts (Table S3). Among the 14 tissue sites of four-year-old ginseng (Fig. 5A), 62 PgMYB transcripts were expressed in all 14 tissue sites, 62 transcripts were expressed in only one of the 14 tissue sites, and 43 transcripts were not expressed in any of the 14 tissue sites. The number of PgMYB transcripts expressed in different tissues showed that PgMYB transcripts were more inclined to be expressed in the fruit peduncle of ginseng. From the expression of PgMYB transcripts in ginseng roots at four different age stages (Fig. 5B), 110 PgMYB transcripts were expressed at all four age stages, 64 transcripts were expressed at specific periods only and 174 transcripts showed no expression at all four age stages. Among the 42 farm cultivars (Fig. 5C), 48 PgMYB transcripts were expressed in all 42 farm cultivars, 17 PgMYB transcripts were expressed only in specific farm cultivars, and 79 transcripts did not show expression in any of the 42 farm cultivars. By dotted line plot, we found (Fig. 6) that the average expression of PgMYB transcripts was higher in fruit peduncle in 14 tissues. However, in seed, the average expression of PgMYB transcripts was lower. In four different-year-old ginseng roots, the average expression of PgMYB was higher in 25-year-old roots, and among the 42 farm cultivars, the average expression content of PgMYB was lower in two farm cultivars, S13 and S40. The above results suggest that although the same transcript is expressed differently in different tissue parts of four-year-old ginseng, different-year-old ginseng roots in different farming cultivars may be due to the temporal and spatial constraints on the expression of the PgMYB transcript.
Numbers of the PgMYB transcripts expressing across different tissues, year-old of ginseng root, and cultivars. A The histogram of PgMYB transcripts expressed in 14 different tissues of 4-year-old ginseng. B The histogram of PgMYB transcripts expressed in 4 different year-old of ginseng roots. C The histogram of PgMYB transcripts expressed in 42 farm’s cultivars of 4-year-old ginseng
The expression range of PgMYB transcripts in Jilin ginseng. A The point plot of all PgMYB transcripts expression 14 different tissues of 4-year-old ginseng. B The point plot of PgMYB transcripts expressed in 4 different year-old of ginseng roots. C The point plot of PgMYB transcripts expressed in 42 farm’s cultivars of 4-year-old ginseng. The point shows the average expression amount of PgMYB gene transcripts, and the line shows the expression range of PgMYB gene transcripts
Expressions and network analysis of the PgMYB transcripts
We analyzed the relationship between the expression of these 420 PgMYB transcripts using WGCNA. Under the set conditions, we generated one module (turquoise) (Fig. 7A), which contains 51 PgMYB transcripts. By visualizing the network results in Cytoscape, we found a tight co-expression relationship between these 51 transcripts (Fig. 7B). Expression pattern analysis of these 51 PgMYB transcripts revealed (Fig. 8) that the expression of the same transcript was relatively stable across samples. In 42 farmer’s cultivar roots and 14 different tissues, the expression of PgMYB59–03, PgMYB107–10, PgMYB66–01, and PgMYB107–13 transcripts were relatively high in four different-year-old ginseng roots, PgMYB59–03, PgMYB107–10, PgMYB86, and PgMYB107–13 had higher expression content. These transcripts with higher expression levels may have important functions in ginseng.
Co-expression network analysis of the PgMYB gene transcripts expressed in 60 different samples. A Weighted network co-expression analysis of 420 PgMYB gene expressions. Turquoise transcripts are assigned the same module, gray transcripts are not assigned to any module as listed. B Co-expression network of 51 PgMYB gene transcripts under turquoise module
Heatmaps analysis spatiotemporal expression patterns of PgMYB transcripts in Panax ginseng. A The PgMYB gene transcripts expressed in the 42 farmer’s cultivars of 4-year-old ginseng roots. B The PgMYB gene transcripts expressed in the 14 different tissues of 4-year-old ginseng. C The PgMYB gene transcripts expressed in the 4 different growing years (5, 12, 18, 25 years-old) of ginseng roots
Response of the PgMYB transcripts to salt stress treatment in ginseng adventitious roots
To investigate the effect of the MYB gene family in Jilin ginseng in response to salt stress treatment, we identified some MYB gene family members that have been validated to play a role in salt stress treatment, OsMYB2 [17], MdMYB4 [38], OsMYB6 [39], TaMYB56-B [40], LcMYB1 [41], TaMYB86B [42], AtMYB49 [43], the MYB transcription factors (PgMYB01, PgMYB71–01, PgMYB71–03, and PgMYB71–05) that may be associated with salt stress were initially screened in ginseng utilizing local Blast. By collating the RT-qPCR results (Fig. 9), we found that the relative expressions of PgMYB71–01 and PgMYB71–05 genes were significantly different from the control under the condition of 70 mM NaCl treatment. Under the condition of 80 mM NaCl treatment, the relative expressions of PgMYB71–01 and PgMYB71–03 genes were highly significantly different from the control. Only the relative expression of PgMYB01 gene was significantly different from the control under the condition of 90 mM NaCl treatment. The relative expressions of PgMYB01 and PgMYB71–05 genes were 5.1-fold and 5.5-fold higher than the control under 100 mM NaCl treatment conditions, which were extremely significantly different from the control. Among the four transcripts tested in response to salt treatment, PgMYB01 and PgMYB71–05 genes were more sensitive to salt stress treatment.
The expressions of PgMYB01, PgMYB71–01, PgMYB71–03, and PgMYB71–05 gene in the ginseng roots treated with and without salt stresses. The 2−△△Ct method was used to evaluate the relative expression, and the expression levels of genes in the control were defined as “1”. The values are presented as the means of three replicates. “*” as significant at P ≤ 0.05, “**” as significant at P ≤ 0.01
Discussion
In recent years, ginseng has been mainly used for medicinal functions, which has gradually entered our lives as food and health products [44]. However, the identification and study of MYB gene family members in Panax ginseng have not been reported. As one of the plants’ largest transcription factor families, MYB transcription factors have been studied in many plants. We screened a total of 117 PgMYB genes from the Jilin ginseng transcriptome database that compared with the model plant Arabidopsis (197) and the food crops rice (155), maize (322) [13, 45], and the annual or perennial herb tomato (127) [46]. The number of MYB transcription factors in ginseng was relatively small. Alternative splicing produces multiple mRNAs from the same gene through variable splice site selection during pre-mRNA splicing, and these mRNAs encode proteins with different functions [47]. Therefore, the functions of these 420 PgMYB transcripts still need to be further verified by relevant experiments. By phylogenetic tree analysis, PgMYB transcripts were divided into 19 subfamilies, and there were differences in the number of transcripts between different subfamilies. Conservative motif and structural domain analysis showed that although the PgMYB transcripts contain the structural domains and motifs of the MYB gene family, the number of these motifs differs somewhat among different subfamilies, thus indicating that the PgMYB transcripts may be functionally similar among the same subfamilies.
A large proportion of genes for core biological functions are shared by all eukaryotes [36]. Most MYB proteins function as transcription factors with varying numbers of MYB structural domain repeats, conferring the ability to bind DNA. At the same time, transcription factors further regulate various functional genes under specific developmental and stress conditions [48]. From the results of GO annotation, we found that 315 PgMYB transcripts (75%) were annotated to the binding function in molecular function, which also indicates that PgMYB mainly plays the function of binding DNA in ginseng. Meanwhile, we noticed that 246 PgMYB transcripts (58.6%) were annotated to biological processes, which implies that PgMYB transcripts are involved in regulating the growth and developmental processes of Jilin ginseng. This result is consistent with previous studies reported [49]. Interestingly, 181 PgMYB transcripts (43.1%) were annotated to cellular components. This is consistent with the report that MYB103, MYB85, MYB52, and MYB54 in Arabidopsis are necessary for their normal secondary wall synthesis [50].
By analyzing the expression patterns of PgMYB in 42 farm cultivars, 14 different tissues, and four different-year-old ginseng roots, we can easily see that although the expression patterns of PgMYB transcripts are not consistent, most of them are still expressed in multiple cultivars, tissues, and year-old, which provides a good reference for Jilin ginseng subsequent developmental biology studies, which provides a good reference. At the same time, we also found that a small proportion of transcripts were specifically expressed, and these specifically expressed PgMYB transcripts provide favorable candidates for functional studies of ginseng.
Previous studies have reported a hierarchical regulatory network of the MYB gene family, and in maize, the transcriptional activator ZmMYB69 can directly target and activate the expression of ZmMYB31 and ZmMYB42, there by inhibiting lignin biosynthesis in maize [51]. Our co-expression analysis of the weighted network of these 420 PgMYB transcripts revealed that a similar hierarchical regulatory network of PgMYB transcripts exists in Jilin ginseng, which consists of 51 PgMYB transcripts that may be involved in the regulation of secondary metabolites in Jilin ginseng. This result also paves the way for the subsequent study on the interaction mechanism of MYB transcription factors in Jilin ginseng.
A wealth of information has accumulated on the role of proteins in regulating important processes in plants, including development, metabolism, and responses to environmental stresses [52]. An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt resistance [53]. Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salt stress resistance in Arabidopsis thaliana [54]. In ginseng, a MYB gene (PgMYB1) was amplified from the hairy roots of ginseng by RT-PCR using a pair of primers corresponding to the conserved sequence of the plant MYB gene, which showed a significant up-regulation of expression in 50 mM NaCl treatment for 48 h [21]. This suggests that MYB transcription factors are widely involved in plants’ salt stress resistance process.
Conclusions
In conclusion, 117 PgMYB genes were screened by combining the transcriptome data of Jilin ginseng. The studies on MYB family members in ginseng were enriched by phylogeny, conserved motifs, and structural domains, GO functional annotation, expression patterns and co-expression network analysis. Treatment of ginseng adventitious roots with different concentrations of NaCl revealed that the expression levels of most salt-resistant related MYB transcripts were higher under high concentrations of NaCl, thus reducing the harm of salt stress treatment on ginseng adventitious roots. This study provides a data resource for the subsequent functional study of MYB gene family in ginseng and provides a genetic reference for salt stress resistance in ginseng.
Availability of data and materials
All data analyzed during this study are included in the supplementary information files, and genotypic data have been deposited in the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) to NCBI under BioProject PRJNA302556.
References
Kiefer D, Pantuso T. Panax ginseng. Amer Fam Phys. 2003;68(8):1539.
Kim NH, Jayakodi M, Lee SC, Choi BS, Jang W, Lee J, et al. Genome and evolution of the shade-requiring medicinal herb Panax ginseng. Plant Biotechnol J. 2018;16(11):1904–17.
Shi W, Wang Y, Li J, Zhang H, Ding L. Investigation of ginsenosides in different parts and ages of Panax ginseng. Food Chem. 2007;102(3):664–8.
Xiao D, Yue H, Xiu Y, Sun X, Wang Y, Liu S. Accumulation characteristics and correlation analysis of five ginsenosides with different cultivation ages from different regions. J Ginseng Res. 2015;39(4):338–44.
Li W, Pang S, Lu Z, Jin B. Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants (Basel). 2020;9(11):1515.
Wang X, Niu Y, Zheng Y. Multiple functions of MYB transcription factors in abiotic stress responses. Int J Mol Sci. 2021;22(11):6125.
Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta. 2012;1819(2):97–103.
Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim Biophys Acta. 2012;1819(2):86–96.
Du H, Zhang L, Liu L, Tang XF, Yang WJ, Wu YM, et al. Biochemical and molecular characterization of plant MYB transcription factor family. Biochemistry (Mosc). 2009;74(1):1–11.
Paz-Ares J, Ghosal D, Wienand U, Peterson P, Saedler H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 1987;6(12):3553–8.
Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol. 2006;60(1):107–24.
Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol. 2001;4(5):447–56.
Katiyar A, Smita S, Lenka S, Rajwanshi R, Chinnusamy V, Bansal K. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics. 2012;13(1):544.
Du H, Yang SS, Liang Z, Feng BR, Lei L, Huang YB, et al. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 2012;12(1):106.
Abbas F, Ke Y, Zhou Y, Yu Y, Waseem M, Ashraf U, et al. Genome-wide analysis reveals the potential role of MYB transcription factors in floral scent formation in Hedychium coronarium. Front Plant Sci. 2021;12:623742.
Xu Q, He J, Dong J, Hou X, Zhang X. Genomic survey and expression profiling of the MYB gene family in watermelon. Hortic Plant J. 2018;4(1):1–15.
Yang A, Dai X, Zhang WH. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot. 2012;63(7):2541–56.
Li XW, Li JW, Zhai Y, Zhao Y, Zhao X, Zhang HJ, et al. A R2R3-MYB transcription factor, GmMYB12B2, affects the expression levels of flavonoid biosynthesis genes encoding key enzymes in transgenic Arabidopsis plants. Gene. 2013;532(1):72–9.
Zhou H, Lin-Wang K, Wang F, Espley RV, Ren F, Zhao J, et al. Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation. New Phytol. 2019;221(4):1919–34.
Chen X, Wang P, Gu M, Lin X, Hou B, Zheng Y, et al. R2R3-MYB transcription factor family in tea plant (Camellia sinensis): genome-wide characterization, phylogeny, chromosome location, structure and expression patterns. Genomics. 2021;113(3):1565–78.
Afrin S, Zhu J, Cao H, Huang J, Xiu H, Luo T, et al. Molecular cloning and expression profile of an abiotic stress and hormone responsive MYB transcription factor gene from Panax ginseng. Acta Biochim Biophys Sin Shanghai. 2015;47(4):267–77.
Liu T, Luo T, Guo X, Zou X, Zhou D, Afrin S, et al. PgMYB2, a MeJA-responsive transcription factor, positively regulates the dammarenediol synthase gene expression in Panax ginseng. Int J Mol Sci. 2019;20(9):2219.
Liu M, Pan Z, Yu J, Zhu L, Zhao M, Wang Y, et al. Transcriptome-wide characterization, evolutionary analysis, and expression pattern analysis of the NF-Y transcription factor gene family and salt stress response in Panax ginseng. BMC Plant Biol. 2022;22(1):320.
Peel GJ, Pang Y, Modolo LV, Dixon RA. The LAP1 MYB transcription factor orchestrates anthocyanidin biosynthesis and glycosylation in Medicago. Plant J. 2009;59(1):136–49.
Omer S, Kumar S, Khan BM. Over-expression of a subgroup 4 R2R3 type MYB transcription factor gene from Leucaena leucocephala reduces lignin content in transgenic tobacco. Plant Cell Rep. 2013;32(1):161–71.
Shingote PR, Kawar PG, Pagariya MC, Kuhikar RS, Thorat AS, Babu KH. SoMYB18, a sugarcane MYB transcription factor improves salt and dehydration tolerance in tobacco. Acta Physiol Plant. 2015;37(10):217.
Wisniewska A, Wojszko K, Rozanska E, Lenarczyk K, Kuczerski K, Sobczak M. Arabidopsis thaliana Myb59 gene is involved in the response to Heterodera schachtii infestation, and its overexpression disturbs regular development of nematode-induced syncytia. Int J Mol Sci. 2021;22(12):6450.
Schaart JG, Dubos C. Romero De La Fuente I, van Houwelingen a, de Vos RCH, Jonker HH, Xu W, Routaboul JM, Lepiniec L, Bovy AG: identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria x ananassa) fruits. New Phytol. 2013;197(2):454–67.
McCarthy RL, Zhong R, Fowler S, Lyskowski D, Piyasena H, Carleton K, et al. The poplar MYB transcription factors, PtrMYB3 and PtrMYB20, are involved in the regulation of secondary wall biosynthesis. Plant Cell Physiol. 2010;51(6):1084–90.
Albert NW, Davies KM, Lewis DH, Zhang H, Montefiori M, Brendolise C, et al. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell. 2014;26(3):962–80.
Rahaie M, Xue GP, Naghavi MR, Alizadeh H, Schenk PM. A MYB gene from wheat (Triticum aestivum L.) is up-regulated during salt and drought stresses and differentially regulated between salt-tolerant and sensitive genotypes. Plant Cell Rep. 2010;29(8):835–44.
Zhang P, Liu X, Yu X, Wang F, Long J, Shen W, et al. The MYB transcription factor CiMYB42 regulates limonoids biosynthesis in citrus. BMC Plant Biol. 2020;20(1):254.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–9.
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–202.
Wang ZH, Wang XF, Lu T, Li MR, Jiang P, Zhao J, et al. Reshuffling of the ancestral core-eudicot genome shaped chromatin topology and epigenetic modification in Panax. Nat Commun. 2022;13(1):1902.
Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–6.
Liu J, Wang Q, Sun M, Zhu L, Yang M, Zhao Y. Selection of reference genes for quantitative real-time PCR normalization in Panax ginseng at different stages of growth and in different organs. Plos One. 2014;9(11):e112177.
Wu R, Wang Y, Wu T, Xu X, Han Z. MdMYB4, an R2R3-type MYB transcription factor, plays a crucial role in cold and salt stress in apple calli. J Am Soc Hortic Sci. 2017;142(3):209–16.
Tang Y, Bao X, Zhi Y, Wu Q, Guo Y, Yin X, et al. Overexpression of a MYB family gene, OsMYB6, increases drought and salinity stress tolerance in transgenic rice. Front Plant Sci. 2019;10:168.
Li T, Sun J, Bi Y, Peng Z. Overexpression of an MYB-related gene FvMYB1 from Fraxinus velutina increases tolerance to salt stress in transgenic tobacco. J Plant Growth Regul. 2016;35(3):632–45.
Cheng L, Li X, Huang X, Ma T, Liang Y, Ma X, et al. Overexpression of sheepgrass R1-MYB transcription factor LcMYB1 confers salt tolerance in transgenic Arabidopsis. Plant Physiol Biochem. 2013;70:252–60.
Song Y, Yang W, Fan H, Zhang X, Sui N. TaMYB86B encodes a R2R3-type MYB transcription factor and enhances salt tolerance in wheat. Plant Sci. 2020;300:110624.
Zhang P, Wang R, Yang X, Ju Q, Li W, Lu S, et al. The R2R3-MYB transcription factor AtMYB49 modulates salt tolerance in Arabidopsis by modulating the cuticle formation and antioxidant defence. Plant Cell Environ. 2020;43(8):1925–43.
Baeg IH, So SH. The world ginseng market and the ginseng (Korea). J Ginseng Res. 2013;37(1):1–7.
Jiang Y, Zeng B, Zhao H, Zhang M, Xie S, Lai J. Genome-wide transcription factor gene prediction and their expressional tissue-specificities in maize. J Integr Plant Biol. 2012;54(9):616–30.
Li Z, Peng R, Tian Y, Han H, Xu J, Yao Q. Genome-wide identification and analysis of the MYB transcription factor superfamily in Solanum lycopersicum. Plant Cell Physiol. 2016;57(8):1657–77.
Syed NH, Kalyna M, Marquez Y, Barta A, Brown JW. Alternative splicing in plants coming of age. Trends Plant Sci. 2012;17(10):616–23.
Ambawat S, Sharma P, Yadav NR, Yadav RC. MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Plants. 2013;19(3):307–21.
Liu J, Osbourn A, Ma P. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol Plant. 2015;8(5):689–708.
Han X, Chen C, Hyun TK, Kumar R, Kim JY. Metabolic module mining based on independent component analysis in Arabidopsis thaliana. Mol Cells. 2012;34(3):295–304.
Qiang Z, Sun H, Ge F, Li W, Li C, Wang S, et al. The transcription factor ZmMYB69 represses lignin biosynthesis by regulating ZmMYB31 and ZmMYB42 in maize. Plant Physiol. 2022;189(4):1916–9.
Li C, Ng CKY, Fan L-M. MYB transcription factors, active players in abiotic stress signaling. Environ Exp Bot. 2015;114:80–91.
Cui MH, Yoo KS, Hyoung S, Nguyen HT, Kim YY, Kim HJ, et al. An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Lett. 2013;587(12):1773–8.
He Y, Li W, Lv J, Jia Y, Wang M, Xia G. Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salinity stress tolerance in Arabidopsis thaliana. J Exp Bot. 2012;63(3):1511–22.
Acknowledgements
The author thanks Shenzhen BGI for its support of RNA-seq services. The author also thanks Jilin Engineering Research Center Ginseng Genetic Resources Development and Utilization for their help in experimental plant materials.
Funding
This work was supported by an award from the Bureau of Science and Technology of Jilin Province (20200801063GH and 20210402043GH), the Department of Education of Jilin Province (JJKH20200318KJ and JJKH20200362KJ), and the Development and Reform Commission of Jilin Province (2018C047–3), National College Students Innovation and Entrepreneurship Project in China.
Author information
Authors and Affiliations
Contributions
Meiping Zhang and Yi Wang designed the experiments of the study. Mingming Liu and Mingzhu Zhao wrote and revised the main manuscript. Ke Li, Shichao Sheng, Mingyu Wang, Panpan Hua, Yanfang Wang, Ping Chen, and Kangyu Wang performed the experiments and contributed to the data analysis. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All plant materials were used following national and international standards and local laws and regulations. Using all plant materials does not pose any risk to other species in nature. No specific permission is required to collect all samples described in this study. The planting area of Panax ginseng samples is the cooperative base of our laboratory, and the field collection does not involve endangered or protected species. All ginseng samples (14 tissues, 42 cultivars’ roots, and 4 different-year-old roots) and ginseng adventitious root materials were stored in Jilin Agricultural University and Jilin Engineering Research Center Ginseng Genetic Resources Development and Utilization. All plant materials are available from the corresponding authors upon request.
Consent for publication
Not applicable.
Competing interests
The authors declare that there is no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Basic information of PgMYB gene family.
Additional file 2: Table S2.
The PgMYB protein sequences for phylogenetic analysis.
Additional file 3: Table S3.
The classification, annotation and GO functional categorization of the PgMYB gene transcripts.
Additional file 4: Table S4.
The expressions of the PgMYB gene transcripts in 14 tissues, 42 cultivars’ roots and 4 aged roots (TPM).
Additional file 5: Table S5.
Weight correlation network analysis concatenation relationships and value of PgMYB gene transcripts.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Liu, M., Li, K., Sheng, S. et al. Transcriptome analysis of MYB transcription factors family and PgMYB genes involved in salt stress resistance in Panax ginseng. BMC Plant Biol 22, 479 (2022). https://doi.org/10.1186/s12870-022-03871-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-022-03871-8