Abstract
Background
Histone methylation and acetylation regulate biological processes in plants through various histone modifications (HMs) gene families. However, knowledge of HMs genes is limited in horticultural deciduous trees, including apple (Malus domestica).
Results
Here, a comprehensive study of identifying and investigating HMs genes was performed using the recently published apple genome. In total, 198 MdHMs were identified, including 71 histone methyltransferases, 44 histone demethylases, 57 histone acetylases, and 26 histone deacetylases. Detailed analysis of the MdHMs, including chromosomes locations, gene structures, protein motif and protein-protein interactions were performed, and their orthologous genes were also predicted against nine plant species. Meanwhile, a syntenic analysis revealed that tandem, segmental, and whole genome duplications were involved in the evolution and expansion of the MdHMs gene family. Most MdHMs underwent purifying selection. The expression profiles of 198 MdHMs were investigated in response to 6-BA treatment and different flowering varieties (easy-flowering ‘Yanfu No.6’ and difficult-flowering ‘Nagafu No.2’) using transcriptome sequencing data, and most MdHMs were involved in flower induction processes. Subsequent quantitative real-time PCR was then performed to confirm the expression levels of candidate MdHMs under different flowering-related circumstances.
Conclusion
MdHMs were involved in, and responsive to, flower induction in apple. This study established an MdHMs platform that provided valuable information and presented enriched biological theories on flower induction in apple. The data could also be used to study the evolutionary history and functional prospects of MdHMs genes, as well as other trees.
Similar content being viewed by others
Background
Histone modifications (HMs), which repressed or promoted gene expression, affected various processes and played important roles during plant growth and development. Methylation, demethylation, acetylation and deacetylation were common histone modifications processes. These modifications depended on four different HMs gene family members, including histone methyltransferases (HMTs), histone demethylases (HDMs), histone acetylases (HATs), and histone deacetylases (HDACs). Similarly, HMTs, HDMs, HATs and HDACs regulated various biological processes in plants [1, 2].
These four gene family contained different subfamilies. HMTs family included two subfamilies, and they were SDG (set domain group) and PRMT (protein arginine methyltransferases). HDMs family also included two subfamilies, HDMA (SWIRM and C-terminal domain) and JMJ (JmjC domain-containing proteins). As for HATs family, four kinds of subfamilies (HAG, HAM, HAC and HAF) were contained. I): HAG types included GCN5-, ELP3-, and HAT1-like histone acetylases domain structure; II): HAM types included a MOZ-YBF2 domain; III): HAC types included a p300/CREB-binding protein structure; IV): HAF types included a TATA binding protein-associated factors TAFII250. HDACs family shared three subfamilies, including HAD (RPD3/HDA1 superfamily), SRT (silent information regulator 2) and HDT (HD2 families) [3,4,5]. Totally, each subfamily contained typical domain or structure.
Apart from their different structures, the number of HMs genes was also different in plants. A total of 136 HMs (47 HMTS, 23 HDMs, 50 HAT, and 16 HDACs) have been identified in sweet orange, and they played important roles in fruit development [6]. Additionally, 125 HMs (32 HATs, 15 HDACs, 52 HMTs and 26 HDMs) were also identified from tomato genome [7]. In total, 35 SDGs members have been identified in the grape genome and some were up-regulated during grape softening [3]. Meanwhile, HMs gene functions were partially characterized, especially in the model plant Arabidopsis. They played important roles in plant growth and development, including in photomorphogenesis, seed germination and dormancy, embryo development, flowering-related processes, fruit development, stress and defense, and hormonal signaling [8,9,10,11,12,13,14,15]. HMs can directly function in regulating flowering through their over expression or down expression. They can also affect the expression of flowering related genes. For example, an Arabidopsis thaliana HDA family member, AtHDA9 (AT3G44680), repressed flowering by affecting the acetylation of AGAMOUS-LIKE 19 (AGL19) [16]. Additionally, AtHDA19 (AT4G38130) influenced flower development together with A-class organ identity gene AP2 (APETALA2), similar as AtHDA6 (AT5G63110), which showed late flowering in the HDA6-RNAi plants [17, 18]. Other genes, such as AtHAM1 (At5g64610), AtHAM2 (At5g09740), AtHAC1 (At1g79000) were also responsible for flowering time [19,20,21]. For example, the artificial microRNA AtHAM1 and AtHAM2 showed earlier flowering time, while overexpression AtHAM1 flowered later and had more rosette leaves [20]. In tomato (Solanum lycopersicum), SlHAG22, SlHAG8 and SlHAG18 were involved in vegetative or reproductive development, and SlSRT2 participated in flowering [7]. Additionally, HM genes can also regulate the expression level of flowering-related genes, such as FLOWERING LOCUS C (FLC), LEAFY (LFY), MADS AFFECTING FLOWERING4 (MAF4) and (MAF5) [20, 22, 23]. For example, the over-expression of HAM1 resulted in a higher H4 hyperacetylation and H4K5ac at FLC in Arabidopsis [20]. In Arabidopsis, an enriched level of histone H3 acetylation and H3K4 trimethylation at FLC and MAFs occurred in the histone deacetylase6 mutant (had6) [23, 24]. Meanwhile, FLOWERING LOCUS T (FT) was also influenced by HMs. The Arabidopsis JmjC family protein T-DNA insertion mutant lines (atjmj4, AT4G2040), showed earlier flowering, which might enrich FT mRNA and H3K4me3 levels within FT chromatin [25]. Among various flowering related genes, FLC and FT were the main well researched genes that associated with HMs [25,26,27]. These indicated that HMs affect or interact with their downstream or upstream flowering genes to control flowering.
Apple (Malus domestica) is an economically important fruit tree in temperate regions worldwide, and flower induction is an important issue, which restricted fruit yield and economic incomes [28,29,30]. Hormones mediated flower induction, with GA (gibberellin) inhibiting flowering and 6BA (6-benzylaminopurine) or sugar promoting flowering, has been characterized and researched in apple [31,32,33]. Additionally, some important gene families, including INDETERMINATE DOMAIN (IDD), SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL), MADs-box, and GIBBERELLIC ACID STIMULATED ARABIDOPSIS (GASA), have also been well identified and reported to regulate flower induction in apple [33,34,35,36,37]. However, less is known about of HMs and their potential involvement in apple flower induction. In 2017, with the publication of a new apple genome [38], it is able for us to systematically identify HMs gene family in apple and help us to make a comprehensive investigation about their characterizations and potential response to flower induction.
In this study, we identified 198 HMs gene members in the apple genome. They were 71 MdHMTs (64 MdSDGs and 7 MdPRMTs), 44 MdHDMs (16 MdHDMAs and 28 MdJMJs), 57 HATs (50 MdHAGs, 2 MdHAMs, 4 MdHACs, and 1 MdHAF) and 26 MdHDACs (16 MdHDAs, 3MdSRTs and 7 MdHDTs). Additionally, their chromosomes locations, gene and protein structures, gene phylogeny, synteny analysis and protein-protein interaction network were also performed. Meanwhile, transcriptomic sequencing of 6BA treated trees and different flowering varieties (Nagafu No.2 and Yanfu No.6) were performed to investigate their potential involvement during apple flower induction. Furthermore, quantitative real-time PCR (qRT-PCR) was employed to investigate the expression levels of candidate MdHMs in different tissues (stem, leaf, flower, fruit and bud) and different flowering circumstances (alternate bearing and sugar-treated trees), and various hormones (GA3, SA, ABA and MeJA) stress treatment. The results revealed valuable information of HMs genes in apple, which might be applicable to other fruit trees.
Methods
Identification and chromosomes location of HMs gene family in apple
To identify HMs gene family members in the apple genome, a HMM file of each domain was obtained from Pfam database (http://pfam.sanger.ac.uk/), as previous studies [6, 7]. These files were then used as a query to search against the apple genome (GDDH13 V1.1) with HMMER3.0 [39]. The detailed accession number of each file was listed as Additional file 1: Table S1. However, there was no available HDT in the Pfam database. Thus, the protein sequences encoded by four Arabidopsis HDT genes, AtHDT1 (At3g44750), AtHDT2 (At5g22650), AtHDT3 (At5g03740), and AtHDT4 (At2g27840), were downloaded from the TAIR database (The Arabidopsis Information Resource; http://www.arabidopsis.org/) and used as a query to search against the Genome Database for Rosaceae [apple genome (GDDH13 V1.1; https://www.rosaceae.org/] to predict candidate MdHDTs family members. Finally, the putative HMs genes, including HMTs (SDGs and PRMTs), HDMs (HDMAs and JMJs), HATs (HAGs, HAMs, HACs, and HAFs) and HDACs (HDAs, SRTs and HDTs) were manually checked to confirm their highly conserved segments. The relative locations of HMs were obtained from the apple genome [38]. They were then named according to their chromosome orders, as previous study [6].
Phylogenetic tree construction, gene structure, protein motif and domain, and orthologous genes analysis
For phylogenetic analysis, MEGA 7.0 [40] was used to investigate the phylogenetic interactions of HMs between apple and Arabidopsis. The Arabidopsis and apple HMs protein sequences were aligned by the ClustalW program with default parameters. The multiple sequence alignment generated files were analyzed and then used to build phylogenetic trees with Maximum Likelihood method, pairwise deletion for sequences analysis, and a bootstrap value of 1000 times. For the gene structural analysis, gene models were downloaded from apple genome database (https://iris.angers.inra.fr/gddh13/) [38]. They were then submitted into Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/) for structural analyses [41]. Additionally, the HMs protein sequences were employed to investigate conserved motifs with MEME Suite platform (http://meme-suite.org/), and 10 motifs were found within each gene family. Protein-protein interactions were analyzed with http://string-db.org/. For orthologous genes identification, each pair of gene was used to BLAST with sequences homology more than 60% and e-value less than 1e-20. Gene Ontology (GO) terms analysis were performed with online database (http://www.geneontology.org).
Tandem duplication and synteny analysis
Circos v. 0.63 (http://circos.ca/) was employed to investigate their tandem duplication and synteny relationships as previous methods [33,34,35,36,37]. Tandem duplication MdHMs genes were identified according to their physical locations on individual chromosomes in the apple genome. Adjacent homologous HMs genes on the same apple chromosome with no more than one intervening gene were considered tandem duplicates. The Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/) was used to identify synteny blocks between Arabidopsis and apple. For orthologous gene pairs, synonymous (Ks) and non-synonymous (Ka) nucleotide substitutions were calculated according to the comparative synteny map between the apple and Arabidopsis thaliana genomes, with ClustalX and PAL2NAL programs for protein and coding sequences alignment. They were calculated with DNASP v5.10 software.
Plant materials and treatment
Trees with different flowering capabilities
In total, 18 six-year-old apple trees of contrast flowering varieties (Yanfu No.6 and Nagafu No.2), which had been planted at the Apple Demonstration Nursery of Yangling Modern Agriculture Technology Park (108°70′E, 34°52’N), were used in this study. They were all grafted T337/ Malus robusta Rehd. Additionally, ‘Yanfu No.6’, a spur mutation of ‘Nagafu No.2’, had a higher flowering rate and greater bud morphological development [34, 35]. Trees were divided into three blocks, with three of each, respectively. Terminal buds were collected from current spurs at 30, 50 and 70 days after full bloom (DAFB) [29, 35]. They were then stored for further use.
Sugar and hormones treated apple trees
An additional 18 uniform ‘Fuji’/ T337/ Malus robusta Rehd were used for sucrose treatment experiments in the same orchard. They were also divided into six blocks. Three of them were sprayed with 15,000 and 20,000 mg L− 1 sucrose at 29 and 36 DAFB [32], and the remaining blocks were sprayed with water as control. For 6BA treatment, 18 similar trees were used, and 300 mg L− 1 6BA was performed at 27 and 30 DAFB. They were all sprayed on the whole trees with a low-pressure hand wand sprayer in a clear morning. Samples were collected at 30, 50 and 70 DAFB and stored for further use. Meanwhile, 100 mM GA3, 300 μM ABA, 100 μM SA and 50 μM MeJA were treated on 2-year-old ‘Nagafu No.2’ trees, as water as control; leaves were collected at 0, 3, 6, and 12 h for each treatment as previous study [36].
Alternate bearing trees
Six14-year-old alternate bearing ‘Fuji’ trees were used in Tiandu Town, Fufeng, Baoji, Shaanxi (107°57′ E, 34°28′ N) were sampled. Samples were collected from trees in their ‘ON’ years (with a higher flowering rate) and ‘OFF’ years (with a low flowering rate) in 2014 at 30, 90, and 150 DAFB in the morning. Terminal buds of current spurs were collected from trees in their ‘ON’ or ‘OFF’ years and stored for further use.
Tissues collection
For the tissue-specific expression analyses, various tissues or organs were collected from ‘Fuji’/T337 M. robusta Rehd. Flowers were collected on April 9, 2015 during the full-bloom period. Additionally, stems were collected from new shoots with diameters of 2–3 mm, while mature leaves were collected from the adjacent buds. Fruits with diameters of 2–3 cm were also collected. All samples were immediately frozen in liquid nitrogen and stored at − 80 °C until used in the gene expression analyses.
RNA extraction and cDNA synthesis
Total RNA was extracted from plant tissue samples using the cetyltrimethyl ammonium bromide method with slight modifications [42]. Briefly, 900 μL extraction buffer (2% cetyltrimethyl ammonium bromide, 2.5% PVP-40, 2 M NaCl, 100 mM Tris-HCl [pH 8.0], 25 mM EDTA [pH 8.0], and 2% b-mercaptoethanol) was preheated at 65 °C and added to 2-mL microcentrifuge tubes just before use. Samples containing 200 mg of bud tissue stored at − 80 °C were ground to a powder, added to the tubes, and mixed with extraction buffer. After shaking and inverting each tube vigorously for 5 min and incubating at 65 °C for 30 min, an equal volume of chloroform:isoamyl alcohol (24:1, v/v) was added. Each tube was shaken and inverted vigorously and then centrifuged at 12,000×g for 10 min at 4 °C. For each sample, the supernatant was collected into a new tube and re-extracted with an equal volume of chloroform:isoamyl alcohol (24:1, v/v). The resulting supernatant was then transferred into a new 2-mL tube and LiCl (3 M final concentration) was added. The mixture was incubated at − 20 °C for 4 h and the RNA was selectively pelleted by LiCl after centrifugation at 18,000×g for 20 min at 4 °C. The pellet was resuspended in 500 μL of SSTE buffer (10mMTris-HCl [pH 8.0], 1 mM EDTA [pH 8.0], 1% SDS, and 1 M NaCl) that had been preheated to 65 °C and an equal volume of chloroform:isoamyl alcohol. The mixture was then centrifuged at 12,000×g for 10 min at 4 °C. The supernatant was transferred to a new microcentrifuge tube, and the RNA was precipitated with 2.5 volumes of cold ethanol at − 80 °C for at least 30 min and centrifuged at 1,2000×g for 20 min at 4 °C. Finally, the pellets were washed with 70% ethanol and resuspended in diethylpyrocarbonate-treated water. Total RNA integrity levels were verified by running the samples on 2% agarose gels. First-strand cDNA was synthesized from 1 μg of total RNA using a PrimeScript RT Reagent kit with gDNA Eraser (Takara Bio, Shiga, Japan) following the manufacturer’s instructions.
Gene expression analysis
The expression levels of the 12 candidate HM genes were analyzed using quantitative real-time PCR (qRT-PCR). Primers were designed to span an intron-exon junction with Primer Premier 6.0 software. And they were designed with the preferred values to specific amplification with high yield as follows. (1) Length of PCR primers (18–24 bp); (2) Melting temperature (60 °C); (3) GC content (40–60%); (4) GC Clamp: more than 3 G’s or C’s should be avoided in the last 5 bases at the 3′ end of the primer. (5) Avoided hairpins, self and cross dimer, and repeats. (6) Avoid template secondary structure and cross homology. The qRT-PCR mix (20 μL) consisted of 2-μL cDNA samples (diluted 1:8), 10 μL 2× SYBR Premix ExTaq II (Takara Bio), 0.8 μL of each primer (10 μM) (Additional file 2: Table S2), and 6.4 μL distilled deionized H2O. Each PCR assay was run on an iCycler iQ5 Real Time PCR Detection System (Bio-Rad, Plano, TX, USA) with an initial denaturation at 95 °C for 3 min, followed by 40 cycles at 94 °C for 15 s, 62 °C for 20 s, and 72 °C for 20 s. The resulting fragments were subjected to melting-curve analysis to verify the presence of gene-specific PCR products. The melting curve analysis was performed directly after real-time PCR and included an initial step of 94 °C for 15 s, followed by a constant increase from 60 °C to 95 °C at a 2% ramp rate. The apple EF-1α gene (GenBank accession No. DQ341381) was used as an internal control to normalize all mRNA expression levels under different treatments and different tissues [34,35,36]. Experiments were performed using three biological replicates with three technical replicates. The 2−ΔΔCt method was used to calculate the relative amount of template present in each PCR amplification mixture [43].
Statistical analysis
Gene expression data of RT-qPCR were subjected to an analysis of variance (ANOVA) at the 5% level with the SPSS 11.5 software package (SPSS, Chicago, IL, USA). Figures were constructed using Origin 8.0 (Microcal Software Inc., Northampton, MA, USA).
Results
Genome-wide identification of HMs gene family in the apple genome
In total, 198 HMs were identified in the apple genome, including 71 HMTs, 44 HDMs, 57 HATs and 26 HDACs (Figs. 1a, 2a). All of the MdHMs were classified into 11 subfamilies according to their different protein domain. For example, the HMTs included 64 SDGs and 7 PRMTs, the 44 HDMs included 16 HDMAs and 28 JMJs, the 57 HATs included 50 HAGs, 2 HAMs, 4 HACs, and 1 HAF gene, and the 26 HDACs included 16 HADs, 3 SRTs, and 7 HDTs. Meanwhile, a detailed GO annotation was provided for all the HMs (Additional file 3: Table S3). These HMs genes were divided into three categories, including biological process, cellular component and molecular function.
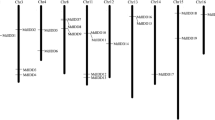
Chromosomes locations of MdHMs gene family on the apple genome. a Apple histone methyltransferase genes MdHMTs (MdSDGs and MdPRMTs) and histone demethylase genes MdHDMs (MdHMAs and MdJMJs); b Apple histone acetyltransferase genes MdHATs (MdHAGs, MdHAMs, MdHACs, and MdHAFs) and histone deacetylase genes MdHDACs (MdHDAs, MdSRTs, and MdHDTs)
Comparison the number of HMs genes among different species and different chromosomes. a Analysis of HMs members within six different species; Md, Malus domesctia; Sl, Solanum lycopersicum; Cs, Citrus sinensis; At, Arabidopsis thaliana; Os, Oryza sativa; Zm, Zea mays. b Comparison of different gene family members on apple genomes; each gene family was present with different color
Chromosome distributed of different HMs
To clearly identify the HMs, each of the HMs were named based on their chromosomal locations (Fig. 1, Table 1), as MdSDGs, MdPRMTs, MdHDMAs, MdJMJs, MdHAGs, MdHAMs, MdHACs, MdHAFs, MdHDAs, MdSRTs, and MdHDTs. All genes were distributed from chromosome 00 to chromosome 17 on the apple genome. Chromosome 15 contained the greatest number of HMs (Fig. 2b), followed by chromosome 12. Chromosome 17 had the lowest number of HMs genes. The 64 MdSDGs genes were distributed on all chromosomes except chromosome 6. Chromosome 15 contained the greatest number of MdSDGs genes. Seven MdPRMTs genes were distributed on four chromosomes (chromosome 2, 8 13 and 15). Additionally, 16 MdHDMAs and 28 MdJMJs were widely distributed throughout the apple genome, except on chromosome 2, 11, 13 and 17. The 50 MdHAGs were distributed throughout 16 of the 18 chromosomes; however, chromosomes 0 and 17 lacked MdHAGs genes. The remaining groups of HATs, including MdHAMs, MdHACs, and MdHAFs, had partial distributions, similar to those of the MdHDACs.
Phylogenetic and synteny analysis of HMs genes between apple and Arabidopsis
To understand their evolutionary relationship among HMs genes, four rooted phylogenetic trees, including HMTs, HDMs, HATs and HDACs genes, were built with Arabidopsis and apple HMs proteins (Fig. 3). All Arabidopsis and apple HMs genes were classified and clustered into different trends. For HMTs, all the SDGs and PRMTs genes were clustered together, with an exception of AtPRMT16 (Fig. 3a). Additionally, HDMAs and JMJs were also clustered with each other (Fig. 3b). For HATs, three HAFs genes (AtHAF1, AtHAF2, and MdHAF01) were clustered and surrounded by HAGs gene members, and other HAMs and HACs were also tightly grouped with themselves (Fig. 3c). For HDACs, three subfamilies (HDAs, HDTs and SRTs) were also clustered (Fig. 3d).
To characterize the expansion patterns of the HMs in the apple genome, a diagram together with Circos algorithm was performed and generated to investigate the duplicated blocks within the apple genome. A total of 67 pairs of HMs were identified from 18 chromosomes (Fig. 4, Additional file 4: Table S4), including one pair of MdHAFs, MdSRTs and MdHAMs, two pairs of MdPRMTs and MdHACs, three pairs of MdHDTs, five pairs of MdHDAs and HDMAs, 10 pairs of MdJMJs, 16 pairs of MdHAGs and 20 pairs of MdSDGs. These paired duplicated genes were all located in different chromosomes, chromosome 15 contained the most HMs genes (Fig. 1). Chromosome 1 had the lowest gene number. Totally, these duplicated genomic regions contributed to expansion of MdHMs family.
Synteny of MdHMs genes in the apple genome. Colored lines with two connected genes within indicated syntenic regions. Detailed information was listed as Additional file 4: Table S4
Additionally, a syntenic map of MdHMs and AtHMs were also created to help better understand their potential evolutionary and functional relationships. As shown in (Fig. 5, Additional file 5: Table S5), 72 orthologous pairs of MdHMs and AtHMs were found in the apple and Arabidopsis genome, including two pairs of HACs, SRTs and HDTs, one pair of HAFs and HAMs, three pairs of HAGs and PRMTs, 13 pairs of HDMAs, 14 pairs of JMJs and 31 pairs of SDGs. The remaining HMs genes did not have ortholog pairs. In addition, to understand the divergence among the orthologous gene pairs of apple and Arabidopsis, the ratio of the non-synonymous to the synonymous substitution rate (Ka/Ks) was used to evaluate the selection pressure during duplication. The Ka and Ks value was smaller in apple than betwen Arabidopsis and apple. However, the Ka/Ks values between gene pairs in apple less than 1, which was similar to apple and Arabidopsis (Additional file 6: Figure S1). The average Ka/Ks ratio between gene pairs in apple was 0.267, which was larger than between gene pairs in apple and Arabidopsis (0.106).
Synteny of HMs genes in apple and Arabidopsis genome. Colored lines with two connected genes within indicated syntenic regions. Detailed information was listed as Additional file 5: Table S5
Structure analysis of MdHMs
As mentioned above and (Additional file 7: Figure S2) shown, different HMs genes had different typical domains. We then investigated the structures of the 11 kinds of HMs gene families to confirm the present of each domain in apple, and a random gene was selected and to analyze their DOMAIN structure (Fig. 6). The HMs proteins shared various structures, with MdSDG08 containing a PWWP, PHD, and SET, MdJMJs containing a JmjN, JmjC, zf-C5H FYRN, and FYRC, MdHDMAs containing a SWIRM, COG3942, and SWIRM-a, MdHAGs containing a NAT-SF, MdHAMs containing a M0Z-SAS, MdHACs containing a ZnF, PHD, HAT, and ZZ, MdHAF containing a DUF, MdHDA containing a HDAC, MdSRT containing a SIR2, MdHDT containing a lambda-1, and MdPRMTs containing a PRMT5. These identified structures were similar to those found previously in the HMs of Citrus and other plants, indicating their conserved evolution.
Gene structures and motifs also play important roles during gene evolution. Therefore, we performed detailed exon-intron structure and protein motif analysis for t seven candidate gene families. Seven individual phylogenetic trees (MdSDGs, MdPRMTs, MdHDMAs, MdJMJs, MdHAGs, MdHDAs and MdHDTs) were built based on protein sequences (Additional file 8: Figure S3, Additional file 9: Figure S4, Additional file 10: Figure S5, Additional file 11: Figure S6, Additional file 12: Figure S7, Additional file 13: Figure S8, Additional file 14: Figure S9 and Additional file 15: Table S6). As shown in Additional file 8: Figure S3, the proteins encoded by the SDGs gene family (MdSDG43, 57, 51, 50, 49, and 28), which shared similar structures, were closely clustered. Additionally, 13 MdSDGs family members (MdSDG25, 26, 29, 59, 45, 35, 19, 24, 14, 39, 47, 03, 61, 29, 43, and 57) shared the greatest number of motifs compared with the other MdSDGs proteins. In addition, MdPRMT05, 02, 07 and 01 also shared similar protein motifs (Additional file 9: Figure S4). MdHDMAs also showed conserved motifs. For example, MdHDMA03, 12, 07, 08, 13, 06, 11, 02, and 01 shared only motif 1, 6, and 7; while the remaining MdHDMAs proteins, except MdHDMA09, shared more motif. MdHDMA14 and MdHDMA9 were encoded by genes having similar structures (Additional file 10: Figure S5). Gene structures and protein motifs of the MdJMJs were similar to MdHDMAs. For example, 9 MdJMJs proteins (MdJMJ17, 08, 03, 22, 06, 10, 14, 09, and 20) shared similar motifs. In addition, their gene structures showed less variability, especially among closely connected genes (MdJMJ9 and MdJMJ20, MdJMJ27 and MdJMJ16, MdJMJ23 and MdJMJ7, and MdJMJ28 and MdJMJ11) (Additional file 11: Figure S6, Additional file 15: Table S6).
The structures of one HATs and two HDACs gene families were also determined. Among the MdHAGs gene family members, most of them shared only one CDS (Additional file 12: Figure S7). For example, MdHAG18, 19, 17, 08, 14, 34, 49, 38, 13, 33, and 01 contained a coherent CDS within their gene structures. Their motifs were also conserved among some closely related genes, as seen with other HMs. As for HDACs and HDTs, they also had similar gene structures and encoded proteins with similar motifs, such as MdHDA15 and MdHDA09, MdHDA10 and MdHDA16, MdHDT01 and MdHDT03, and MdHDT02 and MdHDT04 (Additional files 13-14: Figure S8 and S9).
Analysis of HMs orthologous genes against in other species
BLASTP algorithm was employed to identify MdHMs orthologous genes with other sequenced plant species, and they were identified with e-value lower than 1e-20 and sequence homology more than 60%, as previous reported methods [44]. The 10 candidate plants used were including Arabidopsis, Zea may, Solanum lycopersicum, Oryza sativa, Citrus, Vitis vinifera and Populus trichocarpa, as well as the Rosaceae plants Fragaria vesca, Prunus persica, and Pyrus sorotina. Their orthologous relationships were divided into three kinds [44]: a) genes that existed in apple and were absent from a given species; b) apple genes that had one to one orthologs in a given species; and c) apple genes that had homologs in a given species but not orthologs. As shown in Fig. 7, most MdHMs had homologous genes compared with the 10 candidate species. However, MdHAG16 and MdHAG24 had no homolog in V. vinifera, nor MdSDG23 in O. sativa.
Apple HMs genes orthology against with sequenced species. Blue, a one to one ortholog HM in the candidate species; Gray, MdHMs has orthology in the candidate species but it was not one to one detected; White, no detected. At, Arabidopsis thaliana; Cs, Citrus sinensis; Fv, Fragaria vesca; Os, Oryza sativa; Pt, Populus trichocarpa; Pp, Prunus persica; Sl, Solanum lycopersicum; Vv, Vitis vinfera; Zm, Zea mays
Interactions prediction of MdHMs protein
To further predict their biological interactions, we visualized HM proteins with Cytoscape v3.5.1 [45, 46]. As shown in Additional file 16: Figure S10, Additional file 17: Table S7 members from four HM-related clusters, − HMTs, HDMs, HATs, and HDACs, directly or indirectly interacted with other proteins. Among them, the HMTs interacted with the greatest number of proteins (25), followed by the HDACs (11). The HDMs and HDACs only interacted with five and four proteins, respectively. Some proteins, such as MdSDG29, MdHAM02, MdJMJ01, MdJMJ25, MdHAG28, and MdSDG14, could also directly or indirectly interact with at least three kinds of proteins. Totally, MdHMs regulated downstream genes or were regulated by their up-regulated genes to participate in various processes.
Expression profiles of MdHMs with high-throughput sequencing
To better understand their potential involvement in responses to flower induction, we used published transcriptome data to evaluate the 198 MdHMs expression profiles [31]. FPKM (Fragments Per Kilobase of transcript sequence per Millions mapped reads) was calculated to assess gene expression levels. The resulting p value were then adjusted with Benjamini and Hochberg’s approach for controlling the false discovery rate, and a corrected P value of 0.05 and log2 value (fold change) of 1 were set as the criteria for identifying DEGs. [31]. Treatments with 6-BA increase the ratios of short shoots and result in higher flowering rates. Additionally, ‘Yanfu No.6’ has a higher flowering rate than ‘Nagafu No.2’ [31, 35]. We analyzed the candidate MdHMs expression levels in response to exogenous 6-BA treatments and in two varieties with different flowering capabilities, Nagafu No.2 and Yanfu No.6. As shown in Figs. 8 and 9, respectively. Of the 198 MdHMs, 197 genes, with the exception of MdHAG29, were detected in our transcriptome sequencing (Figs. 8 and 9). Of the 197 detected genes, 28 genes, 7 MdSDGs (MdSDG26, 16, 40, 13, 37, 58, and 32), 1 MdPRMT (MdPRMT4), 3 MdJMJs (MdJMJ02, 01, and 06), 13 MdHAGs (MdHAG18, 17, 14, 46, 31, 38, 48, 13, 33, 26, 10, 45, and 06), 1 MdHAC (MdHAC03), and 3 MdHDAs (MdHDA02, 04, and 06) showed no or very low expression levels (less than 1). The non-existent or low expression levels of these MdHMs indicated that they did not function to great degrees in flower development. On the contrary, the expression levels of six genes, MdJMJ16, MdHAG08, MdHAG01, MdHAM01, MdHDT01, and MdHDT03, were extremely high (greater than 100), indicating that they may have important roles during the flower-induction period (Figs. 8 and 9).
Expression profiles of MdHATs and MdHDACs during flower induction period in two different varieties and 6BA treatment. a MdHAGs gene families; b MdHACs gene families; c MdHAFs gene families; d MdHAMs gene families; e MdHDAs gene families. FPKM values were used to generate their expression profiles. f MdHDTs gene families, g MdSRTs gene families
The different expression patterns of the varieties Nagafu No.2 and Yanfu No.6 were also analyzed. In the easy-flowering variety Yanfu No.6, 27 MdSDGs genes, MdSDG07, 29, 27, 48, 35, 19, 24, 14, 39, 03, 61, 29, 51, 10, 33, 22, 49, 50, 52, 04, 17, 18, 46, 06, 08, 27, and 62, were highly expressed. However, only three MdSDG genes MdSDG55, 59, and 36, and two MdPRMT genes (MdPRMT02 and MdPRMT02) were more highly expressed in variety Nagafu No.2 (Fig. 8). Additionally, 12 MdHDMs, MdHDMA06, 11, and 02, and MdJMJ25, 12, 24, 19, 03, 05, 16, 22, and 10, were higher in ‘Yanfu No.6’ while three MdHDMs, MdHDMA05, MdHDMA04, and MdJMJ28, were higher in ‘Nagafu No.2’.
The different expression patterns of the varieties Nagafu No.2 and Yanfu No.6 were also analyzed. In the easy-flowering variety Yanfu No.6, 27 MdSDG genes, MdSDG07, 29, 27, 48, 35, 19, 24, 14, 39, 03, 61, 29, 51, 10, 33, 22, 49, 50, 52, 04, 17, 18, 46, 06, 08, 27, and 62, were highly expressed. However, only three MdSDG genes MdSDG55, 59, and 36, and two MdPRMT genes (MdPRMT02 and MdPRMT01) were more highly expressed in variety Nagafu No.2 (Fig. 8). Additionally, 12 MdHDMs, MdHDMA06, MdHDMA11, MdHDMA02, MdJMJ25, 12, 24, 19, 03, 05, 16, 22, and 10, were higher in ‘Yanfu No.6’, while three MdHDMs, MdHDMA05, MdHDMA04, and MdJMJ28, were higher in ‘Nagafu No.2’ (Figs. 8 and 9).
qRT-PCR analysis of candidate MdHMs genes
In total, 12 MdHMs (MdHAG07, 08, 24, and 34, and MdSDG07, 27, 29, 48, and 55, MdHDT03, MdHAM01, and MdJMJ28) were selected and their expression levels assessed using qRT-PCR under different flowering-related circumstances and in different tissues (stems, leaves, flowers, fruits, and buds). These candidate MdHMs showed different expression patterns in the five tissues, and 11 of them showed higher levels in leaves and buds, with the exception of MdSDG55, which was higher in flowers (Fig. 10). The 12 candidate MdHMs’ responses to various hormones (GA3, ABA, SA, and MeJA) were also investigated (Additional file 18: Figure S11). MdHAG34 and MdSDG55 were not sensitive to these hormone treatments. The remaining 10 genes were up- or down-regulated at different time points after treatment, indicating that they might have roles in hormone stress responses or apple development.
Exogenous sugar treatments can promote flowering and lead to a higher flowering rate [32]. Here, we analyzed the expression profiles of the genes after sugar treatments. As seen in Additional file 19: Figure S12, these candidate genes were also responsive to sugar-mediated flowering induction during the flower-induction period, especially at 70 DAFB. For example, most candidate genes, such as MdHAG07, MdHAG08, MdSDG29, MdSDG55, MdSDG48, MdHDT03, and MdHAM01, showed different expression patterns at 70 DAFB after the sugar treatment. We further analyzed their expression profiles under different flowering-related circumstances (i.e., sugar treatments and alternate bearing). We investigated their expression levels in alternate-bearing ‘Fuji’ trees. The 12 MdHMs were expressed during the flowering periods of both the ‘ON’ and ‘OFF’ tree buds (Fig. 11). Among them, the MdHAG08 level was higher in the ‘OFF’ year in all three developmental stages, while those of MdHAG07 and MdSDG29 showed the opposite trend. MdHAG24, MdHAG34, and MdHDT03 were expressed higher at 30 DAFB in ‘ON’ trees but decreased at 90 and 150 DAFB. The expression patterns of MdSDG07 and MdJMJ28 also differed. At 30 and 90 DAFB, MdSDG07 was higher in ‘ON’ trees and then decreased, while MdJMJ28 was higher in ‘OFF’ trees and then decreased. Thus, the 12 MdHMs appeared to be involved in sugar- or hormone-mediated flower induction, as well as in alternate bearing.
Discussion
HMs played important roles during plant growth and development processes. Although, great advances have been made in some model plants, less has been reported in fruit trees except water deficit and fruit development traits [38, 47, 48]. Here, 198 MdHMs genes, 71 MdHMTs (64 MdSDGs and 7 MdPRMTs), 44 MdHDMs (16 MdHDMAs and 28 MdJMJs), 57 HATs (50 MdHAGs, 2 MdHAMs, 4 MdHACs, and 1 MdHAF), and 26 MdHDACs (16 MdHDAs, 3 MdSRTs, and 7 MdHDTs), were identified in the apple genome. They were further characterized, including gene phylogeny, protein-protein interactions, and expansion and synteny analyses. In addition, we investigated their potential expression levels and roles in response to flower induction. These results will add to the knowledge in this field.
Comparison HMs genes in apple and other sequenced plant species
Identification of HMs genes, including HMTs (SDGs and PRMTs), HDMs (HDMAs and JMJs), HATs (HAGs, HAMs, HACs, and HAFs), and HDACs (HDAs, SRTs, and HDTs) have been systematically or partially reported in Citrus, S. lycopersicum, Arabidopsis, Z. mays, and O. sativa [3, 6, 7, 49]. Little is known about HMs gene families in the important economic apple trees. With the republication of apple genome, it is useful for us to explore more information for genomic analysis. In present study, we totally identified 198 putative MdHMs (Table 2). They were divided into four classifications (HMT, HDM, HAT, and HDAC), and they belonged to 11 different subfamilies, which were similar to those of other species [6, 7]. Of all the HMs, SDGs were the most conserved among the species. The number of MdSDGs was nearly 1.5-fold greater than the numbers of SlSDGs, CsSDGs, AtSDGs, OsSDGs, and ZmSDGs. Additionally, the number of MdHDMAs genes was nearly two to four times greater than those of other species. The number of HAGs in apple was greatly different from other species, especially Arabidopsis, O. sativa and Z. mays. This great difference was partially diminished when the AT1 domain was used as the query in a BLAST algorithm-based search, which led to 33 HAGs being identified in Arabidopsis [4]. Other genes, such as JMJs, HADs, and HDTs, were also present two times more in apple than in other species (Table 2). We also searched orthologous genes against in other species of HMs genes (Fig. 7), which would be a useful tool for further analysis.
The density of apple HMs was followed by Arabidopsis and citrus (Additional file 20: Table S8). Arabidopsis were the most redundant, while Z. mays were the least dense, which might be the result of its large genome size [50]. The apple genome is nearly 1.7 times larger than that of citrus, but their gene numbers are not positively correlated with genome size. Similar relationships exist in other plants (Table 2). The complex connections between genome size and gene number are not well characterized, partially because of duplication events in the genomes of different species and/or their complicated species characterizations. Meanshile, 198 MdHMs were not equally distributed on the 18 chromosomes of the apple genome (Fig. 1), similar to those of citrus [6]. Additionally, this irregular distribution was noticed for the MdGRAS and MdGASA gene families [33, 35]. Theoretically, apple chromosome 15 was longer and larger than other chromosomes, it could easily contain more genes (Fig. 2) [38].
The gain or loss of an exon or intron is always associated with the diversification of gene families. These events can be caused by chromosomal rearrangements or fusions, and they can result in distinct functional characterizations [51]. In the present study, MdHMs genes with different structures were always distantly clustered, while genes with similar structures were tightly clustered (Additional file 8: Figure S3, Additional file 9: Figure S4, Additional file 10: Figure S5, Additional file 11: Figure S6, Additional file 12: Figure S7, Additional file 13: Figure S8 and Additional file 14: Figure S9). This was also observed in the IDD and GASA gene families in the apple genome, indicating potential relationships among phylogeny, gene structures, and protein motifs [33,34,35]. The typical domains of gene clusters in 12 MdHMs were investigated. Generally, these domains were conserved in apple (Fig. 6). For example, the SET domain is conserved in other plants [6, 7, 52], and in apple, MdSDGs also shared this typical SET domain. Additionally, other dispensable domains were also found in MdSDG family members, as in citrus, which shared 19 different domains among its 40 CsSDGs [6]. All of the MdPRMT shared a typical PRMT5 domain. When compared with citrus and tomato, JmjC and SWIRM were also typical conserved domains in the JMJ and HDMA gene families, respectively (Fig. 6) [6, 7]. Similar typical structures are found in other family members among different species. For example, the AT-N domain is in the HAGs, C-terminal MOZ-SAS is in the HAMs, HD is in the HDAs, and SIR2 is in the SRTs (Fig. 6, Additional file 7: Figure S2). Although their sequence characterizations and structures were varied and numerous, their prerequisite domains were conserved, indicating a common characteristic that was conserved among various species [4, 6, 7].
Evolution and expansion analysis of HM gene family
To better understand their phylogenetic interactions, four phylogenetic trees (HMTs, HDMs, HATs, and HDACs) were constructed using the all of the gene members from apple and the model plant Arabidopsis. Interestingly, one PRMT gene, AtPRMT16 was clustered with other SDG genes (Fig. 3a), which might be caused by their partly matching protein sequences. The HDMs, HATs, and HDACs were also clustered. Among the HDMs, the subfamily HDMA clustered separately with the JMJ subfamily (Fig. 3b). The remaining HATs and HDACs were well organized and clustered in a logical fashion, as previous found in other species [6, 7]. Totally, we firstly analyzed the subfamilies within the four trees, HMTs (SDGs and PRMTs), HDMs (HDMAs and JMJs), HATs (HAGs, HAMs, HACs, and HAFs), and HDACs (HDAs, SRTs, and HDTs). The emergence of four different trees from HM subfamilies was useful to investigate their complex phylogenetic interactions.
Gene duplication contributed to the evolution of species [51]. Additionally, in apple, a recent (more than 50 million years ago) whole-genome duplication event took place, which led to a change of apple chromosomes from 9 (ancestral) to 17 (present) [53]. Using improved sequencing technology, a new version of the apple genome was recently published [38]. In the present reference genome, many identified MdHMs showed duplicated genes according to the Circos diagram (Fig. 4). In tomato, eight SlHAGs gene members, including SlHAG11, 19, 20, 21, 22, 24, 25, and 26, underwent tandem duplications [7]. Additionally, SlHACs, SlSDGs, and other subfamilies were also analyzed for gene duplications, as in our study. In apple, it was reported that MdSPL, MdGASA, and MdGRAS also experienced tandem, segmental duplications or whole genome duplications, similar as the MdHMs family members [33, 35, 36]. This gene duplications or gene expansion was associated with the genome duplication [53].
Previously, the Ka/Ks ratio was determined to be a good indicator of positive selection (Ka/Ks > 1), neutral selection (Ka/Ks = 1), or purifying selection (Ka/Ks < 1) [54, 55]. Interestingly, our duplicated gene pairs within apple, or between apple and Arabidopsis, were all less than 1 (Fig. 7), which was similar to a previous report for WRKY in Brachypodium distachyon [46], indicating their important relationships during evolution. Totally, these duplications were associated with the expansion of MdHMs, led to their diverse structures and functions.
The synteny between duplicated blocks in Arabidopsis and apple was also determined of the HMs. Because Arabidopsis is a model plant and functions of the AtHMs are better understood. A comparative genomic comparison investigation helped us understand information on AtHMs to MdHMs, and possible functions of MdHMs can be well inferred [56, 57]. Here, several orthologous genes were also detected in the syntenic maps, and these orthologous genes were located in different duplicated genomic regions of the Arabidopsis and apple genome (Fig. 6), indicating that these genes were derived from a common ancestor. Previously, AtHMs genes, including, AtSDG8 [9], AtHDA9 [8], AtHDA19 [17], AtHDT1 [58], AtHDT3 [59], AtHDA15 [60], AtHAM1 and 2 [19, 20], AtHAF1 [61], and AtSRT1 and 2 [62, 63], were shown to be involved in flower induction. Therefore, based on the orthologous genes between apple and Arabidopsis, several MdHMs could be inferred according to their Arabidopsis comprasion. However, these need to be confirmed by further experiments.
MdHMs were putatively involved in apple flower induction
Histone modifications related genes in plants have been reviewed [12, 13]. Like transcription factors, the HMs genes were also involved in various biological processes during plant growth and development, especially flower induction [64,65,66]. Various genes and gene families involved in flowering have been well characterized in plants. In apple, the MdMAD-box, MdIDD, MdGASA, and MdGRAS gene families were involved in regulating apple flowering [33,34,35, 37]. However, whether MdHMs respond to flower induction was reported. Here, we proposed that MdHMs were also responsible for flower induction in apple. In the model Arabidopsis, several HM genes, such as AtSDG8 [9], AtHDA9 [8], AtHDA19 [17], AtHDT1 [58], AtHDT3 [59], AtHDA15 [60], AtHAM1 and 2 [19, 20], AtHAF1 [61], and AtSRT1 and 2 [63, 64] have been functionally confirmed, and they are involved in flower development. Thus, we identified candidate apple flowering-related genes by referring to their orthologous genes and their expression patterns. For example, MdHDA13, orthologous to AtHDA9, showed a consistent expression pattern during the flower stages and was expressed higher under higher flowering circumstances (‘Yanfu No.6’ and 6BA treatment). Similarly, MdHDA16, an orthologs of AtHDT15; MdHAM01, an orthologs of AtHAM1; and MdHAF01, an orthologs of AtHAF1, were expressed higher in ‘Yanfu No.6’ than in ‘Nagafu No.2’. However, MdHDT04, an orthologs of AtHDT1, was more highly expressed in ‘Nagafu No.2’ (Figs. 8 and 9). Thus, this comparative analysis of HMs genes in apple and Arabidopsis, together with their expression patterns, provided valuable information for the involvement of MdHMs in regulating flower induction.
Leaves and buds are important organs that influence flower development [28, 29, 67]. Here, 11 of the candidate MdHMs were expressed higher in leaves or buds than in other tested tissues (stems, flowers and fruits), which indicated their involvement in flowering. We analyzed their expression patterns in two varieties Nagafu No.2 and Yanfu No.6. ‘Yanfu No.6’ is a ‘Nagafu No.2’ mutant that has a higher proportion of spurs, shorter shoots, larger buds and a higher flowering rate [34, 35]. Most of the MdHMs were expressed and showed consistent patterns during the three developmental stages. A majority of the MdSDG genes were higher in ‘Yanfu No.6’ during the flower development stages (Fig. 8), indicating that methylation is occurring in ‘Yanfu No.6’ and ‘Nagafu No.2’. Similarly, higher acetylation-related activities occurred in ‘Nagafu No.2’ (Fig. 9). Similar epigenetic interactions were also reported among some somatic mutations [68,69,70]. Therefore, we speculated that the up- or down-regulation of MdHMs contributed to different flowering phenomena, which directly or indirectly affected flowering. The continuous differential expression patterns of MdHMs could partly illustrate their modification processes and affect flowering. We also determined their expression levels in response to sugar treatments and hormonal stresses (Additional files 18-19: Figure S11 and S12). In general, they were also partly involved in sugar-mediated flower induction in apple.
Although crosstalk about hormones or sugar-mediated alternate bearing has been reported in perennial trees, unsloved problems were still remained [29, 71, 72]. Here, we investigated the expressions of 12 candidate MdHMs in alternate bearing apple trees. With less reported literature about HMs and alternate bearing, we could not make better propose about this. But they were indeed induced and showed different expression patterns in ‘ON’ and ‘OFF’ trees at different time points, indicating that they were responsible for different development stages (Fig. 11). Further researches needed to be performed to confirm this.
Conclusions
In this study, we systematically identified HMs genes in the apple genome. Their chromosome locations, gene and protein structures, phylogenetic and synteny relationships, and protein-protein interactions were also characterized. Their expression levels in different flowering ability varieties and 6BA treatment were also investigated using high-throughput RNA sequence data in the apple buds, indicated they were responsible to flower induction. Further some candidate HMs genes were then analyzed by qRT-PCR in different tissues (stems, leaves, flowers, fruits, and buds), in different hormones stresses (GA3, ABA, SA and MeJA), and different flowering related circumstances (sugar treatment and alternate bearing buds). Totally, our identification and characterization of HMs genes in apple provided useful information and enriched biological theories, which could be foundation for further analysis.
Abbreviations
- 6BA:
-
6-benzylaminopurine
- ABA:
-
Abscisic acid
- DAFB:
-
Days after full bloom
- HAT:
-
Hours after treatment
- HATs:
-
Histone acetylases
- HDACs:
-
Histone deacetylases
- HDMs:
-
Histone methylases
- HM:
-
Histone modification
- HMTs:
-
Histone methyltransferases
- MeJA:
-
Methyl jasmonate
- SA:
-
Salicylic acid
References
Zhang X. The epigenetic landscape of plants (vol 320, pg 489, 2008). Science. 2008;321(5886):200.
Klose RJ, Yi Z. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Bio. 2007;8(4):307–18.
Aquea F, Vega A, Timmermann T, Poupin MJ, Arce-Johnson P. Genome-wide analysis of the SET DOMAIN GROUP family in grapevine. Plant Cell Rep. 2011;30(6):1087–97.
Pandey R, Muller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002;30(23):5036–55.
Feng Q, Wang HB, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12(12):1052–8.
Xu JD, Xu HD, Liu YL, Wang X, Xu Q, Deng XX. Genome-wide identification of sweet orange (Citrus sinensis) histone modification gene families and their expression analysis during the fruit development and fruit-blue mold infection process. Front Plant Sci. 2015;6:607. https://doi.org/10.3389/fpls.2015.00607.
Cigliano RA, Sanseverino W, Cremona G, Ercolano MR, Conicella C, Consiglio FM. Genome-wide analysis of histone modifiers in tomato: gaining an insight into their developmental roles. BMC Genomics. 2013;14(1):57.
Tanaka M, Kikuchi A, Kamada H. The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 2008;146(1):149–61.
Cazzonelli CI, Cuttriss AJ, Cossetto SB, Pye W, Crisp P, Whelan J, Finnegan EJ, Turnbull C, Pogson BJ. Regulation of carotenoid composition and shoot branching in Arabidopsis by a chromatin modifying histone methyltransferase, SDG8. Plant Cell. 2009;21(1):39–53.
Choi SM, Song HR, Han SK, Han M, Kim CY, Park J, Lee YH, Jeon JS, Noh YS, Noh B. HDA19 is required for the repression of salicylic acid biosynthesis and salicylic acid-mediated defense responses in Arabidopsis. Plant J. 2012;71(1):135–46.
Gohlke J, Scholz CJ, Kneitz S, Weber D, Fuchs J, Hedrich R, Deeken R. DNA Methylation Mediated Control of Gene Expression Is Critical for Development of Crown Gall Tumors. PLoS Genet. 2013;9(2):e1003267.
Ma XJ, Lv SB, Zhang C, Yang CP. Histone deacetylases and their functions in plants. Plant Cell Rep. 2013;32(4):465–78.
Wang Z, Cao H, Chen FY, Liu YX. The roles of histone acetylation in seed performance and plant development. Plant Physiol Bioch. 2014;84:125–33.
Chinnusamy V, Zhu JK. Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol. 2009;12(2):133–9.
Servet C, Silva NCE, Zhou DX. Histone Acetyltransferase AtGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in Arabidopsis. Mol Plant. 2010;3(4):670–7.
Kim W, Latrasse D, Servet C, Zhou DX. Arabidopsis histone deacetylase HDA9 regulates flowering time through repression of AGL19. Biochem Biophys Res Commun. 2013;432(2):394–8.
Krogan NT, Hogan K, Long JA. APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development. 2012;139(22):4180–90.
Wu K, Zhang L, Zhou C, Yu CW, Chaikam V. HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J Exp Bot. 2008;59(2):225–34.
Latrasse D, Benhamed M, Henry Y, Domenichini S, Kim W, Zhou DX, Delarue M. The MYST histone acetyltransferases are essential for gametophyte development in Arabidopsis. BMC Plant Biol. 2008;8(1):121.
Xiao J, Zhang H, Xing LJ, Xu SJ, Liu HH, Chong K, Xu YY. Requirement of histone acetyltransferases HAM1 and HAM2 for epigenetic modification of FLC in regulating flowering in Arabidopsis. J Plant Physiol. 2013;170(4):444–51.
Heisel TJ, Li CY, Grey KM, Gibson SI. Mutations in HISTONE ACETYLTRANSFERASE1 affect sugar response and gene expression in Arabidopsis. Front Plant Sci. 2013;4(2):245.
Gonzalez D, Bowen AJ, Carroll TS, Conlan RS. The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription. Mol Cell Biol. 2007;27(15):5306–15.
Yu CW, Liu XC, Luo M, Chen CY, Lin XD, Tian G, Lu Q, Cui YH, Wu KQ. HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiol. 2011;156(1):173–84.
Jiang D, Yang W, He Y, Amasino RM. Arabidopsis relatives of the human lysine-specific demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell. 2007;19(10):2975–87.
Jeong JH, Song HR, Ko JH, Jeong YM, Kwon YE, Seol JH, Amasino RM, Noh B, Noh YS. Repression of FLOWERING LOCUS T Chromatin by Functionally Redundant Histone H3 Lysine 4 Demethylases in Arabidopsis. PLoS One. 2009;4(11):e8033.
Zhang XY, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009;10(6):R62.
Yang J, Lee S, Hang RL, Kim SR, Lee YS, Cao XF, Amasino R, An G. OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice. Plant J. 2013;73(4):566–78.
Xing L, Zhang D, Zhao C, Li Y, Ma J, An N, Han M. Shoot bending promotes flower bud formation by miRNA-mediated regulation in apple (Malus domestica Borkh.). Plant Biotechnol J. 2016;14(2):749–70.
Fan S, Zhang D, Lei C, Chen HF, Xing LB, Ma JJ, Zhao CP, Han MY. Proteome analyses using iTRAQ labeling reveal critical mechanisms in alternate bearing Malus prunifolia. J Proteome Res. 2016;15(10):3602–16.
Guitton B, Kelner JJ, Celton JM, Sabau X, Renou JP, Chagne D, Costes E. Analysis of transcripts differentially expressed between fruited and deflowered 'Gala' adult trees: a contribution to biennial bearing understanding in apple. BMC Plant Biol. 2016;16(1):1–22.
Li YM, Zhang D, Xing LB, Fan S, Ma JJ, Zhao CP, Du LS, Han MY. A transcriptome analysis of two apple (Malus×domestica) cultivars with different flowering abilities reveals a gene network module associate with floral transitions. Sci Hortic. 2018;239:269–81.
Du L, Qi S, Ma J, Xing L, Fan S, Zhang S, Li Y, Shen Y, Zhang D, Han M. Identification of TPS family members in apple (Malus x domestica Borkh.) and the effect of sucrose sprays on TPS expression and floral induction. Plant Physiol Biochem. 2017;120:10–23.
Fan S, Zhang D, Gao C, Zhao M, Wu HQ, Li YM, Shen YW, Han MY. Identification, classification, and expression analysis of GRAS gene family in Malus domestica. Front Physiol. 2017.8:253. https://doi.org/10.3389/fphys.2017.00253
Fan S, Zhang D, Xing LB, Qi SY, Du LS, Wu HQ, Shao HX, Li YM, Ma JJ, Han MY. Phylogenetic analysis of IDD gene family and characterization of its expression in response to flower induction in Malus. Mol Gen Genomics. 2017;292(4):755–71.
Fan S, Zhang D, Zhang LZ, Gao C, Xin MZ, Tahir MM, Li YM, Ma JJ, Han MY. Comprehensive analysis of GASA family members in the Malus domestica genome: identification, characterization, and their expressions in response to apple flower induction. BMC Genomics. 2017;18(1):827.
Li J, Hou HM, Li XQ, Xiang J, Yin XJ, Gao H, Zheng Y, Bassett CL, Wang XP. Genome-wide identification and analysis of the SBP-box family genes in apple (Malus x domestica Borkh.). Plant Physiol Bioch. 2013;70:100–14.
Kumar G, Arya P, Gupta K, Randhawa V, Acharya V, Singh AK. Comparative phylogenetic analysis and transcriptional profiling of MADS-box gene family identified DAM and FLC-like genes in apple (Malus x domestica). Sci Rep-Uk. 2016;6:20695.
Daccord N, Celton JM, Linsmith G, Becker C, Choisne N, Schijlen E, van de Geest H, Bianco L, Micheletti D, Velasco R, et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat Genet. 2017;49(7):1099−+.
Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–37.
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–4.
Hu B, Jin JP, Guo AY, Zhang H, Luo JC, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–7.
Gambino G, Perrone I, Gribaudo I. A rapid and effective method for RNA extraction from different tissues of grapevine and other Woody plants. Phytochem Analysis. 2008;19(6):520–5.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(−Delta Delta C) method. Methods. 2001;25(4):402–8.
Grimplet J, Agudelo-Romero P, Teixeira RT, Martinez-Zapater JM, Fortes AM. Structural and functional analysis of the GRAS gene family in grapevine indicates a role of GRAS proteins in the control of development and stress responses. Front Plant Sci. 2016;7:353.
Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–8.
Chen SK, Niu X, Guan YX, Li HF. Genome-wide analysis and expression profiles of the MYB genes in Brachypodium distachyon. Plant Cell Physiol. 2017;58(10):1777–88.
Xu J, Zhou S, Gong X, Song Y, van Nocker S, Ma F, Guan Q. Single-base methylome analysis reveals dynamic epigenomic differences associated with water deficit in apple. Plant Biotechnol J. 2018;16(2):672.
Xu JD, Wang X, Cao HB, Xu HD, Xu Q, Deng XX. Dynamic changes in methylome and transcriptome patterns in response to methyltransferase inhibitor 5-azacytidine treatment in citrus. DNA Res. 2017;24(5):509–22.
Lu FL, Li GL, Cui X, Liu CY, Wang XJ, Cao XF. Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J Integr Plant Biol. 2008;50(7):886–96.
Schnable PS, Ware D, Fulton RS, Stein JC, Wei FS, Pasternak S, Liang CZ, Zhang JW, Fulton L, Graves TA, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326(5956):1112–5.
Xu GX, Guo CC, Shan HY, Kong HZ. Divergence of duplicate genes in exon-intron structure. P Natl Acad Sci USA. 2012;109(4):1187–92.
Zhao Z, Yu Y, Meyer D, Wu C, Shen WH. Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat Cell Biol. 2005;7(12):1256–60.
Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, et al. The genome of the domesticated apple (Malus x domestica Borkh.). Nat Genet. 2010;42(10):833−+.
Li J, Zhang Z, Vang SR, Yu J, Wong GKS, Wang J. Correlation between Ka/Ks and Ks is related to substitution model and evolutionary lineage. J Mol Evol. 2009;68(4):414–23.
Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–5.
Lyons E, Pedersen B, Kane J, Alam M, Ming R, Tang HB, Wang XY, Bowers J, Paterson A, Lisch D, et al. Finding and comparing Syntenic regions among Arabidopsis and the Outgroups papaya, poplar, and grape: CoGe with Rosids. Plant Physiol. 2008;148(4):1772–81.
Ghiurcuta CG, Moret BME. Evaluating synteny for improved comparative studies. Bioinformatics. 2014;30(12):9–18.
Kidner CA, Martienssen RA. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature. 2004;428(6978):81–4.
Colville A, Alhattab R, Hu M, Labbe H, Xing T, Miki B. Role of HD2 genes in seed germination and early seedling growth in Arabidopsis. Plant Cell Rep. 2011;30(10):1969–79.
Liu C, Li LC, Chen WQ, Chen X, Xu ZH, Bai SN. HDA18 affects cell fate in Arabidopsis root epidermis via histone acetylation at four kinase genes. Plant Cell. 2013;25(1):257–69.
Bertrand C, Benhamed M, Li YF, Ayadi M, Lemonnier G, Renou JP, Delarue M, Zhou DX. Arabidopsis HAF2 gene encoding TATA-binding protein (TBP)-associated factor TAF1, is required to integrate light signals to regulate gene expression and growth. J Biol Chem. 2005;280(2):1465–73.
Konig AC, Hartl M, Pham PA, Laxa M, Boersema PJ, Orwat A, Kalitventseva I, Plochinger M, Braun HP, Leister D, et al. The Arabidopsis class II Sirtuin is a lysine Deacetylase and interacts with mitochondrial energy metabolism. Plant Physiol. 2014;164(3):1401–14.
Wang CZ, Gao F, Wu JG, Dai JL, Wei CH, Li Y. Arabidopsis putative Deacetylase AtSRT2 regulates basal defense by suppressing PAD4, EDS5 and SID2 expression (vol 51, pg 1291, 2010). Plant Cell Physiol. 2010;51(10):1820.
Fuchs J, Demidov D, Houben A, Schubert I. Chromosomal histone modification patterns--from conservation to diversity. Trends Plant Sci. 2006;11(4):199–208.
Alvarez ME, Nota F, Cambiagno DA. Epigenetic control of plant immunity. Mol Plant Pathol. 2010;11(4):563–76.
Berr A, Shafiq S, Shen WH. Histone modifications in transcriptional activation during plant development. Biochim Biophys Acta. 2011;1809(10):567–76.
Monerri C, Fortunato-Almeida A, Molina RV, Nebauer SG, Garcia-Luis A, Guardiola JL. Relation of carbohydrate reserves with the forthcoming crop, flower formation and photosynthetic rate, in the alternate bearing 'Salustiana' sweet orange (Citrus sinensis L.). Sci Hortic-Amsterdam. 2011;129(1):71–8.
Petit RJ, Hampe A. Some evolutionary consequences of being a tree. Annu Rev Ecol Evol S. 2006;37:187–214.
El-Sharkawy I, Liang D, Xu KN. Transcriptome analysis of an apple (Malus x domestica) yellow fruit somatic mutation identifies a gene network module highly associated with anthocyanin and epigenetic regulation. J Exp Bot. 2015;66(22):7359–76.
Wang ZG, Meng D, Wang AD, Li TL, Jiang SL, Cong PH, Li TZ. The methylation of the PcMYB10 Promoter is associated with green-skinned sport in max red Bartlett pear. Plant Physiol. 2013;162(2):885–96.
Shalom L, Samuels S, Zur N, Shlizerman L, Doron-Faigenboim A, Blumwald E, Sadka A. Fruit load induces changes in global gene expression and in abscisic acid (ABA) and indole acetic acid (IAA) homeostasis in citrus buds. J Exp Bot. 2014;65(12):3029–44.
Yanik H, Turktas M, Dundar E, Hernanderz P, Dorado G, Unver T. Genome-wide identification of alternate bearing-associated microRNAs (miRNAs) in olive (Olea europaeaL.). Bmc Plant Biol. 2013;13:10.
Funding
This work was supported by China Apple Research System (CARS-27), National Natural Science Foundation of China (31672101), Shaanxi key research and development plan (2017ZDXM-NY-019), Yangling Subsidiary Center Project of the National Apple Improvement Center and Collaborative Innovation of the Center for Shaanxi Fruit Industry Development.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and additional files.
Author information
Authors and Affiliations
Contributions
FS and HM conceived and designed the experiment. FS, GC, WJ and ZD performed the experiment. FS, YY, LY, LC and AN analyzed the data. FS and HM wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. List of Pfam accession number of each HMs gene family (DOCX 13 kb)
Additional file 2:
Table S2. Primer information for gene expression analysis (DOCX 13 kb)
Additional file 3:
Table S3. Table S3. Detailed annotations of MdHMs according to the Gene Ontology (GO) terms (XLS 354 kb)
Additional file 4:
Table S4. Synteny analysis of MdHMs genes (DOCX 26 kb)
Additional file 5:
Table S5. Synteny information of MdHMs and AtHMs genes (DOCX 25 kb)
Additional file 6:
Figure S1. Average Ka, Ks values of duplication gene pairs. (A) gene pairs of apple; (B) gene pairs of apple and Arabidopsis (TIF 367 kb)
Additional file 7:
Figure S2. Diagram of HMs typical conserved domains (TIF 846 kb)
Additional file 8:
Figure S3. Gene structure and protein motifs analysis of MdSDGs gene family members (TIF 3270 kb)
Additional file 9:
Figure S4. Gene structure and protein motifs analysis of MdPRMTs gene family members (TIF 571 kb)
Additional file 10:
Figure S5. Gene structure and protein motifs analysis of MdHDMAs gene family members (TIF 639 kb)
Additional file 11:
Figure S6. Gene structure and protein motifs analysis of MdJMJs gene family members (TIF 1445 kb)
Additional file 12:
Figure S7. Gene structure and protein motifs analysis of MdHAGs gene family members (TIF 1174 kb)
Additional file 13:
Figure S8. Gene structure and protein motifs analysis of MdHDAs gene family members (TIF 1253 kb)
Additional file 14:
Figure S9. Gene structure and protein motifs analysis of MdHDTs gene family members (TIF 429 kb)
Additional file 15:
Table S6. Motif sequences of MdHMs proteins (DOCX 16 kb)
Additional file 16:
Figure S10. Interaction networks analysis of MdHMs genes (TIF 1052 kb)
Additional file 17:
Table S7. Predication protein-protein interaction information according to their orthologous in Arabidopsis (XLS 59 kb)
Additional file 18:
Figure S11. Transcript levels of 12 MdHMs genes following GA3, ABA, SA, and MeJA in by qRT-PCR. Leaves were collected after 0, 3,6 and 12 h after treatment. Each value represents the mean ± standard error of three replicates. Means followed by small letters are significantly different at the 0.05 level (the same below). (TIF 3532 kb)
Additional file 19:
Figure S12. Transcript levels of 12 MdHMs genes following sugar treatment and in Yanfu No.6. Terminal buds were collected from 30, 50, and 70 DAFB. (TIF 734 kb)
Additional file 20:
Table S8. Summary of HMs in different species (DOCX 13 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Fan, S., Wang, J., Lei, C. et al. Identification and characterization of histone modification gene family reveal their critical responses to flower induction in apple. BMC Plant Biol 18, 173 (2018). https://doi.org/10.1186/s12870-018-1388-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-018-1388-0