Abstract
Background
Deoxyxylulose 5-phosphate synthase (DXS) and deoxyxylulose 5-phosphate reductoisomerase (DXR) are the enzymes that catalyze the first two enzyme steps of the methylerythritol 4-phosphate (MEP) pathway to supply the isoprene building-blocks of carotenoids. Plant DXR and DXS enzymes have been reported to function differently depending on the plant species. In this study, the differential roles of rice DXS and DXR genes in carotenoid metabolism were investigated.
Results
The accumulation of carotenoids in rice seeds co-expressing OsDXS2 and stPAC was largely enhanced by 3.4-fold relative to the stPAC seeds and 315.3-fold relative to non-transgenic (NT) seeds, while the overexpression of each OsDXS2 or OsDXR caused no positive effect on the accumulation of either carotenoids or chlorophylls in leaves and seeds, suggesting that OsDXS2 functions as a rate-limiting enzyme supplying IPP/DMAPPs to seed carotenoid metabolism, but OsDXR doesn’t in either leaves or seeds. The expressions of OsDXS1, OsPSY1, OsPSY2, and OsBCH2 genes were upregulated regardless of the reductions of chlorophylls and carotenoids in leaves; however, there was no significant change in the expression of most carotenogenic genes, even though there was a 315.3-fold increase in the amount of carotenoid in rice seeds. These non-proportional expression patterns in leaves and seeds suggest that those metabolic changes of carotenoids were associated with overexpression of the OsDXS2, OsDXR and stPAC transgenes, and the capacities of the intermediate biosynthetic enzymes might be much more important for those metabolic alterations than the transcript levels of intermediate biosynthetic genes are. Taken together, we propose a ‘Three Faucets and Cisterns Model’ about the relationship among the rate-limiting enzymes OsDXSs, OsPSYs, and OsBCHs as a “Faucet”, the biosynthetic capacity of intermediate metabolites as a “Cistern”, and the carotenoid accumulations as the content of “Cistern”.
Conclusion
Our study suggests that OsDXS2 plays an important role as a rate-limiting enzyme supplying IPP/DMAPPs to the seed-carotenoid accumulation, and rice seed carotenoid metabolism could be largely enhanced without any significant transcriptional alteration of carotenogenic genes. Finally, the “Three Faucets and Cisterns model” presents the extenuating circumstance to elucidate rice seed carotenoid metabolism.
Similar content being viewed by others
Background
Plant terpenoids are the most diverse group of secondary metabolites and participate in essential functions, such as photosynthesis (carotenoids, chlorophylls, and prenylquinones), regulation of growth and development (cytokinin, gibberellins, abscisic acid, and strigolactones), respiration (ubiquinone), and secondary roles responding to environmental conditions [1, 2]. Also, a large number of terpenoids have been widely used in the industrial or medicinal fields as flavors, pigments, polymers, or drugs [3]. Terpenoids are generated by the sequential condensation and modification of two building-blocks, isopentenyl pyrophosphate (IPP, C5) and dimethylallyl pyrophosphate (DMAPP, C5), via the methylerythritol 4-phosphate (MEP) pathway in plastids and the mevalonic acid (MVA) pathway in cytosols [3]. In the first step, the pyruvate (C3) and glyceraldehyde 3-phosphate (GAP, C3) derived from glycolysis are condensed into deoxyxylulose 5-phosphate (DXP, C5) by a DXP synthase (DXS), and DXPs are subsequently converted into MEP (C5) by a DXP reductoisomerase (DXR) [4].
Plant DXS and DXR genes have been reported to function in terpenoid metabolism in a species- or organ-specific manner. In Arabidopsis, both AtDXS and AtDXR function as rate-limiting enzymes to enhance the leaf carotenoid and chlorophyll contents [4, 5], but their functions vary in a plant species-specific manner. For example, the heterologous expression of either AtDXS or AtDXR increased the amount of abietane diterpenes (C20) in Salvia sclarea hairy roots [6], but only AtDXS caused an increase in carotenoids and chlorophylls in leaves and roots of Daucus carota [7], and AtDXS in Lavandula latifolia significantly increased the monoterpene content (C10) but did not affect the amount of carotenoids and chlorophylls [8]. In addition, AtDXR affect neither the carotenoid, chlorophyll, nor monoterpene content in leaves of L. latifolia [9] or carrot [7]. On the other hand, overexpression of either the Amomum villosum Lour DXR gene (AvDXR) or the cyanobacteria (Synechosystis sp. strain PCC6803) DXR gene (SyDXR) enhanced the accumulation of carotenoid and chlorophyll in tobacco (Nicotiana tabacum) leaves. Similarly, the two tobacco genes NtDXR1 and NtDXR2 have also been reported to enhance the carotenoid and chlorophyll contents in tobacco leaves [10,11,12]. In this way, the differential roles of DXS and DXR have been elucidated in several plant systems, but the roles of DXS and DXR genes have largely remained unknown in rice plants.
The gene family of DXS enzymes has been classified into three groups. The type I-DXS group plays essential roles in the biosynthesis of housekeeping and photosynthetic terpenoids such as chlorophylls and carotenoids in leaves of Arabidopsis, Medicago (Medicago truncatula), rice and maize (Zea mays), and their gene expressions are dependent on the light condition [13,14,15,16]. The type II-DXS group plays secondary and ecological roles in the production of functional terpene metabolites, such as ginkgolide in ginkgo (Ginkgo biloba), oleoresin in Norway spruce (Picea abies), phytoalexin in rice, apocarotenoids in mycorrhizal roots of Medicago, and carotenoids in yellow kernels of maize [13, 17,18,19,20]. Type III-DXS enzymes have tentative roles specifically in the Poaceae family and a few dicots among angiosperms [13, 15, 21, 22]. The DXR enzyme is encoded by a single gene in most plants, except tobacco (N. tabacum), Hevea brasiliensis, and soybean (Glycine max), and it plays essential roles in the development and survival of plants [23,24,25].
Carotenoids, a large family of tetraterpenoids (C40), are abundant in photosynthetic and non-photosynthetic plant tissues [26], and are also essential nutrients for humans as they are the precursors of pro-vitamin A [27]. Since carotenoids are enriched in photosynthetic green parts but lacking in non-photosynthetic seed endosperms in rice plants [28, 29], several “Golden Rice” varieties have been genetically engineered in which carotenoids, such as β-carotene, are enriched in endosperms [29,30,31,32] to enhance the nutritional value of rice as a staple cereal. In maize plants (Zea mays L.), the expression levels of the three maize DXS genes were examined in different organs, including the mature kernels of two varieties with white and yellow grain [13]. The highest level of ZmDXS2 expression was observed in yellow kernels compared with its expression level in white kernels, whereas ZmDXS1 was not expressed, and ZmDXS3 was expressed in mature kernels of both varieties, suggesting that ZmDXS2 might be important for the carotenoid enrichment in yellow kernels. To increase the accumulation efficiency of carotenoids in rice seed endosperms, the MEP pathway supplying the major building-blocks, IPP/DMAPP, to the carotenoid pathway has been issued, and the heterologous expression of AtDXS has shown the possibility that the increase in DXS enzyme could derive the enhancement of carotenoid accumulation in rice endosperms [33]. However, even though the “Golden rice” varieties have been developed for years, there has been no report of the rate-limiting enzymes of the rice MEP pathway, which function to increase the flux of the isoprene building-blocks IPP/DMAPP into carotenoid metabolism.
In this study, we investigated the differential roles of rice DXS and DXR, the first two enzymes of the MEP pathway, and determined their influences on rice carotenoid metabolism in leaves and seeds. Considering the genome editing era, our studies provide useful information on gene candidates for genome editing.
Results
Molecular characterization of rice genes encoding DXS and DXR
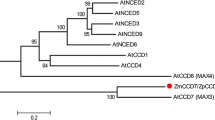
In rice, the three genes that encode DXS proteins are Os05g33840 (XP_015640505), Os06g05100 (XP_015642490), and Os07g09190 (XP_015647944). The predicted amino sequences of three rice DXS genes were phylogenetically analyzed with plant DXS protein sequences and were independently classified as type I, II, or III, corresponding to OsDXS1 (XP_015640505), OsDXS2 (XP_015642490), and OsDXS3 (XP_015647944), respectively (Fig. 1a). The three rice DXSs are closely related to DXS in foxtail millet (Setaria italica) and maize, which belong to the Poaceae family. OsDXS2 showed the highest sequence similarity (87.5%) to ZmDXS2 (NP_001295426), therefore it was chosen to study its influence on rice carotenoid metabolism in leaves and seeds. Similar to DXR in other plants, rice DXR is encoded by the single copy gene OsDXR (XP_015618768, Os01g01710) and phylogenetically categorized into plant DXR clade II, which includes the tree plants gingko, yew, and pine, and the Poaceae family plants sorghum (Sorghum bicolor), maize, foxtail millet, and barley (Hordeum vulgare). On the other hand, rice DXR is not categorized into clade I, which includes herbage plants (Fig. 1b). The predicted amino acid sequence of OsDXS2 has a conserved core domain for thiamine pyrophosphate (TPP)-binding and two residues of histidine and tyrosine that participate in the active site (Additional file 1: Figure S1). In addition, OsDXR has three highly conserved domains, including a proline-rich domain, two NADPH binding sites, and two substrate-binding sites (Additional file 2: Figure S2).
Phylogenetic relationships of plant a deoxyxylulose 5-phosphate synthases (DXSs) and b deoxyxylulose 5-phosphate reductoisomerases (DXRs). The accession numbers of plant DXSs and DXRs are displayed with the abbreviated names of plant species as follows: Aa, Artemisia annua; Am, Amomum villosum Lour; At, Arabidopsis thaliana; Ca, Capsicum annuum; Cr, Catharanthus roseus; Do, Dendrobium officinal; Gb, Ginkgo biloba; Gm, Glycine max; Hb, Hevea brasiliensis; Hv, Hordeum vulgare; La, Lavandula angustifolia; Mt, Medicago truncatula; Nt, Nicotiana tabacum; Os, Oryza sativa; Pa, Picea abies; Pt, Pinus taeda; Rr, Rosa rugosa; Sb, Sorghum bicolor; Si, Setaria italica; Sl, Solanum lycopersicum; Sm, Salvia militorrhiza; Sr, Stevia rebaudiana; Tm, Taxus x media; Tw, Tripterygium wilfordii; Zm, Zea mays. Their deduced amino acid sequences were aligned using the ClustalV algorithm of the MegAlign program (DNASTAR, Inc.). Type II proteins, including OsDXS2, and clade II proteins, including OsDXR, are distinguished by black and gray bars, respectively, and proteins belonging to the Poaceae family including rice are in bold
Expression profile of OsDXSs and OsDXR in different tissues
The endogenous expression patterns of OsDXSs and OsDXR genes were analyzed in leaves and roots of the vegetative stage and in leaves, roots, florets, and seeds of the reproductive stage (Fig. 2). The transcript level of OsDXS1 was significantly higher in the leaves compared with other tissues of both stages, suggesting its relevant roles in photosynthetic tissues. Similarly, the expression of OsDXS2 was also higher in the leaves compared with other tissues at both stages. In the roots of the vegetative stage, the expression levels of OsDXS2, OsDXS1, and OsDXS3 were 0.029, 0.0009, and 0.008, respectively (Fig. 2), and that the expression level of OsDXR was 0.048 in the roots. Considering an OsDXR is a single copy-downstream gene of OsDXS, OsDXS2 was supposed to play important roles as a major DXS enzyme in the roots as a non-photosynthetic tissue. In contrast, OsDXS3 was constitutively expressed at low levels in most tissues, including seeds. As expected as a single copy-downstream gene of OsDXS, OsDXR was strongly expressed in the leaves of both developmental stages, and exhibited the highest expression among the four genes in all tissues, regardless of the developmental stage (Fig. 2).
Spatial and temporal expression of rice genes encoding deoxyxylulose 5-phosphate synthase (DXS) or deoxyxylulose 5-phosphate reductoisomerase (DXR). Endogenous transcript levels of the OsDXS1, OsDXS2, OsDXS3, and OsDXR genes were examined by qRT-PCR using total RNA isolated from various tissues, including leaves (L), roots (R), and florets (F) of vegetative stage and reproductive stage tissues, and seeds (Se) harvested at 40 days after flowering using gene-specific primers as shown in Additional file 4: Table S8. All transcript levels measured in three technical replicates were calculated by the ΔCt equation against the OsUbi5 gene. The results are expressed as the mean ± standard error (SE)
Constitutive overexpression of either OsDXS2 or OsDXR alone or in combination with the bio-fortified trait of β-carotene accumulation in rice endosperm
To investigate the differential roles of OsDXS2 and OsDXR in rice carotenoid metabolism in leaves and seeds (Fig. 3a), PGD1::OsDXS2 and PGD1::OsDXR vectors were generated to constitutively overexpress OsDXS2 (Os07g09190) and OsDXR (Os01g01710) (Fig. 3b). Their expression cassettes, either PGD1::OsDXS2::PinII or PGD1::OsDXR::PinII, were cloned into the pGlb::stPAC vector for co-expression with a stPAC recombinant gene producing β-carotene in rice endosperms to generate PGD1::OsDXS2_Glb::stPAC or PGD1::OsDXR_Glb::stPAC vectors, respectively (Fig. 3b). Finally, three representative lines for each of the four constructs were selected by considering the integration of the transgene and its copy numbers in leaves of T3 plants (Additional file 3: Figure S3).
Schematic representation of the carotenoid pathway, the binary vectors used in this study, and transgene expression in leaves and seeds of rice plants. a Built-in pathway for carotenoid biosynthesis in rice plants; CRTI, Pantoea annatis desaturase; DMAPP, dimethylallyl diphosphate; DXP, deoxyxylulose 5-phosphate; DXR, DXP reductoisomerase; DXS, DXP synthase; IPP, isopentenyl diphosphate; GGPP, geranylgeranyl pyrophosphate; G3P, glyceraldehyde 3-phosphate; LCYB, lycopene β-cyclase; MEP, methylerythritol 4-phosphate; OsDXR, rice DXP reductoisomerase; OsDXS2, rice DXP synthase; PSY, phytoene synthase; stCrtI, rice codon-optimized synthetic gene encoding the Pantoea CrtI gene; stPsy, rice codon-optimized synthetic gene of the Capsicum gene encoding PSY; stPAC, a recombinant gene of stPsy and stCrt linked with 2A, which is the rice codon-optimized foot-and-mouth disease virus 2A peptide. b Diagrams of the four vectors for rice transformation of OsDXS2 and OsDXR with the pGlb::stPAC vector that was previously used to generate a carotenoid-intensifying trait in rice endosperm [32]. Bacterial attachment attB sites needed for Gateway cloning are marked with hatched boxes. BR, right border; BL, left border; PGD1, rice phosphogluconate dehydrogenase promoter; OsDXS2, rice DXP synthase 2 gene; OsDXR, rice DXP reductoisomerase gene; PinII, 3′ region of the potato proteinase inhibitor II gene; BAR, Bialaphos-resistant gene cassette; Glb, rice globulin promoter; stPAC, a recombinant gene of stPsy-2A-stCrt. The primer locations used in vector construction, PCR, and qRT-PCR for the transgene analyses in Additional file 3: Figure S3a, Fig. 3c, and Fig. 3d are indicated as arrows, and their information is listed in Additional file 4: Table S7. c Transgene expression levels of OsDXS2 and OsDXR were examined by qRT-PCR using total RNA isolated from 10-day-old leaves. d Transgene expression levels of OsDXS2, OsDXR, and stPAC were examined by qRT-PCR using total RNA purified from unpolished mature seeds 40 DAF. All results using gene-specific primer pairs F7/R7 for OsDXS2, F8/F7 for OsDXR, and F9/R9 for stPAC were calculated as the mean of three replicates and normalized to the expression of the OsUbi5 gene, which was amplified using a U5F/U5R primer pair. The primers are indicated in Fig. 3b and listed in Additional file 4: Table S7. NT is a non-transgenic wild type of Oryza sativa L. cv. Ilmi, different varieties of transgenic plants are represented in different colors, such as blue, light-blue, orange, light-orange and yellow in the bar graph, and the X-axis labeling consists of three independent-transgenic plant lines
All transgene expressions were examined in leaves and seeds of the T4 generation (Fig. 3c and d). The transcript level of OsDXS2 or OsDXR in both organs relative to the NT rice plant demonstrated their constitutive overexpression, and the stPAC expression in seeds of PGD1::OsDXS2_Glb::stPAC and PGD1::OsDXR_Glb::stPAC lines verified the integration of the β-carotene-producing pathway (Fig. 3d).
No positive effect of OsDXS2 and OsDXR on carotenoid- and chlorophyll metabolism in rice leaves
Firstly, we analyzed the carotenoid and chlorophyll contents in leaves of T4 generation homozygous transgenic lines using high-performance liquid chromatography (HPLC), and the data are shown in Table S1 (Additional file 4). Compared to NT leaf controls, the total content of carotenoids was slightly reduced by 20% in the PGD1::OsDXR lines with the statistical significance, and by 10% in the PGD1::OsDXS2 lines (Fig. 4a). We also measured the contents of chlorophyll a and b in the same tissues, and the data are shown in Table S2 (Additional file 4). The content of total chlorophylls was slightly reduced by 11% in the PGD1::OsDXS2 lines and by 15% in the PGD1::OsDXR lines (Fig. 4b) with the statistical significance. Taken together, the results show that the contents of carotenoids and chlorophylls are slightly reduced rather than increased by the overexpression of OsDXS2 or OsDXR in rice leaves. We also tried to determine if the reduction of carotenoids and chlorophylls affected the leaf phenotypes, but no phenotypic changes were observed, suggesting that the reductions caused by OsDXS2 or OsDXR overexpression did not significantly affect leaf phenotypes.
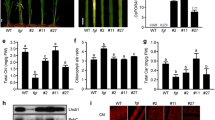
Contents and composition of carotenoids and chlorophylls in the leaves of PGD1::OsDXS2 and PGD1::OsDXR transgenic plants relative to NT plants. a Carotenoid levels determined by HPLC analysis of T4 leaf tissues harvested from three independent transgenic plants for each construct. b Chlorophyll absorbance in the same leaves as in a measured by spectrophotometry. NT represents non-transgenic rice plants (Oryza sativa L. cv. Ilmi). All data are represented as the mean ± SE of three independent measurements. Statistical analyses were performed using a two-tailed Student’s t-test, and significant differences were indicated by a p value (** p < 0.01 and *** p < 0.001). Different varieties of transgenic plants are represented in different colors, such as green and light-green in the bar graph, and the X-axis labeling consists of three independent-transgenic plant lines
The organ-specific differential roles of OsDXS2 as a rate-limiting enzyme in rice seed carotenoid metabolism, compared to OsDXR
We next examined whether the overexpression of either OsDXS2 or OsDXR affected seed carotenoid metabolism (Fig. 5). Compared to NT seeds, although no color or phenotypical changes were observed in the brown seeds (Fig. 5a), total carotenoid content was increased by 26% in the PGD1::OsDXS2 lines and decreased by 11% in the PGD1::OsDXR lines (Fig. 5b and Additional file 4: Table S3) with statistical significance even if the alterations are a little. Also, as shown in Fig. 6, the yellow color in Glb::stPAC seeds [32] was largely intensified to an orange color in the PGD1::OsDXS2_Glb::stPAC lines, but remained just yellow in the PGD1::OsDXR_Glb::stPAC lines (Fig. 6a). Compared to Glb::stPAC seeds, the total carotenoid contents of PGD1::OsDXS2_Glb::stPAC seeds were largely increased by 5.4-fold (Fig. 6b). Specifically, the contents of lycopene, α-carotene and β-carotene increased 8.7-fold, 13.1-fold, and 5.8-fold, respectively, while lutein and zeaxanthin decreased 0.7-fold and 0.4-fold, respectively (Additional file 4: Table S3). In contrast, the overexpression of OsDXR did not enhance carotenoid accumulation in either the PGD1::OsDXR or PGD1::OsDXR_Glb::stPAC seeds (Fig. 6b), indicating the differential roles of OsDXR compared with the positive roles of OsDXS2 in the enhancement of seed carotenoid metabolism.
Color phenotypes and carotenoid levels in the seeds of PGD1::OsDXS2 and PGD1::OsDXR transgenic plants relative to NT plants. a Mature seed colors in homozygous T4 generations were compared with NT (Oryza sativa L. cv. Ilmi) at 40 days after flowering (DAF) among the three independent transgenic plants for each construct before polishing because polished colors are indistinguishable from white. b Carotenoid levels determined by HPLC analysis in the same seeds as in a. All carotenoid content data are displayed as the mean value ± standard error (SE) of three independent measurements. Statistical analyses were performed using a two-tailed Student’s t-test (* p < 0.05). Different varieties of transgenic plants are represented in different colors, such as blue and light-blue in the bar graph, and the X-axis labeling consists of three independent-transgenic plant lines
Color phenotypes and carotenoid levels in the seeds of PGD1::OsDXS2_Glb::stPAC and PGD1::OsDXR_Glb::stPAC relative to Glb::stPAC transgenic plants. a Mature seed colors in the homozygous T4 generation were compared among three independent transgenic plants for each construct with stPAC rice seeds 40 days after flowering (DAF) and after polishing. b Carotenoid levels measured by HPLC analysis in the same seeds as in a. All carotenoid content data are displayed as the mean value ± standard error (SE) of three independent measurements. Statistical analyses were performed using a two-tailed Student’s t-test (** p < 0.01, *** p < 0.001 and **** p < 0.0001). NT is a non-transgenic wild type of Oryza sativa L. cv. Ilmi, different varieties of transgenic plants are represented in different colors, such as orange, light-orange and yellow in the bar graph, and the X-axis labeling consists of three independent-transgenic plant lines
Interestingly, the level of total xanthophylls increased 6.4-fold, whereas both α-carotene and β-carotene contents were greatly enhanced by 315.3-fold in PGD1::OsDXS2_Glb::stPAC seeds compared with NT. This suggests that the hydroxylation step to convert carotenes into xanthophylls, which is one of the rate-limiting steps in carotenoid metabolism, is tightly regulated to maintain a basal level of xanthophylls in rice seeds.
OsDXS2- or OsDXR-mediated transcriptional alteration of intrinsic carotenogenetic genes in rice leaves and seeds
The transcript levels of the intrinsic genes involved in MEP, carotene, and xanthophyll pathways were determined in the leaves and seeds of NT and transgenic plants, and all expression patterns were visualized using a heatmap graph that represented the fold changes (Fig. 7). The expression data are shown in Additional file 4: Table S4-S6. As shown in Fig. 7a and b, the expression of endogenous carotenogenetic genes in leaves was mostly increased, as indicated by the red-color in the heatmap. The expression levels of these genes were further statistically analyzed using a two-tailed Student’s t-test, and the statistical significances were indicated with orange or green asterisks (Fig. 7a and b). The expression of the genes encoding DXS, PSY, and BCH enzymes were significantly increased following the overexpression of either OsDXS2 or OsDXR (* p < 0.05 and ** p < 0.01). Of them, the levels of OsDXS1, OsPSY2, and OsBCH2 were increased by 3.1-fold to 13.9-fold in both OsDXS2 and OsDXR overexpressed transgenic leaves, suggesting that they might be associated with the enhanced activities of OsDXS2 and OsDXR in rice leaves. Interestingly, the endogenous expression of OsDXS1 and OsDXS2 increased up to 6.7-fold and 4.0-fold, respectively, following the overexpression of OsDXR, but the enhanced activity of OsDXS2 did not cause a significant increase in OsDXR expression, even though the expression of the OsPSY2 gene was significantly increased up to 5.7-fold following the overexpression of OsDXS2 (Fig. 7a). These results demonstrate the differential roles of DXS and DXR genes in rice leaves. However, the significant increases in gene expression were not proportional to the metabolic changes, considered with a slight reduction in the amount of carotenoids and chlorophylls in the leaves of the PGD1::OsDXS2 and PGD1::OsDXR plants.
Expression profiles of carotenoid biosynthetic genes in leaves and seeds among three independent transgenic plants for each OsDXS2 and OsDXR construct relative to non-transgenic (NT) and stPAC plants. In the 10-day-old leaves, the expression patterns of 11 rice genes, including five that are involved in the MEP pathway and six carotene biosynthetic genes (a) and 10 rice xanthophyll biosynthetic genes (b) are presented as the fold-change values (2−ΔΔCt) compared to the ΔCt values of NT, and the qRT-PCR data are listed in Additional file 4: Table S4. Statistical analyses of expression levels were performed using a two-tailed Student’s t-test (*p < 0.05 and ** p < 0.01). The t-test results are represented as an orange color (*) and a green color (*) for the significant effects of OsDXS2 and OsDXR enhancements, respectively. Also, the expression patterns of the same gene sets were analyzed in the mature seeds 40 days after flowering (DAF). The expression data of 11 rice genes, including five that are involved in the MEP pathway and six carotene biosynthetic genes (c) and 10 rice xanthophyll biosynthetic genes (d) are presented as the fold-change values (2-ΔΔCt) compared to the ΔCt values of NT, and all qRT-PCR data are listed in Additional file 4: Table S5–S6. The ΔCt values of NT are listed in the gray colored boxes, and all fold-change values (2-ΔΔCt) are presented with the corresponding heatmap graphs. The color key of the heatmap graph is presented under each graph, and the accession numbers of genes and primer sequences are listed in Additional file 4: Table S8
Twelve genes involved in the MEP and carotene biosynthetic pathways were also transcriptionally compared among seeds from five transgenic plants of PGD1::OsDXS2, PGD1::OsDXS2_Glb::stPAC, PGD1::OsDXR, PGD1::OsDXR_Glb::stPAC, and Glb::stPAC, and NT rice plants (Fig. 7c, Additional file 4: Table S5). In contrast to leaves, the expression of the genes encoding DXSs and PSYs were not highly upregulated by the overexpression of either OsDXS2 or OsDXR and altered slightly without statistical significance in the seeds of PGD1::OsDXS2 and PGD1::OsDXR, as shown by the faint pink and blue colors on the heatmap (Fig. 7c). The expression analysis of the biosynthetic genes involved in xanthophyll metabolism in the seeds of PGD1::OsDXS2, PGD1::OsDXS2_Glb::stPAC and Glb::stPAC showed that the expression patterns of OsCYP97B (a P450-type α-carotene hydroxylase gene) and OsBCH1 (a β-carotene hydroxylase-1 gene) were significantly upregulated and downregulated, respectively, even though the expressions only changed by 2-fold (Fig. 7d). Therefore, these small changes in the expression patterns were inconsistent with the 73.8-fold and 315.3-fold increases observed in the carotenoid contents in Glb::stPAC and PGD1::OsDXS2_Glb::stPAC seeds, respectively, compared to NT seeds. In other words, the strong enhancement of carotenoid accumulation is quite dependent on the addition of transgenes rather than changes in endogenous carotenogenic gene expression.
Collectively, the expression of endogenous carotenogenic genes was differentially regulated by either OsDXS2 or OsDXR overexpression in an organ-specific manner, but there was no significant correlation between their expression level and subsequent metabolic changes. These results suggest that the large enhancement of carotenoid accumulation is mediated by the stepwise addition of OsDXS2 and stPAC genes, but not largely dependent on any changes in the expression of intrinsic carotenogenic genes.
Discussion
In this study, we investigated the differential roles of rice DXS and DXR as the first two enzymes of MEP pathway on rice carotenoid metabolism. The content of total seed carotenoids was increased by 26% in the PGD1::OsDXS2 lines compared with NT plants and was considerably increased on average by 3.4-fold (5.4-fold maximum) in the PGD1::OsDXS2_Glb::stPAC lines compared with the Glb::stPAC lines, but in contrast, decreased by 11% in the PGD1::OsDXR lines and by 50% in the PGD1::OsDXR_Glb::stPAC lines compared to NT plants and Glb::stPAC rice lines, respectively (Fig. 5 and Fig. 6). Considering that the phytoene biosynthesis is quite limited in rice endosperm [28], the large enhancement of carotenoid accumulation in PGD1::OsDXS2_Glb::stPAC seeds suggests that OsDXS2 functions as a rate-limiting enzyme supplying IPP/DMAPPs to carotenoid metabolism, even though total carotenoid content in PGD1::OsDXS2 seeds was a little changed to increase just by 26%. However, in rice leaves, total carotenoids were reduced by an average of 10% in the PGD1::OsDXS2 lines and by 20% in the PGD1::OsDXR lines. Further, total chlorophylls were also reduced by an average of 11% in PGD1::OsDXS2 and 15% in PGD1::OsDXR compared to NT plants (Fig. 4). These results suggest that OsDXS2 functions as a rate-limiting enzyme just only in seed carotenoid metabolism but not in leaves, whereas OsDXR is a rate-limiting enzyme in neither leaves nor seeds.
In Arabidopsis leaves, both AtDXS and AtDXR are rate-limiting enzymes in the biosynthetic pathways that produce carotenoid and chlorophyll [4, 5], however, the differential roles of plant DXS and DXR enzymes have also been consistently reported in several plant species. In carrot, the overexpression of AtDXR failed to increase the content of total carotenoids in both tissues, while AtDXS overexpression did increase the content of total carotenoids in both leaves and roots by modifying the expression of PSY1 and PSY2 genes [7]. In lavender, the overexpression of AtDXS did not affect the carotenoid and chlorophyll contents in leaves but did significantly increase the essential oil monoterpene [8]. In contrast, AtDXR did not mediate any changes in essential oil, carotenoid or chlorophyll contents [9]. These studies clearly demonstrate that the rate-limiting functions of the DXR enzyme may not be consistent and the rate-limiting functions of the DXS enzyme could vary between plant species or even different tissues.
In this study, the expression profiling of rice carotenogenic genes involved in the MEP, carotene, and xanthophyll pathways showed that their expression patterns were not proportional to the changes in carotenoid metabolism in the leaves and seeds of rice transgenic plants. The expression of carotenogenic genes, such as OsDXS1, OsPSY1, OsPSY2, and OsBCH2, were upregulated by the enhancement of either OsDXS2 or OsDXR genes in leaves compared to the reduction of the carotenoid and chlorophyll content, and there were only slight changes in the expression of most carotenogenic genes, even though there was a large increase in the seed carotenoid accumulation (Fig. 7). In contrast to our results, the overexpression of a DXS-encoding gene in tomato fruits and potato tubers increased the expression of the endogenous PSY-encoding genes and subsequently caused a large enhancement of carotenoid accumulation [34, 35]. Similar results have been observed in carotenoid metabolism in carrot leaves and roots [7]. These results show that the DXS-mediated increase of carotenoid and chlorophyll contents is correlated with an increase in PSY gene expression. However, a positive correlation was not observed in rice leaves. Specifically, the expression of OsPSY1 and OsPSY2 genes were preferentially upregulated compared to the reduction of carotenoid content in leaves, and were not altered in rice seeds, even though there was an increase in carotenoid content. Similarly, in tomato fruits, the expression of the DXS-, PSY-, and PDS-encoding genes are preferentially downregulated compared to the enhancement of β-carotene [34], and in Bixa orellana L., the carotenoid content was increased by the salt stress treatment, but the expression of carotenogenic genes were non-proportionally downregulated [36]. These results suggest the possibility that the transcriptional expression of the intrinsic carotenogenic genes, such as the DXS- and PSY-encoding genes, might not be proportional to the expression levels of their encoded-proteins, and the protein stability of the intermediate biosynthetic enzymes might be equally as important as the enhanced activity of rate-limiting enzymes for the enhancement of carotenoid accumulations.
In Arabidopsis, AtDXS proteins have been reported to be post-translationally regulated by chloroplast biogenesis 6 (CLB6, a hydroxy-2-methyl-butenyl 4-diphosphate reductase), Hsp100 chaperones, and several Clp proteases [37], and the proteostasis of Arabidopsis PSY proteins are controlled directly by the Clp protease and ORANGE protein [38]. In Catharanthus roseus, the stability of DXR proteins is highly dependent on Clp protease-mediated degradation [39], and the enzyme activity of PDS proteins is more stably sustained in the β-carotene-enhanced tomato fruits, even if its transcripts are downregulated [34]. Similarly, our studies showed that the seed carotenoid accumulations could be largely increased without any proportional upregulation of the intermediate biosynthetic genes, except the enhancement of the rate-limiting enzymes, such as DXS and PSY, suggesting that seed carotenoid metabolism might be preferentially controlled by the stability or activity of the carotenogenic enzymes, rather than their transcript levels.
Collectively, we propose a “Three Faucets and Cisterns Model” to describe rice seed carotenoid metabolism (Fig. 8). The term “Three Faucets” represents the three rate-limiting steps involving DXS, PSY, and BCH enzymes that supply restricted levels of precursors from upstream to downstream pathways. The intermediate carotenoid biosynthetic machinery between two faucets is regarded as the “Cistern” that fills with particular carotenoid metabolites representing IPP/DMAPP, carotenes, and xanthophylls in our model, and whose capacity is assumed to be determined only by the status of active proteins (Fig. 8). By following the “Three Faucets and Cisterns Model,” the overexpression of OsDXS2 in PGD1::OsDXS2 seeds might turn on the first “Faucet” to increase the metabolic flux into the first “Cistern,” and the carotenoid content could slightly increase (by 1.3-fold) as much as the capacity of the second “Faucet” (rice PSY genes), which is maintained at a basal level in rice seeds. Also, by the stepwise addition of OsDXS2 and stPAC, both the first and second “Faucets” are turned on to increase the metabolic flux from the first “Cistern” to the second “Cistern,” and these consecutive metabolic streams could increase protein stability to enhance the capacity of the “Cisterns” and enable the large enhancement of α/β-carotene accumulation by 315.3-fold without any changes in the expression of “Cistern” genes. In the case of Glb::stPAC seeds, the second “Faucet” (stPAC) was turned on without any enhancement of the first “Faucet,” but the carotenoid content was increased by 73.8-fold. In contrast, in PGD1::OsDXR seeds, the enhancement of OsDXR regarded as a component of the first “Cistern” did not increase carotenoid accumulation, clearly demonstrating the differential functions between the “Faucet” and “Cistern.” In other words, the enhancement of the second “Faucet” (stPAC) could increase the metabolic flux from the first “Cistern” to the second “Cistern,” but the increase of a “Cistern” component (OsDXR) might fail to affect the metabolic flux from the upstream steps to the second “Faucet,” therefore any enhancement of carotenoid accumulation was not observed in rice seeds. Finally, the contents of xanthophylls were gradually increased by the stepwise addition of either OsDXS2 or stPAC genes from 1.3-fold to 6.8-fold in rice seeds, but no additional increase was observed in PGD1::OsDXS2_Glb::stPAC seeds, whose carotene content was increased by 4.3-fold compared to Glb::stPAC seeds, suggesting that it might be due to the maximum capacity of the third “Faucet.”
Proposed “Three Faucets and Cisterns Model” as a carotenoid accumulation mechanism for the rice seed system. The rate-limiting key enzymes are displayed as “Faucets,” and the biosynthetic and accumulation machinery are shown as squares representing the “Cisterns.” The metabolite pools of G3P/pyruvate, IPP/DMAPP, carotenes, and xanthophylls are presented as four colors of gray, sky-blue, yellow, and green, respectively. OsDXS2 or stPAC transgenes are signified as blue or red, respectively. Carotene components are composed of lycopene, α-carotenes, and β-carotenes, and xanthophyll components contain lutein, β-cryptoxanthin, zeaxanthin, and violaxanthin
Conclusion
In this study, the differential roles of OsDXS2 and OsDXR in carotenoid metabolism of rice leaves and seeds were investigated, and the changes in expression of carotenogenic genes were measured. The results suggest that OsDXS2 is a rate-limiting enzyme supplying IPP/DMAPPs to carotenoid metabolism in rice seeds but not in rice leaves, whereas no OsDXR has function as a rate-limiting enzyme in leaves or seeds. The expression profiling of the carotenogenic genes showed non-proportional correlations between the changes in gene expression and the metabolic changes, suggesting that the biosynthetic activity of carotenoid intermediate metabolites could be enhanced without any increases in transcript levels. Taken together, these differential roles of OsDXS2 and OsDXR are suggested to occur in a rice plant-specific pattern, and the “Three Faucets and Cisterns Model” was proposed to describe carotenoid metabolism in rice seeds. Finally, our studies provide useful information for designing fine-tuned strategies of carotenoid metabolic engineering in rice plants.
Methods
Plant materials and growth conditions
Mature seeds of Korean rice (Oryza sativa L. cv. Ilmi) were germinated and grown in soil in a greenhouse under the conditions of a16 h light/8 h dark cycle at 28 °C. Rice seeds obtained from the National Institute of Agricultural Sciences (South Korea) were used for Agrobacterium-mediated transformation and expression analyses of DXS genes. Organ-specific samples were harvested at different stages and used for quantitative real-time (qRT)-PCR.
T0 generation transgenic rice plants were first grown in a growth chamber, and then after transplanting them into the soil, T1 and T2 generations were grown in the greenhouse under the same conditions of a 16 h light/8 h dark cycle at 28 °C and in the field during the summer season until the T4 seed generation. All field studies were performed following local legislation with the permissions of the Rural Development Administration (South Korea). After the sterilization of T4 seeds with 70% ethanol and 2% sodium hypochlorite, 10-day-old seedlings were grown in a room maintained at 28 °C with a 12 h light/12 h dark cycle and under 70–90% relative humidity. These seedlings were used for qRT-PCR and metabolite analysis with T4 seeds, which were harvested at full maturity 40 days after flowering (DAF). The seed endosperm color was visually compared after dehusking (TR-200 Electromotion Rice Husker, Kett, Tokyo, Japan) and polishing (Pearlest Polisher, Kett).
Vector construction
The coding regions, including the open reading frames of OsDXS2 (Os07g09190) and OsDXR (Os01g01710), were amplified from the total RNA of 10-day-old seedlings using the gene-specific primer pairs F1/R1 and F2/R2, respectively. Each amplicon was further amplified using the universal attB primer pair and introduced into the pDONR221 vector by recombination using the Gateway® BP Clonase® II Enzyme Mix (Invitrogen, Waltham, MA). The resultant subclones were recombined with the vector p600-PGD1 (Seoul National University, Pyeongchang Korea), which contained a rice phosphogluconate dehydrogenase 1 (PGD1) promoter for constitutive expression in rice plants, [40] using the Gateway® LR Clonase® II Enzyme Mix (Invitrogen). This resulted in the generation of pPGD1::OsDXS2 and pPGD1::OsDXR vectors for rice transformation (Fig. 1b).
To introduce entire PGD1::OsDXS2::PinII or PGD1::OsDXR::PinII cassettes into the region of the PGD1 promoter and the PinII terminator into the stPAC expression cassette, which produces β-carotene under the control of a rice endosperm-specific globulin (Glb) promoter [32], they were individually PCR-amplified with the F3/R3 primer pair and then cloned into a pGlb::stPAC vector using the XhoI site, yielding either pPGD1::OsDXS2_Glb::stPAC or pPGD1::OsDXR_Glb::stPAC for rice transformation (Fig. 1b). All PCRs were performed using a PrimeSTAR® HS DNA Polymerase (Takara, Shiga, Japan). All primers are shown in Fig. 3b and their sequences are listed in Additional file 4: Table S7.
Rice transformation and selection of transformants
For overexpression of OsDXS2 or OsDXR, the pPGD1::OsDXS2, pPGD1::OsDXR, pPGD1::OsDXS2_Glb::stPAC, and pPGD1::OsDXR_Glb::stPAC final vectors, were individually introduced into Escherichia coli DH5α and then Agrobacterium tumefaciens LBA4404 harboring pSB1 plasmids through tri-parental mating [41]. After co-cultivation with embryogenic calli that were differentiated from the mature seeds of rice (O. sativa L. cv. Ilmi), the putative transgenic plantlets were generated using selection media containing phosphinothricin (4 mg/L) and cefotaxime (500 mg/L) under growth chamber conditions, according to a previously published method [42].
Genomic DNA was purified from leaf tissues using a DNeasy Plant Mini Kit (Qiagen), after grinding with a TissueLyser II (Qiagen), and PCR-amplified using a MightyAmp® DNA Polymerase (Takara) following the manufacturer’s instructions. The positive transgenic plants were first screened at the T0 leaf generation by PCR using the primer pairs F4/R4 for OsDXS2, F4/R5 for OsDXR, and F6/R6 for stPAC transgenes. Three selected lines were further verified using the same PCR protocols at the T3 leaf generation after a segregation test on phosphinothricin (4 mg/L) to assess the homozygosity of T-DNA. To examine the transgene copy number in rice genomes, TaqMan real-time PCR was performed using the primer set NF/NR, a customized probe NP labeled with a 6-carboxyfluorescein dye to detect a Nos terminator on the BAR cassette (35S::Bar::Nos), and the customized VIC dye-labeled α-tubulin probe (Os11g14220), as an internal reference (Assay ID: Os03643486_s1; Applied Biosystems, Foster City, CA). The PCR was carried out using a TaqMan Gene Expression Master Mix (Applied Biosystems), and fluorescence was measured with the CFX Connect™ Real-Time System (Bio-Rad, Richmond, CA) relative to the value of 1 copy in the homozygous T3 generation of the stPAC 25 rice line [32] using PCR conditions, as previously described [43]. The sequences of primers and probes used in the genomic DNA analysis are listed in Additional file 4: Table S7.
Quantification of carotenoids and chlorophylls
Leaf and seed samples of rice plants were prepared for carotenoid extraction, as previously described [43]. For HPLC analysis, the analytical samples were prepared by dissolving in dichloromethane/methanol 50:50 (v/v) following the addition of β-apo-8′-carotenal (0.05 mL of 25 μg/mL, Sigma-Aldrich Chemical Co, St. Louis, MO) as an internal standard, separating into layers with hexane (1.5 mL), and desiccating under liquid nitrogen. Carotenoids were then separated with a YMC ODS C-30 column (3 μm, 4.6 × 250 mm; YMC Europe, Germany) by an Agilent 1100 Series HPLC system (Agilent, Santa Clara, CA) equipped with a photodiode array detector under elution conditions, as previously described [43]. The chromatograms were generated at 472 nm for lycopene and 450 nm for the other compounds, including α-carotene, (all-E)-β-carotene, 9Z-β-carotene, 13Z-β-carotene, β-cryptoxanthin, lutein, violaxanthin, and zeaxanthin. The quantification was determined from the HPLC peak areas relative to the external standard calibration curves of the carotenoid standards, which were purchased from CaroteNature (Lupsingen, Switzerland). The amount of β-carotene was determined as the sum of (all-E)-β-carotene, 9Z-β-carotene, and 13Z-β-carotene.
To extract chlorophylls, 10 mg of the fresh leaf powder of rice plants were mixed with 1 mL of 100% methanol and incubated at 70 °C for 30 min with shaking (500 rpm) using a Thermomixer Comfort (model 5355, Eppendorf AG, Hamburg, Germany). After centrifuging at 800 g for 10 min at 4 °C, the absorbance of the supernatant was measured at 666 nm and 653 nm in a spectrophotometer (Optizen Pop, Mecasys Co, Daejeon, Korea). The contents of chlorophyll a and b were calculated using Wellburn’s formula (1994).
Quantitative real-time PCR
Total RNA was purified from the leaf, root, and floret tissues at nine weeks (vegetative stage) and three months (reproductive stage) to compare the expression patterns of OsDXS1, OsDXS2, OsDXS3, and OsDXR genes. Total RNA was also purified from the leaves of 10 day-old seedlings and from the unpolished mature seeds 40 DAF to profile the expression patterns of carotenogenic genes in each transgenic plant. The frozen powder (100 mg) samples were resolved in the PureLink® Plant RNA Reagent (Invitrogen) with DNase I (Qiagen, Hilden, Germany) for removal of remnant genomic DNA contamination. The 1st cDNA was synthesized using the AccuPower® RT Premix (Bioneer, Daejeon, Korea) and mixed with the SYBR Green Real-time PCR master mix (Bio-Rad). All reactions were performed with a CFX Connect™ Real-Time System (Bio-Rad) according to the manufacturer’s instructions and under the following conditions: 1 cycle of 3 min at 95 °C, 40 cycles of 15 s at 95 °C, and 1 cycle of 30 s at 60 °C. The qRT-PCR for transgene expression was performed with the gene-specific primer pairs F7/R7 for OsDXS2, F8/R7 for OsDXR, and F9/R9 for stPAC, as indicated in Fig. 1b and Additional file 4: Table S7. The transcripts of the 22 genes involved in rice carotenogenesis were examined by qRT-PCR using gene-specific primers. Gene names, accession numbers, primer sequences, and product sizes are detailed in Additional file 4: Table S8. To normalize the amount of RNA, all qRT-PCR values were calculated relative to the rice ubiquitin 5 gene (Os01g22490), which was amplified using the U5F/U5R primer pair [44].
Statistical analysis
All experiments were performed with a minimum of three biological replicates, and the results were expressed as the mean ± standard error (SE). Any statistically significant differences between the two groups were determined using a two-tailed Student’s t-test. A p value < 0.05 was considered to be statistically significant.
Availability of data and materials
The material and datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Abbreviations
- BCH:
-
A β-carotene hydroxylase
- CYP97B:
-
A P450-type-carotene hydroxylase gene
- DMAPP:
-
A dimethylallyl pyrophosphate
- DXP:
-
A deoxyxylulose 5-phosphate
- DXR:
-
A deoxyxylulose 5-phosphate reductoisomerase
- DXS:
-
A Deoxyxylulose 5-phosphate synthase
- Glb:
-
A rice endosperm-specific globulin
- IPP:
-
A isopentenyl pyrophosphate
- MEP:
-
A methylerythritol 4-phosphate
- MVA:
-
A mevalonic acid
- PDS:
-
A pytoene desaturase
- PGD1:
-
A rice phosphogluconate dehydrogenase 1
- PSY:
-
A pytoene synthase
- stPAC:
-
A synthetic phytoene synthase:2A:CrtI
References
Pichersky E, Raguso RA. Why do plants produce so many terpenoid compounds? New Phytol. 2016;220(3):692–702.
Yazaki K, Arimura GI, Ohnishi T. 'Hidden' terpenoids in plants: their biosynthesis, localization and ecological roles. Plant Cell Physiol. 2017;58(10):1615–21.
Gershenzon J, Dudareva N. The function of terpene natural products in the natural world. Nat Chem Biol. 2007;3(7):408–14.
Carretero-Paulet L, Cairo A, Botella-Pavia P, Besumbes O, Campos N, Boronat A, Rodriguez-Concepcion M. Enhanced flux through the methylerythritol 4-phosphate pathway in Arabidopsis plants overexpressing deoxyxylulose 5-phosphate reductoisomerase. Plant Mol Biol. 2006;62(4–5):683–95.
Estevez JM, Cantero A, Reindl A, Reichler S, Leon P. 1-Deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J Biol Chem. 2001;276(25):22901–9.
Vaccaro M, Malafronte N, Alfieri M, Tommasi N, Leone A. Enhanced biosynthesis of bioactive abietane diterpenes by overexpressing AtDXS or AtDXR genes in Salvia sclarea hairy roots. Plant Cell Tissue Organ Cult. 2014;119:65–77.
Simpson K, Quiroz LF, Rodriguez-Concepcion M, Stange CR. Differential contribution of the first two enzymes of the MEP pathway to the supply of metabolic precursors for carotenoid and chlorophyll biosynthesis in carrot (Daucus carota). Front Plant Sci. 2016;7:1344.
Munoz-Bertomeu J, Arrillaga I, Ros R, Segura J. Up-regulation of 1-deoxy-D-xylulose-5-phosphate synthase enhances production of essential oils in transgenic spike lavender. Plant Physiol. 2006;142(3):890–900.
Mendoza-Poudereux I, Munoz-Bertomeu J, Arrillaga I, Segura J. Deoxyxylulose 5-phosphate reductoisomerase is not a rate-determining enzyme for essential oil production in spike lavender. J Plant Physiol. 2014;171(17):1564–70.
Hasunuma T, Takeno S, Hayashi S, Sendai M, Bamba T, Yoshimura S, Tomizawa K, Fukusaki E, Miyake C. Overexpression of 1-Deoxy-D-xylulose-5-phosphate reductoisomerase gene in chloroplast contributes to increment of isoprenoid production. J Biosci Bioeng. 2008;105(5):518–26.
Yang J, Adhikari MN, Liu H, Xu H, He G, Zhan R, Wei J, Chen W. Characterization and functional analysis of the genes encoding 1-deoxy-D-xylulose-5-phosphate reductoisomerase and 1-deoxy-D-xylulose-5-phosphate synthase, the two enzymes in the MEP pathway, from Amomum villosum Lour. Mol Biol Rep. 2012;39(8):8287–96.
Zhang H, Niu D, Wang J, Zhang S, Yang Y, Jia H, Cui H. Engineering a platform for photosynthetic pigment, hormone and cembrane-related diterpenoid production in Nicotiana tabacum. Plant Cell Physiol. 2015;56(11):2125–38.
Cordoba E, Porta H, Arroyo A, San Roman C, Medina L, Rodriguez-Concepcion M, Leon P. Functional characterization of the three genes encoding 1-deoxy-D-xylulose 5-phosphate synthase in maize. J Exp Bot. 2011;62(6):2023–38.
Estevez JM, Cantero A, Romero C, Kawaide H, Jimenez LF, Kuzuyama T, Seto H, Kamiya Y, Leon P. Analysis of the expression of CLA1, a gene that encodes the 1-deoxyxylulose 5-phosphate synthase of the 2-C-methyl-D-erythritol-4-phosphate pathway in Arabidopsis. Plant Physiol. 2000;124(1):95–104.
Kim BR, Kim SU, Chang YJ. Differential expression of three 1-deoxy-D-xylulose-5-phosphate synthase genes in rice. Biotechnol Lett. 2005;27(14):997–1001.
Walter MH, Hans J, Strack D. Two distantly related genes encoding 1-deoxy-d-xylulose 5-phosphate synthases: differential regulation in shoots and apocarotenoid-accumulating mycorrhizal roots. Plant J. 2002;31(3):243–54.
Kim SM, Kuzuyama T, Chang YJ, Song KS, Kim SU. Identification of class 2 1-deoxy-D-xylulose 5-phosphate synthase and 1-deoxy-D-xylulose 5-phosphate reductoisomerase genes from Ginkgo biloba and their transcription in embryo culture with respect to ginkgolide biosynthesis. Planta Med. 2006;72(3):234–40.
Okada A, Shimizu T, Okada K, Kuzuyama T, Koga J, Shibuya N, Nojiri H, Yamane H. Elicitor induced activation of the methylerythritol phosphate pathway toward phytoalexins biosynthesis in rice. Plant Mol Biol. 2007;65(1–2):177–87.
Phillips MA, Walter MH, Ralph SG, Dabrowska P, Luck K, Uros EM, Boland W, Strack D, Rodriguez-Concepcion M, Bohlmann J, et al. Functional identification and differential expression of 1-deoxy-D-xylulose 5-phosphate synthase in induced terpenoid resin formation of Norway spruce (Picea abies). Plant Mol Biol. 2007;65(3):243–57.
Floss DS, Hause B, Lange PR, Kuster H, Strack D, Walter MH. Knock-down of the MEP pathway isogene 1-deoxy-D-xylulose 5-phosphate synthase 2 inhibits formation of arbuscular mycorrhiza-induced apocarotenoids, and abolishes normal expression of mycorrhiza-specific plant marker genes. Plant J. 2008;56(1):86–100.
Carretero-Paulet L, Cairo A, Talavera D, Saura A, Imperial S, Rodriguez-Concepcion M, Campos N, Boronat A. Functional and evolutionary analysis of DXL1, a non-essential gene encoding a 1-deoxy-D-xylulose 5-phosphate synthase like protein in Arabidopsis thaliana. Gene. 2013;524(1):40–53.
Peng G, Wang C, Song S, Fu X, Azam M, Grierson D, Xu C. The role of 1-deoxy-d-xylulose-5-phosphate synthase and phytoene synthase gene family in citrus carotenoid accumulation. Plant Physiol Biochem. 2013;71:67–76.
Budziszewski GJ, Lewis SP, Glover LW, Reineke J, Jones G, Ziemnik LS, Lonowski J, Nyfeler B, Aux G, Zhou Q, et al. Arabidopsis genes essential for seedling viability: isolation of insertional mutants and molecular cloning. Genetics. 2001;159(4):1765–78.
Jung KH, Lee J, Dardick C, Seo YS, Cao P, Canlas P, Phetsom J, Xu X, Ouyang S, An K, et al. Identification and functional analysis of light-responsive unique genes and gene family members in rice. PLoS Genet. 2008;4(8):e1000164.
Devi K, Dehury B, Phukon M, Kumar Modi M, Sen P. Novel insights into structure–function mechanism and tissue-specific expression profiling of full-length dxr gene from Cymbopogon winterianus. FEBS Open Bio. 2015;5:325–34.
Howitt CA, Pogson BJ. Carotenoid accumulation and function in seeds and non-green tissues. Plant Cell Environ. 2006;29(3):435–45.
Beyer P. Golden Rice and 'Golden' crops for human nutrition. New Biotechnol. 2010;27(5):478–81.
Burkhardt PK, Beyer P, Wunn J, Kloti A, Armstrong GA, Schledz M, von Lintig J, Potrykus I. Transgenic rice (Oryza sativa) endosperm expressing daffodil (Narcissus pseudonarcissus) phytoene synthase accumulates phytoene, a key intermediate of provitamin a biosynthesis. Plant J. 1997;11(5):1071–8.
Ye X, Al-Babili S, Kloti A, Zhang J, Lucca P, Beyer P, Potrykus I. Engineering the provitamin a (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science. 2000;287(5451):303–5.
Paine JA, Shipton CA, Chaggar S, Howells RM, Kennedy MJ, Vernon G, Wright SY, Hinchliffe E, Adams JL, Silverstone AL, et al. Improving the nutritional value of Golden Rice through increased pro-vitamin a content. Nat Biotechnol. 2005;23(4):482–7.
Ha SH, Liang YS, Jung H, Ahn MJ, Suh SC, Kweon SJ, Kim DH, Kim YM, Kim JK. Application of two bicistronic systems involving 2A and IRES sequences to the biosynthesis of carotenoids in rice endosperm. Plant Biotechnol J. 2010;8(8):928–38.
Jeong YS, Ku HK, Kim JK, You MK, Lim SH, Kim JK, Ha SH. Effect of codon optimization on the enhancement of the β-carotene contents in rice endosperm. Plant Biotechnol Rep. 2017;11:171–9.
Bai C, Capell T, Berman J, Medina V, Sandmann G, Christou P, Zhu C. Bottlenecks in carotenoid biosynthesis and accumulation in rice endosperm are influenced by the precursor-product balance. Plant Biotechnol J. 2016;14(1):195–205.
Enfissi EM, Nogueira M, Bramley PM, Fraser PD. The regulation of carotenoid formation in tomato fruit. Plant J. 2017;89(4):774–88.
Morris WL, Ducreux LJ, Hedden P, Millam S, Taylor MA. Overexpression of a bacterial 1-deoxy-D-xylulose 5-phosphate synthase gene in potato tubers perturbs the isoprenoid metabolic network: implications for the control of the tuber life cycle. J Exp Bot. 2006;57(12):3007–18.
Sankari M, Hridya H, Sneha P, Doss CGP, Christopher JG, Mathew J, Zayed H, Ramamoorthy S. Implication of salt stress induces changes in pigment production, antioxidant enzyme activity, and qRT-PCR expression of genes involved in the biosynthetic pathway of Bixa orellana L. Funct Integr Genomics. 2019;19(4):565–74.
Guevara-Garcia A, San Roman C, Arroyo A, Cortes ME, de la Luz G-NM, Leon P. Characterization of the Arabidopsis clb6 mutant illustrates the importance of posttranscriptional regulation of the methyl-D-erythritol 4-phosphate pathway. Plant Cell. 2005;17(2):628–43.
Welsch R, Zhou X, Yuan H, Alvarez D, Sun T, Schlossarek D, Yang Y, Shen G, Zhang H, Rodriguez-Concepcion M, et al. Clp protease and OR directly control the proteostasis of phytoene synthase, the crucial enzyme for carotenoid biosynthesis in Arabidopsis. Mol Plant. 2018;11(1):149–62.
Han M, Heppel SC, Su T, Bogs J, Zu Y, An Z, Rausch T. Enzyme inhibitor studies reveal complex control of methyl-D-erythritol 4-phosphate (MEP) pathway enzyme expression in Catharanthus roseus. PLoS One. 2013;8(5):e62467.
Park SH, Yi N, Kim YS, Jeong MH, Bang SW, Choi YD, Kim JK. Analysis of five novel putative constitutive gene promoters in transgenic rice plants. J Exp Bot. 2010;61(9):2459–67.
Komari T, Hiei Y, Saito Y, Murai N, Kumashiro T. Vectors carrying two separate T-DNAs for co-transformation of higher plants mediated by Agrobacterium tumefaciens and segregation of transformants free from selection markers. Plant J. 1996;10(1):165–74.
Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6(2):271–82.
Ha SH, Kim JK, Jeong YS, You MK, Lim SH, Kim JK. Stepwise pathway engineering to the biosynthesis of zeaxanthin, astaxanthin and capsanthin in rice endosperm. Metab Eng. 2019;52:178–89.
Jain M, Nijhawan A, Tyagi AK, Khurana JP. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun. 2006;345(2):646–51.
Acknowledgments
Not applicable.
Funding
This work was supported by grants from the Next-Generation BioGreen 21 Program (PJ01334601 and PJ01368801 to SH Ha) funded by the Rural Development Administration in the Republic of Korea. The funders have no role in the study design, data analysis and interpretation, and manuscript writing.
Author information
Authors and Affiliations
Contributions
SHH coordinated this project and supervised the manuscript. MKY designed this study with SHH and wrote the manuscript. YJL, YAJ, and SHL constructed all rice transgenic plants in this study, JKK and SAB analyzed the contents of carotenoids and chlorophylls, and MKY and YJL performed all expression profiling analyses of carotenogenic genes. All authors have read and approved the final manuscript. SHH is the corresponding author and is responsible for all contact and correspondence.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Figure S1.
Alignment of deduced amino acid sequences among plant type II deoxyxylulose 5-phosphate synthase (DXS) proteins.
Additional file 2: Figure S2.
Alignment of deduced amino acid sequences among plant deoxyxylulose 5-phosphate reductoisomerase (DXR) proteins.
Additional file 3: Figure S3.
Genomic DNA analyses to verify the integration and copy number of transgenes in the rice genome.
Additional file 4: Table S1.
Carotenoid content and composition in the leaves of transgenic rice plants. Table S2. Chlorophyll content in the leaves of transgenic rice plants. Table S3. Carotenoid content and composition in mature seeds of transgenic rice plants. Table S4. Expression profiles of structural genes related to the biosynthesis of carotenoids in rice leaves. Table S5. Expression profiles of structural genes related to the supplement of substrates into the biosynthesis of rice seed carotenes. Table S6. Expression profiles of structural genes related to the xanthophyll biosynthesis in rice seeds. Table S7. The primer list used in vector construction and transgene analysis. Table S8. The primer list used in expression analysis of rice genes.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
You, M., Lee, Y., Kim, J. et al. The organ-specific differential roles of rice DXS and DXR, the first two enzymes of the MEP pathway, in carotenoid metabolism in Oryza sativa leaves and seeds. BMC Plant Biol 20, 167 (2020). https://doi.org/10.1186/s12870-020-02357-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-020-02357-9