Abstract
Background
Leaf shape can affect plantlet development and seed yield in sesame. The morphological, histological and genetic analyses of a sesame mutant cl1 (cl) with curly leaf and indehiscent capsule traits were performed in this study. In order to clone the cl1 gene for breeding selection, genome re-sequencing of the 130 individuals of cl1 × USA (0)-26 F2 population and a bulked segregation analysis (BSA) pool was carried out. The genome re-sequencing data of the 822 germplasm with normal leaf shape were applied.
Results
For cl1 mutant, the adaxial/abaxial character of the parenchyma cells in the leaf blades is reduced. Results proved that the leaf curling trait is controlled by a recessive gene (Sicl1). Cross- population association of the F2 population of cl1 × USA (0)-26 indicated that the target cl locus was located on the interval C29 between C29_6522236 and C29_6918901 of SiChr. 1. Further regional genome variants screening determined the 6 candidate variants using genomic variants data of 822 natural germplasm and a BSA pool data. Of which, 5 markers C29_6717525, C29_6721553, C29_6721558, C29_6721563, and C29_6721565 existed in the same gene (C29.460). With the aid of the validation in the test F2 population of cl1 × Yuzhi 11 and natural germplasm, the integrated marker SiCLInDel1 (C29: 6721553–6721572) was determined as the target marker, and C29.460 was the target gene SiCL1 in sesame. SiCL1 is a KAN1 homolog with the full length of 6835 bp. In cl1, the 20 nucleic acids (CAGGTAGCTATGTATATGCA) of SiCLInDel1 marker were mutagenized into 6 nucleic acids (TCTTTG). The deletion led to a frameshift mutation and resulted in the earlier translation termination of the CL gene. The Sicl1 allele was shortened to 1829 bp. SiCL1 gene was expressed mainly in the tissues of stem, leaf, bud, capsule and seed.
Conclusions
SiCL1 encodes a transcription repressor KAN1 protein and controls leaf curling and capsule indehiscence in sesame. The findings provided an example of high-efficient gene cloning in sesame. The SiCL1 gene and the cl1 mutant supply the opportunity to explore the development regulation of leaf and capsule, and would improve the new variety breeding with high harvest mechanization adaption in sesame.
Similar content being viewed by others
Background
Sesame (Sesamum indicum L., 2n = 26) is an important and specific oilseed crop for the high nutrition and oil quality [1]. Sesame is cultivated mainly in the developing countries for its low yield and labor-consuming production process. Therefore, enhancing the yield is one of the key objectives of sesame breeding and production [2, 3]. Sesame yield is affected by many factors, such as plant type, leaf type, capsule number per plant, seed number per capsule, seed size and seed shattering [2, 4,5,6].
An optimal leaf shape is important to avoid of self-shading and maintains the ideal plant architecture for high yield [7]. Sesame is a kind of high-height crop. Leaf shape can dramatically affect the yield level. A natural sesame mutant (herein named cl1) with curly leaves and indehiscent capsules was found in 1940’s [8, 9]. The mutagenesis of cl1 could affect the plant development and the yield, and the trait is controlled by a regress gene allele [1, 10, 11]. According to the investigation results in 2014 (planted at Pingyu experimental station, China), the cl1 mutant lines only formed 34–37 capsules per plant. The average seed weight per cl1 plant was 4.82 g and obviously lower than that (10.17 g) of var. Yuzhi 11 (unpublished data, Haiyang Zhang). To explore the genetic mechanism, Uzun et al. [12] has identified an AFLP (amplified fragment length polymorphisms) molecular marker linked to the mutated trait using BSA (bulked segregation analysis) approach in sesame. However, the target gene for the curly leaf mutation in cl1 has not been detected mainly for the lack of accurate methods for gene location and validation.
The development of high-throughput sequencing and the initiation of the Sesame Genome Project impel the gene cloning and genomics research in sesame [3]. About one hundred of associated markers and QTLs related to dozens of agronomic traits in sesame have been identified [4, 13,14,15,16,17,18]. Recently, the gene SiDt controlling the inflorescence termination was determined using the ultra-dense SNP map via the fine mapping method [5]. The linkage mapping and GWAS (genome wide association studies) analysis are the two main methods to determine the gene/QTL loci for key agronomic traits in crops. In recent years, integration of linkage mapping and association mapping have been successfully applied in rice [19], cotton [20], rapeseed [21], sunflower [22], soybean [23, 24] for detection of QTLs and candidate genes linked to specific complex agricultural traits. Therefore, in the present study, we sequenced the 130 F2 individuals derived from a cross between the curly leaf mutant cl1 and the wild type, and identified the target gene SiCL1 alleles in sesame using the cross-population association mapping and the genome variants screening. Meanwhile, the mutagenesis character and the expression profiles of the gene SiCL1 alleles were systematically analyzed in sesame. The findings supplied the highly-efficient detection method of candidate genes for key agronomic traits and further studies of curly leaf genes in sesame.
Results
Characterization of mutant cl1 and the wild type
Systematical phenotypic observation showed that there existed the significant difference in leaf and capsule tissues between cl1 and the wild type (WT, Yuzhi 11) (Fig. 1). Compared with the wild type (in Fig. 1a), the mutant cl1 displayed the curly leaf shape character during the entire development life. Some humps presented on the veins of the back blade of leaves. Meanwhile, the tip of capsules was compact and the capsule peel became thick (Fig.1a). After mature or desiccated, the capsules of cl1 were still indehiscent and the carpels did not split naturally (data not shown). Cross- sections observation of leaf blades indicated that the lower epidemis in cl1 did not as smooth as that in the WT. The abaxial parenchyma cell (PC (ab)) and adaxial parenchyma cell (PC (ad)) tissues tended to be horizontally arranged in leaf veins. Especially, for two-pair-leaf seedlings of cl1, the adaxial/abaxial character was reduced.
Phenotype comparison of mutant cl1 and the wild type (WT). a: Morphological comparison of leaf and capsule in cl1 and the wild type (WT, var. Yuzhi 11). In mutant cl1, the positive leaf blade is curly, while humps present on the veins of back blade. The capsule tip constrains, and the capsule peel becomes thick. b: Cross- section of leaf blades of the one-pair-leaf-old seedlings of cl1 and Yuzhi 11. c: Cross- section of leaf blades of the two-pair-leaf-old seedlings of cl1 and Yuzhi 11. The PC (ab) and PC (ad) tissues in veins arrange horizontally, and the adaxial/abaxial character is reduced. UE: upper epidemis. LE: lower epidemis. PC (ad): adaxial parenchyma cell. PC (ab): abaxial parenchyma cell. VB: vascular bundle cells. Bar = 100 μm
Inheritance of curly leaf trait in mutant cl1 and the wild type
In order to clarify the inheritance of cl1 genotype in sesame, we constructed F1 hybrids and F2 populations of cross combination of ‘Yuzhi 11 ×mutant cl1’, and a mapping population of ‘mutant cl1 ×USA (0)-26’ (Table 1). The F1 generations from direct crosses and reciprocal crosses between CL and cl types only displayed the normal leaf shape with dehiscent capsule. In cl1 backcrosses of the two combinations, the segregation ratio of the curly and the normal leaf phenotypes fitted the expected 1 (CL):1 (cl) ratio (Table 1). In addition, the segregation ratio of leaf shape phenotypes in the F2 populations fitted the expected ratio of 3 (CL):1 (cl). Chi-square tests (P > 0.05) proved that the segregation of the curly leaf trait in mutant cl1 fitted the Mendelian inheritance mode, as the χ2 value for trait segregation in the above two F2 populations was 0.67 and 1.29, respectively. The results confirmed that the leaf curling trait is controlled by a recessive gene allele. Thus, we annotated the curly leaf locus in mutant cl1 and Yuzhi 11 (WT) as Sicl1 and SiCL1, respectively.
Identification of CL1 gene locus using association mapping and genome variants screening
To efficiently locate the gene locus of curly leaf trait, we carried out the cross-population association mapping and the genome wide screening strategy. The two parents (i.e., USA (0)-26 and cl1) and their 130 F2 individuals were sequenced using an Illumina sequencing approach. In total, 847.46 Gb of raw data was obtained with the average genome coverage of 18.1 fold per sample (Table 2). The mapped reads of each sample were aligned to the updated sesame reference genome (based on GCA_001692995.1 version) for SNP calling. A total of 598,322 variants were detected between the two parents.
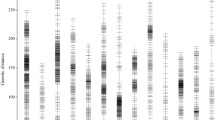
The joint calling indicated that 425,661 SNP/InDel variants were located on 13 chromosomes (Fig. 2). The cross-population association mapping showed that an interval on LG 8 displayed the lowest P value. Based on the specific variant with the lowest P value (C29_6721563, 2.38 × E− 91), up- and down-stream 200 Kb flanking sequences of C29_6721563 was determined as the target interval linked to the curly leaf shape (cl). The interval C29 between C29_6522236 and C29_6918901 markers contained 90 variants with the P value variation of 3.97 × E− 19 to 3.00 × E− 94 (Additional file 1: Table S1). According to the interval site of Scaffold C29 and the chromosome nomination described by Zhao et al. [25, 26], the LG 8 of the cross population corresponded to the SiChr. 1 of S. indicum.
Genomic association mapping of Sicl1 gene locus in sesame. Mahattan plot of SNP/InDel association analysis is performed using the F2 population data. An interval in SiChr. 1 shows the lowest P value. All the variants in the plot are screened using the genome variants data. The 6 green dots are retained as the candidate markers linked to the curly leaf trait
To screen the target SNP/InDel marker, we filtered the located SNP/InDel variants in the interval C29 using the regional genome variants data of 822 sesame accessions (M1-M822) (Additional file 2: Table S2) (Fig. 2). The results showed that 6 variant loci (the green dots in Fig. 2) were retained in the population. At the same time, we screened the plotted variant sites using the BSA pool data for cl1 (cl). The reads of the BSA pool was 38.78 Gb (deposited in NCBI dataset). As a result, 42 SNP/InDel sites were still retained in the interval C29 (Additional file 3: Table S3).
Subsequently, we compared the above two groups of the screening results (shown in Additional file 1: Table S1 and Additional file 3: Table S3, respectively). Interestingly, we found that the 6 variants screened using the variants data of 822 germplasm existed in the 42 candidate variants filtered through the BSA pool data. Of the 6 variants, 5 sites belonged to InDel mutation type (Additional file 4: Table S4). C29_6674693 was located in the intergenic region, while the other five sites of C29_6717525, C29_6,721,553, C29_6721558, C29_6721563, and C29_6721565 were individually distributed in the 5′ flanking, exon or intron regions of C29.460. Therefore, based on the integrated sequences of the four markers (C29_6721553, C29_6721558, C29_6721563, and C29_6721565) in gene C29.460, we designed the primer pair of SiCLInDel 1 and performed the verification of these candidate markers using the test population (Additional file 5: Table S5). As a result, the SiCLInDel 1 allele of gene C29.460 entirely accorded with the phenotype segregation in the 1000 F2 individuals and the phenotype of the 500 sesame germplasm materials with normal leaf shape (partially shown in Additional file 6: Figure S1). Therefore, C29.460 was regarded as the candidate CL gene (i. e., SiCL1) for the curly leaf shape in sesame.
Structure analysis of SiCL1 gene and SiCL1 homolog in sesame
With the aid of the reference genome information, we designed the primer pairs and amplified the entire cDNA and DNA sequences of SiCL1 (C29.460) (Fig. 3) (Additional file 5: Table S5, Additional file 7: Table S6). Sanger sequencing and gene alignment results proved that the full length of SiCL1 in Yuzhi 11 is 6835 bp and comprised 6 exons and 5 introns (Fig. 3a). During sequencing, we found that the SiCL1 gene had two transcripts of SiCL1–1 and SiCL-2, respectively. SiCL1–1 encoded 440 amino acids, while SiCL-2 encoded 439 amino acids for the deletion of one amino acid (Ser393) (Fig. 3b). Structure analysis validated the presence of a splicing site at the position of 4886- 4888 bp (NCBI accession no. MG763174).
Sequence comparison of SiCL1 alleles in sesame. a: Gene structure comparison of SiCL1 and Sicl1. In the second exon, 20 nucleic acids (CAGGTAGCTATGTATATGCA, 1131–1150 bp in SiCL1) are mutated into 6 nucleic acids (TCTTTG, 1131–1136 bp in Sicl1). Blue line refers to SiCL1 gene. Red line refers to Sicl1 gene. Green vertical line indicates a splicing site. b: cDNA sequence alignment of SiCL1–1, SiCL1–2 and Sicl1 alleles and the corresponding amino acids. The dots after the red letter ‘A’ in Sicl1 gene sequences refer to the spliced fragment. The red letters in Sicl1 protein sequence refer the new amino acids formed by the frameshift mutation. The red asterisk refers to the terminator. The blocked sequences in SiCL1–1 and SiCL1–2 refer to a splicing site
In mutant cl1, the 20 nucleic acids of CAGGTAGCTATGTATATGCA (1131- 1150 bp) SiCLInDel 1 marker adjacent to the 2nd exon region of C29.460 allele were mutagenized into 6 nucleic acids of TCTTTG. Interestingly, one splicing site (at the position 1134 bp) existed in the above 20 nucleic acids of SiCL1 allele. The sequence deletion mutation resulted in a series of change in gene structure as follows (Fig. 3b). With the deletion of the SiCLInDel 1 marker and the splicing site, a frameshift mutation formed at the position 883 bp of the cDNA of Sicl1, and led to the substitution of the five amino acids from Arg295-Lys299 to Asn295-Cys299 during translation. Consequently, the new terminator TAA substituted the serine at the residue 300 and resulted in the translation termination (Fig. 3a, b). Then the gene length of Sicl1 shortened to 1829 bp. The mRNA was 900 bp and only encoded 299 amino acids (Fig. 3b). Of note, a conserved MYB domain (Pro271-Thr323) was detected in SiCL1. The above frameshift mutation and the terminator formation coincidentally occurred in the conserved region.
Non-redundant (NR) protein annotation indicated that the SiCL1 protein belonged to a transcription repressor KAN1 protein. In the sesame genome, there was a homolog gene (named SiKAN1-like) to SiCL1 with the gene resemblance of 70.1% (data not shown). Interspecies comparative analyses indicated that SiCL1 had the high resemblance with the transcription repressor KAN1/ KAN1-like protein in other ten plants (Fig. 4). The resemblance rate varied from 32.8% (ZmKAN1) to 70.1% (SiKAN1-like). The conservative MYB domain existed in these SiCL1 homologs. Of the 22 homologs in the other 10 plants, KAN1 homologs of potato (Solanum tuberosum) and tobacco (Coffea canephora) displayed the closer relationship with SiCL1 protein (Fig. 5).
Protein sequence comparison of SiCL1 and KAN homologs in sesame and other four plant species. The blocked region represents the highly conserved MYB domain in members of the GARP gene family. The proteins includes SiCL1 protein and 15 homologs in ten plants, i.e., SiKAN1-like (XP_011099920.1) in Sesamum indicum, AtKAN1 (AAL05436.1), AtKAN2 (AAL05437.1), AtKAN3 (AAL05438.1) and AtKAN4 (AAL05439.1) in Arabidopsis thaliana; ZmMWP (NP_001147322.1), ZmKAN1 (NP_001295437.1), ZmKAN3 (NP_001307939.1), ZmKAN4 (XP_008661896.1) and ZmKAN5 (NP_001132886.1) in Zea mays; RLL1 (XP_015610960.1), OsKAN2 (XP_015648273.1), OsKAN3 (XP_015648944.1), OsKAN4 (XP_015629095.1) and OsKAN5 (XP_015623607.1) in Oryza sativa. Identical residues are shaded in black; conserved residues are shaded in gray; residues with low identity are shaded in light gray. The black dot indicates amino acid gap
Phylogeny analyses of SiCL1 protein and KANAD1 family in sesame and other ten plants. Numbers above the branches indicate the support with 1000 bootstrap replications. The cluster includes SiCL1 protein and 23 homologs in ten plants, i.e., SiKAN1-like (XP_011099920.1) in Sesamum indicum, AtCL1 (AAL05436.1), AtKAN2 (AAL05437.1), AtKAN3 (AAL05438.1) and AtKAN4 (AAL05439.1) in Arabidopsis thaliana; ZmMWP (NP_001147322.1), ZmCL1 (NP_001295437.1), ZmKAN3 (NP_001307939.1), ZmKAN4 (XP_008661896.1) and ZmKAN5 (NP_001132886.1) in Zea mays; RLL1 (XP_015610960.1), OsKAN2 (XP_015648273.1), OsKAN3 (XP_015648944.1), OsKAN4 (XP_015629095.1) and OsKAN5 (XP_015623607.1) in Oryza sativa; MeCL1 (XP_021601546.1) in Manihot esculenta; NtCL1 (XP_016437587.1) in Nicotiana tabacum; HbCL1 (XP_021642823.1) of Hevea brasiliensis; InCL1 (XP_019172029.1) in Ipomoea nil; CcCL1 (CDP01671.1) in Coffea canephora; and StCL1 (XP_015166410.1) in Solanum tuberosum
Expression profiles of SiCL1 alleles in sesame
To explore the expression profiles of SiCL1 and Sicl1 in sesame, we monitored and compared the transcription level of SiCL1 alleles in root, leaf, stem, bud and capsule tissues of the WT (Yuzhi 11) and mutant cl1, respectively ((Fig. 6) using qPCR assay. In Yuzhi 11, SiCL1 gene was expressed in root, leaf, stem, bud and capsule tissues. The maximum and the minimum amounts of CL1 transcript presented in 0 DAP (day after pollination) capsule and root, respectively. The transcript amount in 0d capsule was 61.04 times than that of root, followed was the bud sample (43.68 times). Similarly, the relatively high expression level of Sicl1 gene was observed in bud and 0 DAP (day after pollination) capsules. The amount of the Sicl1 transcript reached 136.04 times than that of the root sample.
Discussion
Leaf shape takes the function of capturing light energy and synthesizing organic compounds through photosynthesis and finally affects the seed yield in sesame [2, 7]. To our knowledge, there are more than six leaf curling types (such as gentle curling, up-curling, up-curling with indehiscent capsule (cl1 type), curling and shrinking, down-curling, and whole curling) found in sesame mutant library [27] (Haiyang Zhang, the Sesame Genome, ISBN 978–3–319-98098-0, in press). In this study, the morphological and genetic analyses of mutant cl1 were systematically performed. The target gene SiCL1 controlling the leaf shape and capsule indehiscence was cloned in sesame. Compared with the normal sesame varieties, cl1 mutant presented the lower yield level [1, 10, 11]. Meanwhile, the leaf curling trait of cl1 mutant was synchronized with the capsule indehiscence. Even though the cl1 mutant presents the negative effect of cl1 gene on the growth and development and seed yield in sesame, the mutation characteristics and the SiCL1 gene supply the opportunity to explore the development regulation of leaf and capsule, and would impel the harvest mechanization in sesame. Meanwhile, the integrity strategy of cross-population association mapping and the regional genome variants screening provided an example of high-efficient gene cloning in sesame for the first time.
Traditionally, candidate genes/QTLs linked to the specific trait are identified using mapping populations in crops [26, 28, 29]. Recently, combined linkage- association mapping strategies have been applied for detecting the genes or QTLs of quantitative traits in crops [30, 31]. In sesame, the first ultra-dense SNP genetic map was applied for the cloning of SiDt gene [5]. About 30,193 SNPs were located in the 13 linkage groups of the SNP map. The main objective of constructing this SNP map with high-resolution markers was to assist in the fine genome assembly and the genomics analysis in sesame. Nevertheless, constructing an ultra-dense linkage map was time-consuming and lavish to clone a functional gene of a quality trait. In this paper, we carried out the common genome joint calling in the 130 individuals of the F2 population. A total of 425,697,661 variants were directly plotted in the 13 chromosomes (Fig. 2). Based on the accurate association analysis of the variants, an ~ 400 Kb physical region with significantly high P value was determined in linkage group 8. Compared with the traditional linkage mapping method, cross-population association mapping analysis is fast and efficient, as the procedures of constructing a genetic map could be ignored [31]. The association analysis of all the variants with phenotype information in the mapping population could directly performed using GLM (general linear model) model in TASSELE5.0. In addition, candidate-gene analyses within linked regions have been greatly accelerated by the availability of the complete sequence of the genome [32]. To accurately carry out the association mapping, we used the fine sesame genome (Yuzhi 11, GCA_001692995.1), SNP genetic map [5], and the chromosome set annotation system described by Zhao et al. [25] in this study. The high-quality genome supplied the reliable genomic information.
As to the genome screening procedure, we applied both the genome variants data of sesame germplasm and the variants data of a BSA pool to screen the candidate variants in the interval C29. The genome variants data of sesame germplasm contained 8,837,186 SNP/InDel variants (data not shown) and was applied in this study. As shown in Additional file 1: Table S1 and Additional file 4: Table S4, 84 of 90 variants in C29 were filtered. However, for the BSA with 50 individuals with cl1 genotype, the 191,008 bp distance of the interval between C29_6721553 and C29_6912561 markers presented the significant linkage (Additional file 3: Table S3). For screening the target variant in the tight linkage region, genome variants data is more useful than BSA pool data.
Previous studies showed that leaf curling or rolling was affected by the polarity of cells and complicated development controls [33,34,35]. Three gene families, i.e., HD-ZIP III family, KANAD1 family and YABBY family are found to regulate the establishment of polarity of leaf in plants [35]. For example, the leaf rolling gene SHALLOT-LIKE1 (SLL1)/RL9 in rice encodes a transcription factor of KANADI family and regulates the leaf abaxial cell development [36]. In this study, cross-section observation reflected that the polarity of the abaxial parenchyma cells (PC (ab)) and adaxial parenchyma cell (PC (ad)) was affected in cl1 (Fig. 1). The phenotype of leaf curling indeed accorded with the function of the target gene SiCL1 in sesame. Analysis results reflected that SiCL1 belonged to the KAN1/ KAN1-like protein family, and was the closest to CcKAN1 (CDP01671.1) (Figs. 4 and 5). Similar to the KAN homologs, SiCL1 contained the conserved MYB domain in GARP family (Figs. 3 and 5). As the conserved MYB region was mutated, the gene lost the polarity control. Therefore, we inferred that the frameshift mutation and translation termination resulted in the function loss of SiCL in sesame. The results supplied the foundation for elucidating the regulation of the molecular mechanism on leaf curling and capsule indehiscence traits in sesame.
Conclusions
For cl1 sesame mutant, curling leaf and indehiscent capsule traits are controlled by a same recessive gene (Sicl1). The adaxial/abaxial character of the parenchyma cells in the leaf blades is reduced. The target gene SiCL1 is a transcription repressor KAN1 homolog with the full length of 6835 bp. The 20 nucleic acids (CAGGTAGCTATGTATATGCA) were mutagenized into 6 nucleic acids (TCTTTG) and eternally resulted in a frameshift mutation and earlier translation termination of the CL gene in cl1. The findings provided an example of high-efficient gene cloning in sesame and supply the opportunity to explore the development regulation of leaf and capsule and to improve the new sesame variety breeding with high harvest mechanization adaption.
Methods
Plant materials and cross populations
A mutant cl1 with leaf curling and capsule indehiscence traits was a kind of gift from Mr. Ray Langham, USA in 1980’s. A line (N49) of mutant cl1 with the curly leaf phenotype was chosen and self-pollinated more than six generations before the genetic analysis. Yuzhi 11, USA (0)-26, and 500 germplasm accessions (M1-M500) with normal leaf shape (listed in Additional file 2: Table S2) were randomly chosen from the sesame germplasm bank to investigate the inheritance of the curly leaf and indehiscent capsule traits in sesame.
The mutant cl1 and the wild type (Yuzhi 11) were cultured at the Yuanyang experiment station for trait investigation and histological analysis during 2012–2014. Two cross combinations of cl1 (cl, P1) and USA (0)-26 (CL, P2), and Yuzhi 11 (CL, P1) and cl1 (cl, P2), respectively, were cultured for phenotypic segregation analysis in 2012 and 2013 (Table 1).
The 130 F2 individuals derived from the cross between cl1 and USA (0)-26 and the two parents were chosen for genome re-sequencing and gene locus detection. A total of 1231 F2 individuals of the cross between cl1 and Yuzhi 11 and 500 natural sesame germplasm (M1-M500) were cultured at Sanya experimental station for the target gene validation. Fifty individuals of cl genotype were chosen from the test F2 population and were sequenced as a BSA pool. All the above materials were available from Henan Sesame Research Center, Henan Academy of Agricultural Sciences (HSRC, HAAS) (Zhengzhou, China).
The leaf shape and capsule indehiscence traits in each sample were assessed during seedling, flowering and (or) maturation stages. Chi-square tests (P = 0.05) were used to determine the segregation significance for the curly leaf trait. Young leaf tissues of the above accessions and population progeny were collected, immersed in liquid nitrogen and frozen at − 70 °C for genomic DNA extraction. To clone the cDNA sequences and assay the expression pattern of SiCL1 alleles, the young roots, stem segments, young leaves, buds, 0d capsules and 5d capsules of the cl1 line N49 and Yuzhi 11 (wild type) at flowering stage were harvested and immersed in liquid nitrogen for RNA extraction.
Histological analysis of mutant cl1 and the wild type
Before producing paraffin sections, the fresh of leaf tissues of CL and cl genotypes were fixed in formalin-glacial acetic acid-alcohol (FAA) solution containing 3.8% formalin, glacial acetic acid and 70% alcohol (V:V:V = 1:1:18) for 24 h, and then air-extracted using pump. The samples were dehydrated with a series of ethanol and stained with 1% safranin for 24 h. After infiltrated using a series of ethanol and dimethybenzene solution for 5 h, the samples were immersed in a series of dimethybenzene solution for overnight. After embedded in paraffin, cross section (10 μm) were cut and stained with 0.1% fast green. The specimens were observed and photographed under the Leica DM6000B microscope equipped a DFC500 camera (Leica, Germany). Image quality was optimized in Adobe Photoshop 7.0 (Adobe, USA).
Genomic DNA extraction, library construction and sequencing
The two parents (i.e., USA (0)-26 and cl1) and their 130 F2 individuals and a BSA pool were sequenced using an Illumina HiSeq 2500 sequencing approach. Genomic DNA was extracted from young leaves of each sample using DNeasy Plant Mini Kits (QIAGEN, Hilden, Germany) and fragmented by sonication. Standard paired-end (PE) libraries were constructed according to Illumina guidelines. The libraries were prepared and sequenced on an Illumina HiSeq 2500 platform (Illumina, San Diego, USA) according to the procedures described by Zhang et al. [5].
Sequencing data analysis and variants detection in population
Raw reads obtained from the Illumina HiSeq 2500 platform were filtered using Trimmomatic 0.33 [37]. Based on the ultra-dense SNP genetic map [5] and chromosome annotation results [25], the genome data of Yuzhi 11 (PRJNA315784) (version 2.0) was re-assembled and applied as the reference genome in this study [38]. Alignment of re-sequencing data to the reference genome was performed using BWA 0.7.15 with the default settings described by Li and Durbin [39]. Putative SNPs and InDels were screened using Genome Analysis Tool Kit (GATK3.7) packages according to GATK joint calling best practice [40].
All the variants from all the 132 sequencing samples were filtered according to the following high quality (high-confidence) criterion: minimal variant count ≥100, minimum frequency of 0.1 (the minimum frequency of the minority polymorphisms for the site).
Statistical significance and candidate gene location
Association analysis of all the variants with phenotype information in the mapping population was performed using GLM (general linear model) model in TASSELE5.0. The target interval was defined as the two-side flanking regions (400 Kb distance) of the SNP/InDel variant with the lowest association P value. Distribution of the genome scaffolds in 13 chromosomes was numbered according to the chromosome annotation described by Zhao et al. [25]. Home-made scripts were used to screen the specific SNPs and InDels of the target interval. The genomic variants data of 822 sesame accessions (partially listed in Additional file 2: Table S2 at www.sesamum.org) and the SNP/InDel database of the population BSA pool (uploaded to NCBI database) of cl genotype were applied for the target variant detection.
The candidate SNP/InDel sites were transformed into PCR-based markers using the Primer Premier 5.0 program (http://www.premierbiosoft.com/products/products.html) according to the method of Wei et al. [41]. PCR reaction was performed in a 10 μL reaction mixture containing 1 × Buffer, 2.0 mmol L− 1 MgCl2, 0.1 mmol L− 1 dNTPs, 1 μmol L− 1 of each primer, 0.5 U Taq polymerase (Takara, Dalian, China), and 80 ng template DNA. Amplification was performed on a PTC-225 machine (MJ Research, Waltham, MA) using the following conditions: 94 °C for 3 min, 30 cycles of 30s at 94 °C, 30s at 55 °C, and 30s at 72 °C, with a final 6 min extension at 72 °C. All the PCR products were electrophoresed in 8% non-denaturing polyacrylamide gels and visualized via silver staining [42].
Cloning and annotation of Sicl1 gene
To clone the gDNA and cDNA sequences of SiCL1 alleles in mutant cl1 and the wild genotype, the primer pairs were designed using Primer Premier 5.0 (Additional file 5: Table S5, Additional file 7: Table S6). Total RNA was extracted from tissues using Trizol Reagent (Ambion, Life Technologies, USA) following the manufacturer’s instructions. First-strand cDNA synthesis was performed using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Germany) according to the manufacturer’s instruction. PCR was performed in a 50 μL reaction mixture containing 1 × Buffer, 2.0 mmol·L− 1 MgCl2, 0.1 mmol·L− 1 dNTPs, 1 μmol·L− 1 of each primer, 1.0 U Taq polymerase and 100~ 200 ng template DNA or 5 × diluting cDNA. Standard PCR reactions were carried out on a Eppendorf Mastercycler (Eppendorf, Germany) under the following conditions: 94 °C for 4 min; 35 cycles of 30s at 94 °C, 2 min at 55–58 °C and 1 min at 72 °C, with a final 8 min extension at 72 °C. PCR products were individually gel purified for Sanger sequencing. Non- redundant (NR) protein and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations for candidate genes in the interval were obtained using BLASTP and BLAST2GO, respectively (http://www.genome.jp/kegg/pathway.html).
Sicl1 homolog detection and phylogenetic analysis
BLASTN was applied to screen SiCL1 homolog(s) in Yuzhi 11 reference genome. Gene alleles in sesame accessions were screened using MEGA 5.2 according to the Muscle method [43].
The amino acid sequences of SiCL1 and Sicl1 were aligned with the homologs in Arabidopsis thaliana, Zea mays, Oryza sativa, Vitis vinifera, Manihot esculenta, Nicotiana tabacum, Hevea brasiliensis, Ipomoea nil, Coffea canephora, and Solanum tuberosum, respectively, using DNAMAN (http://www.lynnon.com/pc/framepc.html). All the above homologs information was downloaded from NCBI dataset. A neighbor joining phylogenetic tree was constructed based on the above orthologs using the MEGA 5.2 program [43].
RNA extraction and expression profile assay of SiCL1 allele
Total RNA was extracted from tissues using Trizol Reagent (Ambion, Life Technologies, USA) following the manufacturer’s instructions. The primer pairs for quantitative real time PCR (qRT-PCR) analyses were designed with Primer Premier 5.0 program (http://www.premierbiosoft.com/products/products.html). The forward sequence (RT-CL1_F) was 5′CCTAACCCTCCATTCCCATT 3′ (Tm = 58.0 °C), and the reverse sequence (RT-CL1_R) was 5′GATACGACGACAGCCCACTAA 3′ (Tm = 57.8 °C). The amplicon size was 271 bp. Real-time PCR reaction was performed on a Mastercycler realplex (Eppendorf, Germany). The sesame β-tubulin gene was used as an endogenous reference gene [5]. Transcript amount of target genes was normalized against the β-tubulin gene and analyzed using ∆∆Ct method according to Wei et al. [44].
Abbreviations
- AFLP:
-
Amplified fragment length polymorphisms
- BSA:
-
Bulked segregation analysis
- CL:
-
Curly leaf
- DAP:
-
Day after pollination
- FAA:
-
Formalin-glacial acetic acid-alcohol
- GWAS:
-
Genome wide association study
- InDel:
-
Insertion-deletion
- LG:
-
Linkage group
- PC (ab):
-
Abaxial parenchyma cell
- PC (ad):
-
Adaxial parenchyma cell
- QTL:
-
Quantitative trait loci
- SNP:
-
Single nucleotide polymorphism
References
Ashri A. Sesame breeding. In: Janick J, editor. Plant Breeding Reviews, vol. 16. Israel: john Wiley & sons; 1998. p. 179–228.
Langham DR, Wiemers T. Progress in mechanizing sesame in the US through breeding. In: Juanick J, Whipkey A, editors. Trends in new crops and new uses. Alexandria: ASHS Press; 2002. p. 157–73.
Zhang H, Miao H, Wang L, Qu L, Liu H, Wang Q, Yue M. Genome sequencing of the important oilseed crop Sesamum indicum L. Genome Biol. 2013;14(1):401.
Wu K, Liu H, Yang M, Tao Y, Ma H, Wu W, Zuo Y, Zhao Y. High-density genetic map construction and QTLs analysis of grain yield-related traits in sesame (Sesamum indicum L.) based on RAD-seq techonology. BMC Plant Biol. 2014;14(1):274.
Zhang H, Miao H, Li C, Wei L, Duan Y, Ma Q, Kong J, Xu F, Chang S. Ultra-dense SNP genetic map construction and identification of SiDt gene controlling the determinate growth habit in Sesamum indicum L. Sci Rep. 2016;6:31556.
Wei X, Zhu X, Yu J, Wang L, Zhang Y, Li D, Zhou R, Zhang X. Identification of sesame genomic variations from genome comparison of landrace and variety. Front Plant Sci. 2016;7:1169.
Stewart DW, Costa C, Dwyer LM, Smith DL, Hamilton RI, Ma BL. Canopy structure, light interception, and photosynthesis in maize. Agron J. 2003;95(6):1465–74.
Langham DG. Genetics of sesame III. Open sesame and mottled leaf. J Hered. 1946;37:149–52.
Langham DG, Rodriguez M. Abrete sesamo: Ajonjoli (Sesamum indicum) que no pierde semillas. Circ. 1946;17:153–9.
Langham DR. Shatter resistance in sesame. In: L. Van Zanten (ed.), Sesame improvements by induced mutations, Proc. Final FAO/IAEA Co-ord. Res. Mtng, IAEA-TECDOC-1195, 2001, pp:51–61.
Diouf M, Boureima S, Diop T, ÇAğIrgan MI. Gamma rays-induced mutant spectrum and frequency in sesame. Turk J Field Crops. 2010;15(1):99–105.
Uzun B, Lee D, Donini P, Cagirgan MI. Identification of a molecular marker linked to the closed capsule mutant trait in sesame using AFLP. Plant Breed. 2003;122(1):95–7.
Zhang H, Miao H, Wei L, Li C, Zhao R, Wang C. Genetic analysis and QTL mapping of seed coat color in sesame (Sesamum indicum L.). PLoS ONE. 2013;8(5):e63898.
Li C, Miao H, Wei L, Zhang T, Han X, Zhang H. Association mapping of seed oil and protein in Sesamum indicum L. using SSR Markers. PLoS ONE. 2014;9(8):e105757.
Wei X, Liu K, Zhang Y, Feng Q, Wang L, Zhao Y, Li D, Zhao Q, Zhu X, Zhu X, Li W, Fan D, Gao Y, Lu Y, Zhang X, Tang X, Zhou C, Zhu C, Liu L, Zhong R, Tian Q, Wen Z, Weng Q, Han B, Huang X, Zhang X. Genetic discovery for oil production and quality in sesame. Nat Commun. 2015;6:8609.
Wang L, Xia Q, Zhang Y, Zhu X, Zhu X, Li D, Ni X, Gao Y, Xiang H, Wei X, Yu J, Quan Z, Zhang X. Updated sesame genome assembly and fine mapping of plant height and seed coat color QTLs using a new high-density genetic map. BMC Genomics. 2016;17(1):31.
Mei H, Liu Y, Du Z, Wu K, Cui C, Jiang X, Zhang H, Zheng Y. High-density genetic map construction and gene mapping of basal branching habit and flowers per leaf axil in sesame. Front Plant Sci. 2017;8:636.
Dossa K, Diouf D, Wang L, Wei X, Zhang Y, Niang M, Fonceka D, Yu J, Mmadi MA, Yehouess LW, Liao B, Zhang X, Cisse N. The emerging oilseed crop Sesamum indicum enters the “omics” era. Front Plant Sci. 2017;8:1154.
Deborah DA, Vemireddy LR, Roja V, Patil S, Choudhary GP, Noor S, Srividhya A, Kaliappan A, Rani BS, Satyavathi VV, Anuradha G, Radhika K, Yamini KN, Kadambari GM, Nagireddy RK, Siddiq EA, Nagaraju J. Molecular dissection of QTL governing grain size traits employing association and linkage mapping in basmati rice. Mol Breeding. 2017;37(6):77.
Zhao Y, Wang H, Chen W, Li Y. Genetic structure, linkage disequilibrium and association mapping of Verticillium wilt resistance in elite cotton (Gossypium hirsutum L.) germplasm population. PLoS ONE. 2014;9(1):e86308.
Shi J, Zhan J, Yang Y, Ye J, Huang S, Li R, Wang X, Liu G, Wang H. Linkage and regional association analysis reveal two new tightly-linked major-QTLs for pod number and seed number per pod in rapeseed (Brassica napus L.). Sci Rep. 2015;5:14481.
Cadic E, Coque M, Vear F, Grezes-Besset B, Pauquet J, Piquemal J, Lippi Y, Blanchard P, Romestant M, Pouilly N, Rengel D, Gouzy J, Langlade N, Mangin B, Vincourt P. Combined linkage and association mapping of flowering time in sunflower (Helianthus annuus, L.). Theor Appl Genet. 2013;126(5):1337–56.
Korir PC, Zhang J, Wu K, Zhao T, Gai J. Association mapping combined with linkage analysis for aluminum tolerance among soybean cultivars released in yellow and Changjiang River valleys in China. Theor Appl Genet. 2013;126(6):1659.
Zhao X, Han Y, Li Y, Liu D, Sun M, Zhao Y, Lv C, Li D, Yang Z, Huang L, Teng W, Qiu L, Zheng H, Li W. Loci and candidate gene identification for resistance to Sclerotinia sclerotiorum in soybean (Glycine max L. Merr.) via association and linkage maps. Plant J. 2015;82(2):245–55.
Zhao R, Miao H, Song W, Chen C, Zhang H. Identification of sesame (Sesamum indicum L.) chromosomes using the BAC-FISH system. Plant Biol. 2018;20(1):85–92.
Sim SC, Durstewitz G, PlieskeJ WR, Ganal MW, Deynze AV, Hamilton JP, Buell CR, Causse M, Wijeratne S, Francis DM. Development of a large SNP genotyping array and generation of high-density genetic maps in tomato. PLoS ONE. 2012;7(7):e40563.
Wang H, Zhang H, Ma Q, Wei L, Ju M, Li C, Duan Y, Miao H. Optimization of EMS mutagenesis condition and screening of mutants in sesame. J Henan Agri Sci. 2017;46(1):36–41.
Wang S, Chen J, Zhang W, Hu Y, Chang L, Fang L, Wang Q, Lv F, Wu H, Si Z, Chen S, Cai C, Zhu X, Zhou B, Guo W, Zhang T. Sequence-based ultra-dense genetic and physical maps reveal structural variations of allopolyploid cotton genomes. Genome Biol. 2015;16(1):108.
Verma P, Goyal R, Chahota RK, Sharma TR, Abdin MZ, Bhatia S. Construction of a genetic linkage map and identification of QTLs for seed weight and seed size traits in lentil (Lens culinaris Medik.). PLoS ONE. 2015;10(10):e0139666.
Pagny G, Paulstephenraj PS, Poque S, Sicard O, Cosson P, Eyquard J, Caballero M, Chague A, Gourdon G, Negrel L, Candresse T, Mariette S, Decroocq V. Family-based linkage and association mapping reveals novel genes affecting Plum pox virus infection in Arabidopsis thaliana. New Phytol. 2012;196(3):873–86.
Li N, Shi J, Wang X, Liu G, Wang H. A combined linkage and regional association mapping validation and fine mapping of two major pleiotropic QTLs for seed weight and silique length in rapeseed (Brassica napus L.). BMC Plant Biol. 2014;14(1):1–14.
Carlson CS, Eberle MA, Kruglyak L, Nickerson DA. Mapping complex disease loci in whole-genome association studies. Nature. 2004;429(6990):446–52.
Xiang JJ, Zhang GH, Qian Q, Xue HW. SEMI-ROLLED LEAF1 encodes a putative glycosylphosphatidylinositol-anchored protein and modulates rice LEAF rolling by regulating the formation of bulliform cells. Plant Physiol. 2012;159(4):1488–500.
Liu X, Ming L, Kai L, Ding T, Sun M, Li Y, Shen Y, Du G, Cheng Z. Semi-rolled leaf2 modulates rice leaf rolling by regulating abaxial side cell differentiation. J Exp Bot. 2016;67(8):2139–50.
Zhao S, Zhao L, Liu F, Wu Y, Zhu Z, Sun C, Tan L. NARROW AND ROLLED LEAF 2 regulates LEAF shape, male fertility, and seed size in rice. J Integr Plant Biol. 2016;58(12):983–96.
Zhang GH, Xu Q, Zhu XD, Qian Q, Xue HW. SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell. 2009;21(3):719–35.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20.
Miao H, Zhang H. The Genome of Sesamum indicum L. International Plant and Animal Genome Conference XXIV, 2016, January, 7-13th. San Diego, USA.
Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinfomatics. 2009;25(14):1754–60.
Poplin R, Ruano-Rubio V, DePristo MA, Fennell TJ, Carneiro MO, Van der Auwera GA, Kling DE, Gauthier LD, Levy-Moonshine A, Roazen D, Shakir K, Thibault J, Chandran S, Whelan C, Lek M, Gabriel S, Daly MJ, Neale B, MacArthur DG, Banks E. Scaling accurate genetic variant discovery to tens of thousands of samples. BioRxiv. 2017. https://doi.org/10.1101/201178.
Wei L, Miao H, Li C, Duan Y, Niu J, Zhang T, Zhao Q, Zhang H. Development of SNP and InDel markers via de novo transcriptome assembly in Sesamum indicum L. Mol Breeding. 2014;34(4):2205–17.
Liang H, Wang C, Li Z, Luo XZ, Zou G. Improvement of the silver-stained technique of polyacrylamide gel electrophoresis. Hereditas. 2008;30(10):1379–82.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9.
Wei L, Miao H, Zhao R, Han X, Zhang T, Zhang H. Identification and testing of reference genes for sesame gene expression analysis by quantitative real-time PCR. Planta. 2013;237(3):873–89.
Acknowledgements
We would like to thank Prof. Derald Langham and Prof. Ray Langham of the United States for their kind donation of the curly leaf mutant (cl1 type) to Henan Sesame Research Center, Henan Academy of Agricultural Sciences in 1980’s.
Funding
This work was financially supported by the earmarked fund for China Agriculture Research System (CARS-14), the National Natural Science Foundation of China (31471537), the Plan for Scientific Innovation Talent of Henan Province (184200510002), the Key Project of Science and Technology in Henan Province (151100111200), the Importing International Agricultural Sciences and Technology Program (2016-X05), the Henan Province Specific Professor Position Program (SPPP2016), the Distinguished Professor Program of Institutions of Higher Learning in Henan Province (DPPIHL2017), and the Innovation Scientists and Technicians Troop Construction Projects of Henan Province (ISTTCPHP2016).
Availability of data and materials
The cDNA sequence of Sicl1 gene has been submitted to NCBI dataset (NCBI accession no. MG763174). The BSA data of cl1 has been submitted to NCBI dataset under the BioProject PRJNA433597 with SRA accession number SRR6704961.
Author information
Authors and Affiliations
Contributions
ZH conceived and designed the experiments, and guided manuscript correction for publishing. MH guided the experiments, performed the data analysis and drafted the manuscript. WL performed the genetic experiments and drafted the manuscript. LC and DY conducted the main experiments and participated in the data analysis. XF conducted the cross-section. QW and CS participated in the genetic experiments, molecular markers screening and gene cloning. ZR and JM participated in the manuscript correction and data analysis. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing financial interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1 Genomic variants screening for the curly leaf trait using genome variants data of sesame accessions. (XLSX 14 kb)
Additional file 2:
Table S2. Variants of the candidate interval C29 linked to the leaf curling trait in 822 sesame accessions with normal leaf shape and a cl1 mutant line (www.sesamum.org). (XLSX 14 kb)
Additional file 3:
Table S3. Genomic variants screening for the curly leaf trait in sesame using BSA pool data. (XLSX 50 kb)
Additional file 4:
Table S4 Genome information of the six candidate variants for the curly leaf trait. *F refers to forward direction. R refers to reverse direction. - indicates that no genes are found in the 10 Kb flanking region of the variant site. **KEGG refers to Kyoto Encyclopedia of Genes and Genomes. C29.460 is the retained gene linked to the curly leaf trait in the interval C29 in this study. (XLSX 11 kb)
Additional file 5:
Table S5. Information of primer pairs of the candidate markers for curly leaf trait. (XLSX 10 kb)
Additional file 6:
Figure S1. Amplification of the SiCLInDel 1 marker in the test F2 population and sesame germplasm. M: DNA marker; Lane 1–10: F2 individuals with curly leaf phenotype; Lane 11–20: F2 individuals with normal leaf phenotype; Lane 21–40: sesame germplasm materials (M1-M20) with normal leaf phenotype. (TIF 396 kb)
Additional file 7:
Table S6. Information of primer pairs of the cDNA sequences of SiCL1 alleles in sesame. Genome information of the six candidate variants for the curly leaf trait. Note: The italic letters in the sequences are the 5′ and 3′ UTR regions of SiCL1 alleles. (XLSX 13 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhang, H., Miao, H., Wei, L. et al. Identification of a SiCL1 gene controlling leaf curling and capsule indehiscence in sesame via cross-population association mapping and genomic variants screening. BMC Plant Biol 18, 296 (2018). https://doi.org/10.1186/s12870-018-1503-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-018-1503-2