Abstract
Background
Piriformospora indica, an endophytic fungus of Sebacinales, colonizes the roots of many plant species including Arabidopsis thaliana. The symbiotic interaction promotes plant performance, growth and resistance/tolerance against abiotic and biotic stress.
Results
We demonstrate that exudated compounds from the fungus activate stress and defense responses in the Arabidopsis roots and shoots before the two partners are in physical contact. They induce stomata closure, stimulate reactive oxygen species (ROS) production, stress-related phytohormone accumulation and activate defense and stress genes in the roots and/or shoots. Once a physical contact is established, the stomata re-open, ROS and phytohormone levels decline, and the number and expression level of defense/stress-related genes decreases.
Conclusions
We propose that exudated compounds from P. indica induce stress and defense responses in the host. Root colonization results in the down-regulation of defense responses and the activation of genes involved in promoting plant growth, metabolism and performance.
Similar content being viewed by others
Background
The mutualistic interaction between beneficial root-colonizing fungi or bacteria starts with the recognition of both partners before a physical contact is established. Mutual recognition of diffusible signals released by the roots and microbes [arbuscular mycorrhizal (AM), rhizobia-legume root endosymbionts, beneficial endophytes] initiates a signal exchange which prepares the partners for the interaction. Root-derived flavonoids activate the release of factors from the microbes, which induce calcium spiking in root hairs [1]. Downstream of calcium spiking, reprogramming of gene expression in the roots induces mycorrhiza or nodule formation or the establishment of a beneficial mutualistic interaction [2,3]. The symbiotic signals of mycorrhizal fungi, the Myc factors, and those from rhizobial bacteria, Nod factors, are lipo-chitooligosaccharides. They are perceived by lysin-motif (LysM) receptors which induce a signaling pathway leading to either mycorrhiza or nodule formation. Myc factors from Glomus intraradices reprogram root gene expression and induce root branching and mycorrhization in Medicago truncatula ([4]; and ref. therein). Interestingly, LysM receptors are also involved in the perception of chitooligosaccharides, fungal cell wall compounds that induce defense responses and resistance to pathogens. This raises the question of how plants (legumes) discriminate between beneficial and pathogenic microorganisms (cf. [5]). Furthermore, for the establishment of a mutualistic interaction, the beneficial fungi have to overcome the defense machinery of the host to develop within the host. Kloppholz et al. [6] showed that the AM fungus G. intraradices uses the effector protein SP7 to short-circuit the plant defense program. SP7 is secreted and interacts with the pathogenesis-related transcription factor ERF19 in the plant nucleus. ERF19 is highly induced in roots by the fungal pathogen Colletotrichum trifolii as well as by several fungal extracts, but only transiently during mycorrhiza colonization. When constitutively expressed in roots, SP7 leads to higher mycorrhization while reducing the levels of C. trifolii-mediated defense responses. Therefore, SP7 is an effector that contributes to develop the biotrophic status of AM fungi in roots by counteracting the plant immune program. These examples show that the symbionts cross-talk via chemical mediators which are released into the rhizosphere, and these compounds can be effective prior to the physical contact of the symbionts.
We study the beneficial interaction between the root-colonizing fungus Piriformospora indica and the model plant Arabidopsis thaliana. The endophyte colonizes the roots of many plant species, and - similar to AM fungi - promotes plant growth, biomass and seed production and confers resistance to abiotic and biotic stress ([7,8]; and references therein). P. indica is a member of Sebacinales, grows inter- and intracellularly and forms pear shaped spores, which accumulate within the roots and on the root surface. After the establishment of a beneficial interaction barely any defense or stress genes are activated and no reactive oxygen species (ROS) are produced by the host against P. indica [8,9]. Prior to the establishement of a symbiotic interaction and a physical contact between the two partners, P. indica releases exudate compounds, which induces appropriate responses in the host. For instance, a fungal compound induces cytoplasmic calcium ([Ca2+]cyt) elevation in the roots of Arabidopsis and Nicotiana tabacum, which is important for establishing the proper host response to the microbe. [Ca2+]cyt elevation is followed by a nuclear Ca2+ response in the root cells [3]. Rafiqi et al. [10] presented a list of putative effector molecules which were identified in the P. indica genome and which might be secreted in order to modulate host cell’s function and structure and to promote microbial growth on plant tissue. Finally, P. indica releases small molecular compounds into the medium and the root environment which prevent growth of pathogenic fungi and thereby restrict their growth also in the roots [11].
We have established standardized co-cultivation conditions of P. indica and Arabidopsis seedlings on Petri dishes which allow us to investigate the information exchange and the establishment of the mutualistic interaction between the two partners [12]. Here, we report that the seedlings respond to the presence of the fungus as early as two days after co-cultivation although the two organisms have not yet established a physical contact. After six days the hyphae and roots have contact to each other and the first hyphae are detectable within the exodermis of the roots. We report that both roots and leaves respond to the presence of P. indica already two days after co-cultivation. The response pattern is quite different four days later, when the hyphae have contact to the roots.
Results
Co-cultivation conditions of P. indica and Arabidopsis
An agar plaque with P. indica mycelium and an Arabidopsis seedling were transferred to a nylon membrane on solidified PNM medium on a Petri dish, with a distance of 3 cm. As control, an agar plaque without fungal hyphae was used (Additional file 1: Figure S1A). Under these co-cultivation conditions, the fungal mycelium and the roots start to grow but they have no contact to each other within the first two days of co-cultivation (Additional file 1: Figures S1B; S2A, B). At this time point, both organisms are separated by at least two cm. Therefore, any communication between the two organisms is only possible via exudated soluble compounds into the medium or through the gas phase. After six days of co-cultivation the growing roots and hyphae have reached each other and a physical contact has been established (Additional file 1: Figures S2C, D1, D2). Light and fluorescent microscopical analyses demonstrate that the mycelium penetrates the epidemal layers of the root. Formation of the first fungal spores around the roots becomes also visible (Additional file 1: Figures S2D1, D2). We measured defense and symbiotic responses of the seedlings during the first 14 days of co-cultivation (0, 1, 2, 4, 6, 10, 14 days). After 2 days of co-cultivation, a strong difference in the responses of P. indica-exposed and mock-treated control was detectable. After 6 days of co-cultivation, the response pattern was different from that observed at the earlier time point, and did not change much after longer co-cultivation (14 days). We reasoned that the early changes are induced by chemical mediators from the fungus, and that the later changes occur once a physical contact between the two symbionts is established. Therefore, we analysed the response of the roots to the presence of P. indica after two and six days of co-cultivation in more details.
Stomata aperture
Although a physical contact between the two partners has not yet been established after two days, the leaves of the seedlings respond to the presence of the fungus by closing the stomata (Figure 1). Prior to expose to P. indica, 14.6 ± 1.1% of the stomata in the leaves were closed. Almost identical results were obtained for seedlings exposed to an agar plaque without the fungus for either two or six days (two days: 13.9 ± 3.3%; six days: 12.9 ± 3.7%). In contrast, two days after exposure of the seedlings to the P. indica-containing plaque, 76.7 ± 2.9% of the stomata were closed. Longer co-cultivation resulted in re-opening of the stomata, and after six days, only 17.5 ± 1.2% of the stomata remained closed (Figure 1). This demonstrates that regulation of stomata opening in the leaves in response to the root-colonizing fungus P. indica is a sensitive marker for the interaction of the two partners. To clarify whether the fungal signal(s) is an exudated compound in the medium or a gas, we co-cultivated Arabidopsis seedlings with P. indica on split Petri dishes. Exudated compounds from the fungus in the medium cannot reach the roots, while communication via gases or volatiles is possible. The number of closed stomata in Arabidopsis seedling was not significantly different two days after co-cultivation of the symbionts on the split Petri dishes compared to the mock-treated control (control: 18.00 ± 1.65%; split Petri dishes: 18.87 ± 2.17%) which excludes gases and volatiles as chemical mediators.
Epidermis with stomata of Arabidopsis leaves two or six days after exposure of the seedlings to an agar plaque without (control) or with P. indica (+ P. indica ). Guard cells were visualized under the fluorescent microscope (450-520 nm) after stained with calcoflour white (the upper level). The lower panel shows the % closed stomata. Based on 3 independent biological experiments with 10 leaves from individual seedlings each. Bars represent SEs. Asterisks indicate significant differences, as determined by Student’s t-test (**P < 0.01).
H2O2 production
High doses of the fungus did not stimulate H2O2 production in roots and shoots [9] which has been confirmed for roots exposed to P. indica for six days (Figure 2). In contrast, two days after co-cultivation, we observed a higher H2O2 level in the leaves of P. indica-exposed seedlings compared to the mock-treated controls (Figure 2). This suggests that exudated compounds from the fungus trigger ROS production, and this stimulatory effect is no longer detectable six days after co-cultivation. Separation of the mycelium from the roots in split Petri dishes prevented the stimulation of H2O2 production after two days of co-cultivation (control: 0.0033 ± 0.0014 μg/mg dry weight; + P. indica: 0.0027 ± 0.0013 μg/mg dry weight), which again supports the involvement of a diffusible compound in the medium.
H 2 O 2 levels in leaves of A. thaliana seedlings two or six days after exposure to an agar plaque without or with P. indica . The amount of μg H2O2 per mg dry tissue was determined as described in METHODS. Based on 3 independent biological experiments with 10 leaves from individual seedlings. Bars represent SEs. Asterisks indicate significant differences, as determined by Student’s t-test (**P < 0.01).
Regulation of NTR2.5 in the leaves in response to P. indica
NRT2.5 belongs to the nitrate transporter family and is preferentially, but not exclusively, expressed in leaves. The protein plays an essential role in plant growth promotion by the rhizospheric bacterium strain Phyllobacterium brassicacearum STM196 [13,14]. The regulation of its mRNA level in the leaves appears to be very sensitive to signals from the roots. Figure 3 demonstrates that the mRNA level for NRT2.5 in the roots is ~ 4-6-fold up-regulated by P. indica, two and six days after co-cultivation. Furthermore, while no significant response can be detected in the leaves two days after co-cultivation, a ~4-fold up-regulation is observed six days after co-cultivation of the seedlings with P. indica. This shows that signals from the fungus are transferred to the leaves, although the response is slower than this for stomata closure (Figure 1) and ROS production (Figure 2). The NRT2.5 mRNA levels in the roots and leaves on split Petri dish experiments were not up-regulated in comparison to the mock-treated controls (data not shown) which again demonstrates that the NTR2.5 response is mediated by fungus-derived non-gaseous chemical mediators.
NRT2.5 induction in the roots and shoots of Arabidopsis seedlings which were exposed to P. indica for either two or six days. The fold change relative to the mock-treatment is presented. Based on 3 independent biological experiments with 3 technical replicates each. Bars are SEs; they represent the sum of the SEs of the individual values. Asterisks indicate significant differences (six day shoot value vs. two day shoot value), as determined by Student’s t-test (**P < 0.01).
Phytohormone levels in Arabidopsis roots and shoots two and six days after co-cultivation with P. indica
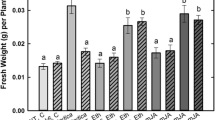
Beneficial plant-microbe interactions are associated with changes in phytohormone levels [15-17]. In order to test whether co-cultivation of Arabidopsis roots with P. indica affects the phytohormone levels, the amounts of jasmonic acid (JA) and its active form JA-isoleucine (JA-Ile), 12-oxo-phytodienoic acid (OPDA), abscisic acid (ABA) and salicylic acid (SA) were determined in the roots and shoots of seedlings either exposed to P. indica or mock-treated. Interestingly, we observed the strongest up-regulation of the phytohormone levels in both roots and shoots two days after co-cultivation. The phytohormone levels decreased significantly in both roots and shoots after six days of co-cultivation (Figure 4). Since the hormones are involved in various types of stress and defense responses, the results indicate that exudated compounds from the fungus induce stress hormones in the roots and systemically also in the leaves. Their level declines as soon as a physical contact between the two organisms is established.
Phytohormone levels in roots and leaves of Arabidopsis seedlings after exposure to P. indica for two or six days. The roots and shoots of the seedlings were harvested at day 0, 2 and 6 after exposure to the P. indica plug or an agar plug without mycelium. SA, ABA, JA, cis-OPDA and JA-Ile levels were determined. The values are means ± SEs of 4 independent biological experiments with 5 replications in each experiment.
Transcriptome analyses for Arabidopsis roots two and six days after exposure to P. indica
Roots exposed to P. indica for two and six days were harvested for RNA extraction and expression profiling. Root material exposed to agar plaques served as control. Only genes from P. indica-exposed material which showed a > 3-fold difference to the agar control were analysed in this study. The comparative transcriptome analysis [18] uncovered that 75 genes were up-regulated and 14 genes down-regulated after two days, whereas 50 genes were up-regulated and 4 genes down-regulated after six days (Figure 5; Figure 6; Additional file 1: Table S1A, C). Categorization of the genes using the Mapman software revealed a huge difference between the two datasets.
Venn datagram of the number of genes which are up- or down-regulated in Arabidopsis roots exposed to P. indica for either two or six days. Numbers of genes regulated only after 2 d of interaction are shown in red colour; those regulated only after 6 d are shown in blue; number of genes regulated at both time points are shown in green. The results are based on 3 independent biological experiments.
Number of genes of the MAPMAN categories which are either up-regulated (blue) or down-regulated (red) in Arabidopsis roots 2 or 6 or [ 2 , 6 ] days after co-cultivation with P. indica. 2 days: genes which are regulated only after 2 days of interaction; 6 days: genes which are regulated only after 6 days of interaction; [2 and 6 days]: common genes which are regulated at both time points. The results are based on 3 independent biological experiments. For detailed information, cf. Additional file 1: Table S1.
Thirthy-five stress- and defense-related genes are only up-regulated during the early time point of co-cultivation and thus appear to respond to chemical mediators released by the fungus (cf. Discussion; Figure 5; Figure 6; Additional file 1: Table S1A). This includes genes for defense-related cell wall proteins and transcription factors, subtilase At1g32940 [19], a protease inhibitor, chitinase, germin-like protein, PAD3, CYP71B6, galactinol synthase 4, glycosyltransferase 73D1, leucine-rich repeat proteins, glutathione-S-transferases (GST) and glutaredoxin 480. Furthermore, phytohormone-related genes such as CYP81D8 (At4g37370), CYSTEINE PROTEINASE1 (At4g36880), GH3.4 (At1g59500), TOUCH3 (At2g41100) and those with Ca2+-related functions [CIPK13 (At2g34180) and At4g33050] are also up-regulated two days after co-cultivation (cf. Discussion). In contrast, genes involved in developmental and DNA modifications, such as HISTONE H1-3, PYRIDOXINE BIOSYNTHESIS1.1 and CML38 are down-regulated.
The number of defense- and stress-related genes is much less after six days of co-cultivation (cf. Discussion).
The majority of the identified genes are regulated by P. indica at both time points (Figure 5; Additional file 1: Table S1B). Closer inspection of the expression levels of these genes also confirms a decline in the degree of defense processes from the 2nd to the 6th day after co-cultivation (cf. Discussion). Examples are genes for the root-specific proline-rich extensin At1g26240, PHOSPHOLIPASE A 2A (At2g26560), GERMIN-LIKE PROTEIN19, CYP81F2, chitinase At2g43570, the disease resistance protein At2g15120, ENDOPEPTIDASE INHIBITOR1 (At2g43510), the Ca2+-binding proteins At5g26920 and At5g39670, the transferase At5g42830, the NAC domain transcription factor JUNGBRUNNEN1, ERD11, ACIREDUCTONE DIOXYGENASE3 and GLUTHATIONE S-TRANSFERASE TAU10 (cf. Discussion). The lower expression level during later stages of co-cultivation indicates that the gene products are less required once a physical contact has been established between the two symbionts.
For 33 randomly chosen genes from the three categories (Additional file 1: Table S1A-C), the microarray results were confirmed by qRT-PCR analyses. Additional file 1: Table S1D demonstrates that most of the results confirmed the microarray data.
To clarify the nature of the fungal signal(s) which modifies the root transcriptome pattern under short term co-cultivation (2 days), we performed co-cultivation experiments on split Petri dishes as described above. The transcriptome pattern of the randomly chosen 33 genes was studied using real-time PCR (Additional file 1: Table S2), but no significant difference was observed to the mock-treated control (Additional file 1: Table S2). This demonstrates again that gases and volatiles do not play a role in changing the gene expression patterns in Arabidopsis roots. Apparently, diffusible compounds released by the hyphae are required for the observed reprogramming of the root transcriptome.
Discussion
Diffusible compounds released by microbes trigger plant responses before physical cell-to-cell contact occurs [1,20-22]. Several lines of evidence demonstrate that P. indica releases compounds which induce defense processes in Arabidopsis roots. The identified genes which are up-regulated after two days of co-cultivation and their role in plant/microbe interaction support this idea. Since the mycelium has not yet reached the roots, plant responses must be induced by either chemical mediators secreted into the medium or gaseous compounds. The split Petri dish experiments support the first possibility, although it cannot be excluded that gaseous compounds also participate in the communication. We also failed to identify major volatile organic compounds which are released into the air in the P. indica/Arabidopsis root symbiosis (D. Tholl and R. Oelmüller, unpublished).
Exudate compounds from both fungal mycelium and roots are well characterized mediators of early communication in mycorrhizal symbiosis [23-25]. The exudate from AM fungi induces also nitric oxide (NO) accumulation in Medicago truncatula roots [26]. NO is involved in control of stomata closure ([27]; and ref. therein), therefore, fungus-induced and plant-released NO could be involved in the regulation of stomata aperture. The early plant responses in the leaves (stomata closure and ROS production) could be caused by NO of plant origin, which is synthesized in response to chemical mediators released from P. indica before a physical contact has been established.
Stomata closure is a typical ABA-mediated stress response, which might be induced by exudated signals from P. indica. Many bacterial pathogens invade plants primarily through stomata on the leaf surface. Sawinski et al. [28] showed that microbial invasion is restricted or prevented by stomata closure upon perception of MAMPs, and this represents an important layer of active immunity at the preinvasive level. The signaling pathways leading to stomatal closure triggered by biotic and abiotic stresses employ several common components, such as ROS, Ca2+, kinases and hormones, suggesting considerable interaction between MAMP- and ABA-induced stomatal closures. Entry of the foliar pathogen Pseudomonas syringae pathovar tomato DC3000 into the plant corpus occurs also through stomatal openings, and consequently a key plant innate immune response is the transient closure of stomata. Kumar et al. [29] showed that root colonization by the rhizobacteria Bacillus subtilis FB17 restricts the stomata-mediated pathogen entry of PstDC3000 in Arabidopsis and root binding of FB17 invokes ABA and SA signaling to close the stomata. These results emphasize the importance of rhizospheric processes and environmental conditions as an integral part of the plant innate immune system against foliar bacterial infections, and similar processes may occur in the system described here.
We have previously demonstrated that colonization of Arabidopsis roots by P. indica does not result in H2O2 production [3,8]. Like the regulation of stomata closure, ROS production is fast in response to fungal signals. ROS is also produced during early stages of symbiotic interactions of bacteria and mycorrhizal fungi with roots [30,31]. Here, we demonstrate an early production of ROS before a physical contact between the two symbionts has been established. This is likely initiated by exudated compounds from the fungus. They can function as PAMPs, similar to PAMPs released by pathogenic fungi which activate ROS production via activation of the root NADPH oxidase or apoplastic peroxidases, or by gaseous compounds. Our results with split Petri dishes argue against a role of gaseous compounds in this response (Additional file 1: Table S2). These ROS could activate the observed defense responses at the mRNA level, both locally and systemically, two days after co-cultivation of the two symbionts. Fungi also contain NADPH oxidases [32]. Epichloe festuca-synthesized ROS regulate hyphal tip growth, thereby restricting growth of the fungus and preventing excessive colonization and host defense gene activation [31,32]. Accumulation of ROS, the oxidative damage to lipids and the membrane electrolyte leakage is lower in AM plants than in non-mycorrhizal plants [33,34], presumably due to the efficient up-regulation of ROS scavenging systems.
Six, but not two days after co-cultivation, we observed the up-regulation of the NRT2.5 mRNA level in the leaves, indicating a slow root-to shoot signal transduction process in the presence of the fungus. Like P. indica, Arabidopsis growth is stimulated by the Phyllobacterium brassicacearum STM196 strain, and this is associated with the up-regulation of NRT2.5 and NRT2.6 [14]. The nrt2.5 and nrt2.6 mutations abolished plant growth and root responses to STM196. Thus, NRT2.5 and NRT2.6, which are preferentially expressed in leaves, play an essential role in plant growth promotion by the rhizospheric bacterium STM196. Members of the NRT2 family have also been described to be involved in plant defense responses: NRT2.1 in the priming against Pseudomonas syringae pv tomato [35] and NRT2.6 in the resistance against Erwinia amylovora [36]. Both genes are required for STM196-induced plant growth promotion, and thus represent new genes in beneficial biotic interactions. Furthermore, these genes participate in a pathway that alters the classically described regulation of shoot - root biomass allocation and root development through the plant nitrogen status. The exact role of these genes in the P. indica/Arabidopsis symbiosis remains to be determined, however, NRT2.5 is a sensitive leaf marker for P. indica colonization of the roots.
Phytohormones play important roles in almost all types of plant-microbe interactions. We demonstrate that the defense-related phytohormones JA, Ja-Ile, ABA, SA and OPDA are strongly up-regulated during early phases of co-cultivation of P. indica with Arabidopsis roots. Since no physical contact has been established at this time point, their up-regulation must be induced by exudated signals from the fungus (Figure 4). Mukherjee and Ané [37] reported that ethylene inhibits induced symbiotic gene expression and root development in response to germinating spore exudates in mono- and dicots. We observed a quite strong up-regulation of ABA in both roots and leaves in response to secreted fungal compounds (Figure 4). It is consistent with the observed closure of the stomata at this time point. Herrera-Medina et al. [38] reported lower colonization of the roots of the ABA-deficient mutant sitiens in tomato. Furthermore, the arbuscules were also less developed in the mutant, and both lesions could be restored by exogenous application of ABA to the sitiens mutant. It appears that ABA is essential for full AM colonization and arbuscule development (cf. [38]). ABA may down-regulate arbuscular formation directly [39], e.g. by stimulating genes involved in defense and cell wall modification [21], or indirectly by stimulating ethylene production [39]. Garrido et al. [40] showed significant differences in gene expression in mycorrhizal roots of wild-type (WT) and ABA-deficient tomato mutants, and these differences corresponded to the ABA content in the roots. Our data support the important role of ABA in beneficial plant/microbe interactions. Up-regulation of components involved in ABA processes has also been reported by Schäfer et al. [41] in the P. indica/barley interaction.
JA, JA-Ile and OPDA are well characterized hormones involved in pathogen attack [42]. Their participation in beneficial plant-microbe interactions is quite controversial (cf. [43]). We observed a strong up-regulation of all these hormonal compounds during early phases of the co-cultivation which is consistent with the observation that JA-regulated stress genes are also up-regulated during the early co-cultivation period. Regvar et al. [44], Isayenkov et al. [45] and Landgraf et al. [46] showed a promotion and Ludwig-Müller et al. [47] a reduction of AM colonization in response to JA or JA-Ile in different systems. Tejeda-Sartorius et al. [48] showed that AM colonization was reduced in a JA-deficient tomato mutant [49], and the lesion could be restored by methyl JA application. In contrast, Herrera-Medina et al. [50] showed that the JA-insensitive jai-1 tomato mutant showed increased colonization and the WT tomato was less colonized after methyl JA application. Nicotiana attenuata plants silenced for COI1 expression showed elevated AM colonization [51]. In spite of quite different results, it appears that JA plays a crucial role in beneficial plant-microbe interactions. JA exogenously applied to the growth medium also decreases the number of nodules induced by Sinorhizobium meliloti on Medicago truncatula roots [52]. JA decreases the responsiveness of Ca2+ spiking to Nod factor application and high concentrations of JA inhibited spiking [52], and this might affect root colonization. Application of JA and methyl JA to roots induced the expression of Nod genes [53] and the production of Nod factors [54]. This suggests that JA is not exclusively involved in the activation of defense responses. The lower level of JA, JA-Ile and OPDA six days after co-cultivation indicates that these compounds play a less dominant role once the partners have recognized themselves as friends. This resembles reports by Kouchi et al. [55] who showed that during early phases of colonization of Lotus japonicus roots by Mesorhizobium loti JA-biosynthesis genes are up-regulated. After initiation of nodule formation, they were repressed again.
SA is mainly required for the plant’s defense against biotrophic pathogens (cf. [56]). We observed a strong response in both roots and shoots, but it is not different from the JA, JA-Ile and OPDA responses (Figure 4). An increase in the SA level has also been reported during early stages of AM colonization [57], and this might be important for root colonization by AM fungi [58]. The transient increase in the SA level induces SA-responsive defense genes in Medicago truncatula roots at early stages of AM colonization [59], similar to the result described here. Tobacco plants with higher SA levels showed reduced root colonization at early time points, but this effect disappeared during later phases of the interaction [50]. How the defense responses induced by the elevated phytohormone levels are down-regulated when a physical contact between the two symbionts has been established remains to be determined. JA signaling might counteract SA signaling at early stages of the recognition of the two symbionts.
Many genes involved in plant defense are regulated during the co-cultivation of Arabidopsis roots with P. indica, however there are clear differences between the early and later time points. Many defense related genes are regulated two and six days after co-cultivation, although their stimulation is lower at the later time point. 35 genes which were up-regulated after 2 days co-cultivation with P. indica are stress and defense genes. The germin-like protein 4 (At1g18970) exhibits superoxide dismutase activity and its homologs in barley and wheat are important resistance component against Blumeria graminis [59]. The defense-related WRKY54 [60], WRKY70 (At3g56400) and MYB51 (At1g18570) transcription factor genes are involved in basal resistance, stress tolerance [60] or secondary metabolite synthesis [61]. The oxygenic stress-inducible aspartyl protease At3g59080 [62], the HOPZ-ACTIVATED RESISTANCE1 leucine-rich repeat protein (ZAR1, At3g50950) [63], the protease YLS5, the leucine-rich repeat protein kinase At1g51890, the VQ motif protein At4g20000, the WD40 protein (At5g42010, TAIR homepage) and PAD3 (At3g26830, CYP71B15) for camalexin biosynthesis (cf. [64]) participate in different aspects of plant immunity or are induced by pathogen treatments. Several glutathione-S-transferase (GST) genes are also up-regulated at the early time point of interaction. GSTF3 (At2g02930) responds to Fusarium sporotrichioides infection [65] and GSTL1 (At5g02780) to a wide range of chemicals and abiotic stress treatments [66]. GST2, a Ca2+-ATPase (At3g63380) is activated by fungal and nematode stimuli and stress (TAIR homepage). Phytohormone-related genes are also up-regulated by chemical mediators from P. indica. The antranilate synthase subunit α1 is important for JA-mediated regulation of auxin biosynthesis and transport during lateral root formation [67], GH3.4 (At1g59500) plays an important role in auxin homeostasis [68], the JA-regulated CYP81D8 (At4g37370) product is involved in phenylpropanoid biosynthesis [69,70], CYSTEINE PROTEINASE1 (At4g36880) responds to gibberellin [71], and TOUCH3 (At2g41100) to SA [72,73]. We conclude that many genes which were up-regulated in response to the fungal exudates, code for defense and stress proteins, compounds involved in signaling leading to defense gene activation or control phytohormone homeostasis.
14 genes which are down-regulated two days after co-cultivation with P. indica are involved in developmental processes and DNA metabolism. HISTONE H1-3 (At2g18050) encodes a linker histone protein whose expression is stimulated by dehydration and ABA [74]. PYRIDOXINE BIOSYNTHESIS1.1 (At2g38230) controls plant growth, development and stress tolerance [75]. At4g12550 is an auxin-induced gene in roots. CML38 (At1g76650) is involved in Ca2+ signaling and important for Ca2+-mediated developmental and stress responses and epidermal development or morphology [76]. The plastid-localized CCL protein (At3g26740) is controlled by the circadian clock during the day [77].
Only ten stress- and defense-related genes are up-regulated six days after co-cultivation. Among them are ALCOHOL DEHYDROGENASE1 (ADH1), which is up-regulated in roots by osmotic stress [78] and ABA [79], the ethylene-responsive transcription factor gene ERF105 (At5g51190) which responds to chitin treatment [80], and INDOLE GLUCOSINOLATE O-METHYLTRANSFERASE1 (At1g21100) involved in hydroxylation reactions of the glucosinolate indole ring [81]. The L-ascorbat oxidase At4g39830 gene is inducible by pathogens [82] and MILDEW RESISTANCE LOCUS6 mediates defense response to fungi and cell death [83]. Genes related to developmental processes code for the AAA-ATPase (At5g40010) which participates in plastidial transport [84], for the CAFFEOYL-COA 3-O-METHYLTRANSFERASE (At1g67980) which catalyses lignin monomer biosynthesis [85], and the CATION/H+ EXCHANGER17 (At4g23700) which regulates cation and pH homeostasis [86].
The group of common genes which are regulated at both time points includes the NAC domain transcription factor gene JUNGBRUNNEN1 which is induced by H2O2 [87], GDSL LIPASE1 (At5g40990) that plays an important role in plant immunity [88], ERD11 (At1g02930) and the GLUTHATIONE S-TRANSFERASE TAU10 (At1g74590) which are induced by oxidative stress and bacterial infections (TAIR homepage). ACIREDUCTONE DIOXYGENASE3 (At2g26400) which functions in H2O2 and SA signaling, is induced by hypoxia and involved in systemic acquired resistance (TAIR homepage). The oxidoreductase At4g10500 gene is induced strongly when Arabidopsis seedlings are grown on a P. indica lawn [9]. Also At5g38900 (DSBA oxidoreductase) and At2g18690 have been described to be involved in defense against pathogenic fungi. All these genes are stronger up-regulated in Arabidopsis roots before a physical contact has been established between the two symbionts, which suggest that they are induced by P. indica-released chemical mediators.
Comparison of transcripts in rice roots, which were colonized by AM Glomalean fungi with those colonized by pathogens (Magnaporthe grisea and Fusarium moniliforme) showed that over 40% of the genes were differentially regulated by both the symbiotic and at least one of the pathogenic microbes. Güimil et al. [89] proposed that the common genes may play a role in compatibility. Furthermore, 34% of the mycorrhiza-associated rice genes were also associated with mycorrhiza in dicots, revealing a conserved pattern of response between the two angiosperm classes. Campos-Soriano and Segundo [90] hypothesized that increased demands for sugars by the fungus might be responsible for the activation of the host defense responses which will then contribute to the stabilization of root colonization by the AM fungus. However, the precise role of defense responses in mutualistic interactions is not clear. Excess root colonization might change a mutualistic association into a parasitic association (cf. [31]). This argues in favor of a role of plant defense compounds in restricting root colonization, thereby stabilizing the symbiotic interaction. Studies with the P. indica/Arabidopsis symbiosis support the idea [16,91]. However, inoculation with G. intraradices stimulated growth and biomass production in WT rice plants and plants overexpressing defense genes. The fungus activates basal defense response in mycorrhizal rice roots, including PR proteins and antioxidant enzymes. Although constitutive expression of defense genes occurred in the roots of the overexpressor lines, the symbiotic efficiency of G. intraradices in these plants was not affected. These results suggest that AM fungi have evolved the capacity to circumvent defense mechanisms that are controlled by the plant’s immune system [92]. Similar observations have been described for the P. indica/Arabidopsis interaction [93]. The authors demonstrate that a broad-spectrum suppression of innate immunity is required for colonization of Arabidopsis roots by P. indica.
Conclusions
In conclusion, our data (Figure 7) suggest that P. indica releases chemical compounds prior to a physical contact which activate stress and defense processes in the host (2 days). Apparently, pre-contact signaling molecules prepare the plant for the symbiotic interaction, and activation of defense may be the first line of recognition. The plant response is not restricted to roots, but also detectable in the leaves. Once root colonization has taken place (6 days) defense responses are down-regulated and genes involved in promoting plant growth, metabolism and performance are activated.
Methods
Growth conditions of A. thaliana and fungi
A. thaliana WT (ecotype Columbia-0) seeds were surface-sterilized and placed on Petri dishes containing MS nutrient medium [94]. After cold treatment at 4°C for 48 h, plates were incubated for 10 days at 22°C under continuous illumination (65 μmol m−2 sec−1). P. indica was cultured for three weeks at 22-24°C on Aspergillus-minimal medium [95,96 (Section A)].
Co-cultivation of seedlings with P. indica
Twelve day-old (48 h cold treatment and 10 days of illumination) Arabidopsis seedlings of equal sizes were selected for co-cultivation experiments. They were transferred to PNM plates with a nylon membrane on the top [96 (Section C-Method 1)] and exposed to a fungal plug 5 mm in diameter or a KM plug of the same size without fungal hyphae (control). The plugs were placed 3 cm away from the closest root part (Additional file 1: Figure S1A). The experimental setup was identical on the split Petri dishes, except that the fungal (or control) plaques were placed on one side and the seedlings on the other side of the Petri dish. The light intensity (80 ± 5 μmol m−2 sec−1) was checked every third day to ensure that both P. indica- and mock-treated seedlings receive equal amounts of light.
Gene expression
Total RNA was isolated separately from roots and shoots of WT seedlings after two and six days of co-cultivation (or mock-treatment) with P. indica using RNeasy Plant Mini Kit (Qiagen). After reverse-transcription, cDNA was synthesized from 1 μg total RNA using the Omniscript RT Kit (Qiagen) and oligo (dT)20 in 20 μl reaction volume. Real-time quantitative PCR was performed with gene-specific primers (Additional file 1: Table S3) and performed using the CFX connect Real-time system and the CFX manager software version 3.1 (Bio-Rad). For the amplification of the PCR products, iQ SYBR Supermix (Bio-Rad) was used according to the manufacturer’s instructions in a final volume of 20 μl. The iCycler was programmed to 95°C 2 min, 35 × (95°C 30 s, 55°C 40 s, 72°C 45 s), 72°C 10 min followed by a melting curve (55-95°C in increasing steps of 0.5°C). All reactions were repeated twice. The mRNA levels for each cDNA probe were normalized with respect to the GAPC2 message levels. Fold induction values were calculated with the ΔΔCP equation of Pfaffl [97]. The ratio of a target gene was calculated in the P. indica-treated sample versus the mock-treated control in comparison to the GAPC2 reference gene.
Microarray analyses, data processing
Microarray hybridizations for P. indica-exposed and mock-treated Arabidopsis roots were performed with the Arabidopsis Genome Array ATH1 (Affymetrix, USA) at the Kompetenzzentrum für Fluoreszente Bioanalytik, Regensburg, Germany. The hybridization signal data were analyzed with ROBIN (http://mapman.gabipd.org/web/guest/robin-download) and MapMan (http://mapman.gabipd.org/web/guest/robin-download) programs. Statistical analysis for t-test and subsequent calculation of false discovery rate were performed according to ROBIN program. The microarray data given in the Supplementary Material are based on 3 biological independent experiments. The results have been submitted to GEO (http://www.ncbi.nlm.nih.gov/geo, submission number GSE58771). The NRT2.5 data shown here are based on Real-time PCR, since the gene was not present on all microarray chips.
Visualization of the cellular pathways and functional categories of the expression data of Arabidopsis roots after two and six days of co-cultivation with P. indica was carried out using the MapMan and Pegman package according to Ath_AFFY_ATH1_TAIR8_Jan 2010 (http://mapman.gabipd.org) [98]. The visualization Mapman tool was used to identify similarities and differences of different pathways involved in biotic and abiotic stress responses [98]. Wilcoxon test was used to visualize significantly expressed genes in Pegman. Venn diagrams were calculated using the expression log values of Mapman package [99]. Specifically expressed genes were determined by Venn diagram with a 3-fold change threshold. Also differentially regulated gene patterns were considered by Venn diagram according to comparative analysis of microarrays in the GEO microarray and NASC data sets.
Microscopy of roots and stomata staining
The roots of Arabidopsis seedlings exposed to P. indica for two or six days were stained with trypan blue and the colonization was analysed by light and fluorescent microscopy as described in Vahabi et al. [100]. Hyphae and spores in the roots could only be detected six days after co-cultivation of the two partners (Additional file 1: Figure S1B, S2C, D1, D2). For stomata staining, detached Arabidopsis leaves were stained using 1 ml calcoflour staining solution (10 mM calcoflour in 50% glycerol, 100 μm Tween 20) for 5 min, and the epidermal layers were analysed under a light and fluorescent microscope (450–520 nm). Opened and closed stomata from 5 areas in 10 leaves from different seedlings were counted. The data are averages of three independent biological experiments. Stomata are considered as closed when no open space can be seen between the two guard cells (Figure 1).
H2O2 and ROS measurements
Arabidopsis seedlings co-cultivated with P. indica for two and six days were stained with 3,3′-diaminobenzidine (DAB) as described by Daudi et al. [101]. As a result of staining a brown precipitate upon oxidation was formed, which is insoluble in aqueous and organic solvents [102,103]. For the detection/quantification of H2O2 inside the plant material, 100 mg of stained tissue was washed with acetone three times, ground to a fine powder and - after drying - dissolved in 1 ml DMSO at 90°C for 1 h. The supernatant was separated from the precipitate by centrifugation at 10,000 rpm for 5 min and was further used for spectrophotometric measurements at 270 nm (Perkin Elmer, Lambda 12) as described by Greenfield et al. [104]. The poly-DAB concentration of the plant tissue was correlated to the H2O2 concentration using a standard curve which was generated by the application of four different concentrations of H2O2 (0.1, 1, 10, 100 μg).
Phytohormone measurement
100 mg of leaf material was frozen in liquid nitrogen and kept at -80°C. After grinding with mortar and pestle, the leaf material was extracted with 1,2 ml of methanol containing 24 ng of 9,10-D2-9,10-dihydrojasmonic acid, 24 ng D4-salicylic acid (Sigma-Aldrich, Germany), 24 ng D6-abscisic acid (Santa Cruz Biotechnology, Santa Cruz, USA), and 4,8 ng of JA-13C6-Ile conjugate as internal standards. JA-13C6-Ile conjugate was synthesized as described by Kramell et al. [105] using 13C6-Ile (Sigma-Aldrich, Germany). The homogenate was mixed for 30 min and centrifuged at 14,000 rpm for 20 min at 4°C. The supernatant was collected. The homogenate was re-extracted with 500 μl methanol, mixed well, centrifuged and supernatants were pooled. The combined extracts were evaporated in a speed-vac at 30°C and re-dissolved in 250 μl methanol. Chromatography was performed on an Agilent 1200 HPLC system (Agilent Technologies). Separation was achieved on a Zorbax Eclipse XDB-C18 column (50 x 4.6 mm; 1.8 μm; Agilent). Formic acid (0.05%) in water and acetonitrile were employed as mobile phases A and B, respectively. The elution profile was: 0-0.5 min, 5% B; 0.5-9.5 min, 5-42% B; 9.5-9.51 min 42-100% B; 9.51-12 min 100% B and 12.1-15 min 5% B. The mobile phase flow rate was 1.1 ml/min. The column temperature was maintained at 25°C. An API 3200 tandem mass spectrometer (Applied Biosystems) equipped with a Turbospray ion source was operated in negative ionization mode. The instrument parameters were optimized by infusion experiments with pure standards, where available. The ionspray voltage was maintained at −4500 eV. The turbo gas temperature was set at 700°C. Nebulizing gas was set at 60 psi, curtain gas at 25 psi, heating gas at 60 psi and collision gas at 7 psi. Multiple reaction monitoring (MRM) was used to monitor analyte parent ion → product ion: m/z 136.9 → 93.0 [collision energy (CE) - 22 V; declustering potential (DP) - 35 V] for SA; m/z 140.9 → 97.0 (CE - 22 V; DP - 35 V) for D4-SA; m/z 209.1 → 59.0 (CE - 24 V; DP - 35 V) for JA; m/z 213.1 → 56.0 (CE - 24 V; DP - 35 V) for 9,10-D2-9,10-dihydrojasmonic acid; m/z 263.0 → 153.2 (CE - 22 V; DP - 35 V) for ABA; m/z 269.0 → 159.2 (CE - 22 V; DP - 35 V) for D6-ABA; m/z 322.2 → 130.1 (CE - 30 V; DP - 50 V) for JA-Ile conjugate; m/z 328.2 → 136.1 (CE - 30 V; DP - 50 V) for JA-13C6-Ile conjugate. Both Q1 and Q3 quadrupoles were maintained at unit resolution. Analyst 1.5 software (Applied Biosystems) was used for data acquisition and processing. Linearity in ionization efficiencies were verified by analyzing dilution series of standard mixtures. Phytohormones were quantified relative to the signal of their corresponding internal standard. For quantification of 12-oxophytodienoic acid, cis-OPDA, 9,10-D2-9,10-dihydro-JA was used as the internal standard applying an experimentally determined response factor of 1.
Availability of supporting data
All the supporting data are included as Additional file 1.
Abbreviations
- Pi :
-
Piriformospora indica
- ROS:
-
Reactive oxygen species
- MAMP:
-
Microbe-associated molecular pattern
- AM:
-
Arbuscular mycorrhiza
- JA:
-
Jasmonic acid
- SA:
-
Salicylic acid
- ABA:
-
Abscisic acid
- OPDA:
-
12-oxo-phytodienoic acid
- JA-Ile:
-
Jasmonic acid-isoleucine
- WT:
-
Wild-type
References
Kosuta S, Chabaud M, Lougnon G, Gough C, Dénarié J, Barker D, et al. A diffusible factor from arbuscular mycorrhizal fungi induces symbiosis-specific MtENOD11 expression in roots of Medicago truncatula. Plant Physiol. 2003;131:952–62.
Oldroyd GE, Downie JA. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol. 2008;59:519–46.
Vadassery J, Ranf S, Drzewiecki C, Mithöfer A, Mazars C, Scheel D, et al. A cell wall extract from the endophytic fungus Piriformospora indica promotes growth of Arabidopsis seedlings and induces intracellular calcium elevation in roots. Plant J. 2009;59:193–206.
Satoh M, Tokaji Y, Nagano AJ, Hara-Nishimura Hayashi M, Nishimura M, Ohta H, Masuda S: Arabidopsis mutants affecting oxylipin signaling in photo-oxidative stress responses. Plant Phys Biochem 2013, pii:S0981-9428 (13) 00418-X.
Gough C, Cullimore J. Lipo-chitooligosaccharide signaling in endosymbiotic plant-microbe interactions. Mol Plant-Microbe Interact. 2011;24:867–78.
Kloppholz S, Kuhn H, Requena N. A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr Biol. 2011;21:1204–9.
Vadassery J, Tripathi S, Prasad R, Varma A, Oelmüller R. Monodehydroascorbate reductase 2 and dehydroascorbate reductase 5 are crucial for a mutualistic interaction between Piriformospora indica and Arabidopsis. J Plant Physiol. 2009;166:1263–74.
Camehl I, Drzewiecki C, Vadassery J, Shahollari B, Sherameti I, Forzani C, et al. The OXI1 kinase pathway mediates Piriformospora indica-induced growth promotion in Arabidopsis. PLoS Pathog. 2011;7:e1002051.
Vahabi K, Camehl I, Sherameti I, Oelmüller R. Growth of Arabidopsis seedlings on high fungal doses of Piriformospora indica has little effect on plant performance, stress, and defense gene expression in spite of elevated jasmonic acid and jasmonic acid-isoleucine levels in the roots. Plant Signal Behav. 2013;8pii:e26301.
Rafiqi M, Jelonek L, Akum NF, Zhang F, Kogel KH. Effector candidates in the secretome of Piriformospora indica, a ubiquitous plant-associated fungus. Front Plant Sci. 2013;4:228.
Sun C, Yongqi S, Vahabi K, Lu J, Bhattacharya S, Dong S, et al. The beneficial fungus Piriformospora indica protects Arabidopsis from Verticillium dahliae infection by downregulation plant defense responses. BMC Plant Biol. 2014;14:268.
Peskan-Berghöfer T, Shahollari B, Giong PH, Hehl S, Markert C, Blanke V, et al. Association of Piriformospora indica with Arabidopsis thaliana roots represents a novel system to study beneficial plant-microbe interactions and involves early plant protein modifications in the endoplasmic reticulum and at the plasma membrane. Physiol Plant. 2004;122:465–77.
Kechid M, Desbrosses G, Rokhsi W, Varoquaux F, Djekoun A, Touraine B. The NRT2.5 and NRT2.6 genes are involved in growth promotion of Arabidopsis by the plant growth-promoting rhizobacterium (PGPR) strain Phyllobacterium brassicacearum STM196. New Phytol. 2013;198:514–24.
Mantelin S, Desbrosses G, Larcher M, Tranbarger TJ, Cleyet-Marel JC, Touraine B. Nitrate-dependent control of root architecture and N nutrition are altered by a plant growth-promoting Phyllobacterium sp. Planta. 2006;223:591–603.
Gutjahr C, Parniske M. Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu Rev Cell Dev Biol. 2013;29:593–617.
Camehl I, Sherameti I, Venus Y, Bethke G, Varma A, Lee J, et al. Ethylene signalling and ethylene-targeted transcription factors are required to balance beneficial and nonbeneficial traits in the symbiosis between the endophytic fungus Piriformospora indica and Arabidopsis thaliana. New Phytol. 2010;185(4):1062–73.
Vadassery J, Ritter C, Venus Y, Camehl I, Varma A, Shahollari B, et al. The role of auxins and cytokinins in the mutualistic interaction between Arabidopsis and Piriformospora indica. Mol Plant-Microbe Interact. 2008;21:1371–83.
Nicolaï M, Roncato MA, Canoy AS, Rouquié D, Sarda X, Freyssinet G, et al. Large-scale analysis of mRNA translation states during sucrose starvation in Arabidopsis cells identifies cell proliferation and chromatin structure as targets of translational control. Plant Physiol. 2006;141(2):663–73.
Wang D, Guo Y, Wu C, Yang G, Li Y, Zheng C: Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genomics 2008, doi:10.1186/1471-2164-9-44
Garcia-Brugger A, Lamotte O, Vandelle E, Bourque S, Lecourieux D, Poinssot B. Early signaling events induced by elicitors of plant defenses. Mol Plant-Microbe Interact. 2006;19:711–24.
Garcia-Garrido J, Ocampo J. Regulation of the plant defence response in arbuscular mycorrhizal symbiosis. J Exp Bot. 2002;53:1377–86.
Olah B, Brière C, Bécard G, Dénarié J, Gough C. Nod factors and a diffusible factor from arbuscular mycorrhizal fungi stimulate lateral root formation in Medicago truncatula via the DMI1/DMI2 signaling pathway. Plant J. 2005;44:195–207.
Chabaud M, Genre A, Sieberer BJ, Faccio A, Fournier J, Novero M, et al. Arbuscular mycorrhizal hyphopodia and germinated spore exudates trigger Ca2+ spiking in the legume and nonlegume root epidermis. New Phytol. 2011;189:347–55.
Nagahashi G, Douds DJ. Separated components of root exudate and cytosol stimulate different morphologically identifiable types of branching responses by arbuscular mycorrhizal fungi. Mycol Res. 2007;111:487–92.
Gadkar V, David-Schwartz R, Nagahashi G, Douds Jr DD, Wininger S, Kapulnik Y. Root exudate of pmi tomato mutant M161 reduces AM fungal proliferation in vitro. FEMS Microbiol Lett. 2003;223:193–8.
Calcagno C, Novero M, Genre A, Bonfante P, Lanfranco L. The exudate from an arbuscular mycorrhizal fungus induces nitric oxide accumulation in Medicago truncatula roots. Mycorrhiza. 2012;22:259–69.
Simontacchi M, García-Mata C, Bartoli CG, Santa-María GE, Lamattina L. Nitric oxide as a key component in hormone-regulated processes. Plant Cell Rep. 2013;32:853–66.
Sawinski K, Mersmann S, Robatzek S, Böhmer M. Guarding the green: pathways to stomatal immunity. Mol Plant-Microbe Interact. 2013;26:626–32.
Kumar AS, Lakshmanan V, Caplan JL, Powell D, Czymmek KJ, Levia DF, et al. Rhizobacteria Bacillus subtilis restricts foliar pathogen entry through stomata. Plant J. 2012;72:694–706.
Fester T, Hause G. Accumulation of reactive oxygen species in arbuscular mycorrhizal roots. Mycorrhiza. 2005;15:373–9.
Tanaka A, Christensen MJ, Takemoto D, Park P, Scott B. Reactive oxygen species play a role in regulating a fungus-perennial ryegrass mutualistic interaction. Plant Cell. 2006;18:1052–66.
Takemoto D, Tanaka A, Scott B. NADPH oxidases in fungi: diverse roles of reactive oxygen species in fungal cellular differentiation. Fungal Genet Biol. 2007;44:1065–76.
Estrada B, Aroca R, Barea JM, Ruiz-Lozano JM. Native arbuscular mycorrhizal fungi isolated from a saline habitat improved maize antioxidant systems and plant tolerance to salinity. Plant Sci. 2013;201–202:42–51.
Evelin H, Kapoor R. Arbuscular mycorrhizal symbiosis modulates antioxidant response in salt-stressed Trigonella foenum-graecum plants. Mycorrhiza. 2014;24(3):197–208.
Camañes G, Pastor V, Cerezo M, García-Andrade J, Vicedo B, García-Agustín P, et al. A deletion in NRT2.1 attenuates Pseudomonas syringae-induced hormonal perturbation, resulting in primed plant defenses. Plant Physiol. 2012;158:1054–66.
Dechorgnat J, Patrit O, Krapp A, Fagard M, Daniel-Vedele F. Characterization of the Nrt2.6 gene in Arabidopsis thaliana: a link with plant response to biotic and abiotic stress. PLoS One. 2012;7:e42491.
Mukherjee A, Ané JM. Germinating spore exudates from arbuscular mycorrhizal fungi: molecular and developmental responses in plants and their regulation by ethylene. Mol Plant-Microbe Interact. 2011;24:260–70.
Herrera-Medina MJ, Steinkellner S, Vierheilig H, Ocampo Bote JA, García Garrido JM. Abscisic acid determines arbuscule development and functionality in the tomato arbuscular mycorrhiza. New Phytol. 2007;175:554–64.
Martín-Rodríguez JA, León-Mocillo R, Vierheilig H, Ocampo JA, Ludwig-Müller J, García-Gardio JM. Ethylene-dependent/ethylene-independent ABA regulation of tomato plants colonized by arbuscular mycorrhizal fungi. New Phytol. 2011;190:193–205.
Garrido JM, Morcillo RJ, Rodríguez JA, Bote JA. Variations in the mycorrhization characteristics in roots of wild-type and ABA-deficient tomato are accompanied by specific transcriptomic alterations. Mol Plant-Microbe Interact. 2010;23:651–64.
Schäfer P, Pfiffi S, Voll LM, Zajic D, Chandler PM, Waller F, et al. Manipulation of plant innate immunity and gibberellin as factor of compatibility in the mutualistic association of barley roots with Piriformospora indica. Plant J. 2009;59:461–74.
Ballaré CL. Jasmonate-induced defenses: a tale of intelligence, collaborators and rascals. Trends Plant Sci. 2011;16:249–57.
Gutjahr C, Paszkowski U. Weights in the balance: jasmonic acid and salicylic acid signaling in root-biotroph interactions. Mol Plant-Microbe Interact. 2009;22:763–72.
Regvar M, Gogala N, Zalar P. The effect of jasmonic acid on mycorrhizal Allium sativum. New Phytol. 1996;134:703–7.
Isayenkov S, Mrosk C, Stenzel I, Strack D, Hause B. Suppression of allene oxide cyclase in hairy roots of Medicago truncatula reduces jasmonate levels and the degree of mycorrhization with Glomus intraradices. Plant Physiol. 2005;139:1401–10.
Landgraf R, Schaarschmidt S, Hause B. Repeated leaf wounding alters the colonization of Medicago truncatula roots by beneficial and pathogenic microorganisms. Plant Cell Environ. 2012;35:1344–57.
Ludwig-Müller J, Bennett R, García-Garrido J, Piché Y, Vierheilig H. Reduced arbuscular mycorrhizal root colonization in Tropaeolum majus and Carica papaya after jasmonic acid application cannot be attributed to increased glucosinolate. J Plant Physiol. 2002;159:517–23.
Tejeda-Sartorius M, Martinez de la Vega O, Délano-Frier J. Jasmonic acid influences mycorrhizal colonization in tomato plants by modifying the expression of genes involved in carbohydrate partitioning. Physiol Plant. 2008;133:339–53.
Li C, Liu G, Xu C, Lee GI, Bauer P, Ling HQ, et al. The tomato suppressor of prosystemin-mediated responses2 gene encodes a fatty acid desaturase required for the biosynthesis of jasmonic acid and the production of a systemic wound signal for defense gene expression. Plant Cell. 2003;15:1646–61.
Herrera-Medina MJ, Gagnon H, Piché Y, Ocampo JA, García-Garrido JM, Vierheilig H. Root colonization by arbuscular mycorrhizal fungi is affected by the salicylic acid content of the plant. Plant Sci. 2003;164:993–8.
Riedel T, Groten K, Baldwin IT. Symbiosis between Nicotiana attenuata and Glomus intraradices: Ethylene plays a role, jasmonic acid does not. Plant Cell Environ. 2008;31:1203–13.
Sun J, Cardoza V, Mitchell D, Bright L, Oldroys G, Harris J. Crosstalk between jasmonic acid, ethylene and Nod factor signaling allows integration of diverse inputs for regulation of nodulation. Plant J. 2006;46:961–70.
Rosas S, Soria R, Correa N, Abdala G. Jasmonic acid stimulates the expression of nod-genes in rhizobium. Plant Mol Biol. 1998;38:1161–8.
Mabood F, Souleimanov A, Khan W, Smith DL. Jasmonates induce Nod factor production by Bradyrhizobium japonicum. Plant Physiol Biochem. 2006;44:759–65.
Kouchi H, Shimomura K, Hata S, Hirota A, Wu GJ, Kumagai H, et al. Large-scale analysis of gene expression profiles during early stages of root nodule formation in a model legume Lotus japonicus. DNA Res. 2004;11(4):263–74.
Lu H. Dissection of salicylic acid-mediated defense signaling networks. Plant Signal Behav. 2009;4:713–7.
Blilou I, Ocampo J, García-Garrido J. Resistance of pea roots to endomycorrhizal fungus or Rhizobium correlates with enhanced levels of endogeneous salicylic acid. J Exp Bot. 1999;50:1663–8.
Liu J, Blaylock L, Endre G, Cho J, Town C, VandenBosch K, et al. Transcript profiling coupled with spatial expression analysis reveals genes involved in distinct developmental stages of the arbuscular mycorrhizal symbiosis. Plant Cell. 2003;15:2106–23.
Christensen AB, Thordal-Christensen H, Zimmermann G, Gjetting T, Lyngkjaer MF, Dudler R, et al. The germinlike protein GLP4 exhibits superoxide dismutase activity and is an important component of quantitative resistance in wheat and barley. Mol Plant-Microbe Interact. 2004;17(1):109–17.
Li J, Besseau S, Törönen P, Sipari N, Kollist H, Holm L, et al. Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 2013;200:457–72.
Frerigmann H, Gigolashvili T. MYB34, MYB51, and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol Plant. 2014;7(5):814–28.
Stanley Kim H, Yu Y, Snesrud EC, Moy LP, Linford LD, Haas BJ, et al. Transcriptional divergence of the duplicated oxidative stress-responsive genes in the Arabidopsis genome. Plant J. 2005;41:212–20.
Lewis JD, Wu R, Guttman DS, Desveaux D. Allele-specific virulence attenuation of the Pseudomonas syringae HopZ1a Type III Effector via the Arabidopsis ZAR1 Resistance Protein. PLoS Gen. 2010;6(4):e100894.
Beets CA, Huang JC, Madala NE, Dubery I. Activation of camalexin biosynthesis in Arabidopsis thaliana in response to perception of bacterial lipopolysaccharides: a gene-to-metabolite study. Planta. 2012;236:261–72.
Asano T, Kimura M, Nishiuchi T. The defense response in Arabidopsis thaliana against Fusarium sporotrichioides. Proc Natl Acad Sci U S A. 2012;10:61.
Dixon DP, Davis BG, Edwards R. Functional divergence in the glutathione transferase superfamily in plants. Identification of two classes with putative functions in redox homeostasis in Arabidopsis thaliana. J Biol Chem. 2002;277:30859–69.
Sun J, Xu Y, Ye S, Jiang H, Chen Q, Liu F, et al. Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell. 2009;21:1495–511.
Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, et al. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell. 2005;17(2):616–27.
Manabe R, Tsutsui K, Yamada T, Kimura M, Nakano I, Shimono C, et al. Transcriptome-based systematic identification of extracellular matrix proteins. Proc Natl Acad Sci U S A. 2008;105(35):12849–54.
Ehlting J, Sauveplane V, Olry A, Ginglinger JF, Provart NJ, Werck-Reichhart D: An extensive (co-)expression analysis tool for the cytochrome P450 superfamily in Arabidopsis thaliana. BMC Plant Biol 2008, doi:10.1186/1471-2229-8-47.
Bouquin T, Meier C, Foster R, Nielsen ME, Mundy J. Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiol. 2001;127(2):450–8.
Sistrunk ML, Antosiewicz DM, Purugganan MM, Braam J. Arabidopsis TCH3 encodes a novel Ca21 binding protein and shows environmentally induced and tissue-specific regulation. Plant Cell. 1994;6:1553–65.
Bodenhausen N, Reymond P. Signaling pathways controlling induced resistance to insect herbivores in Arabidopsis. Mol Plant-Microbe Interact. 2007;20(11):1406–20.
Ascenzi R, Gantt JS. A drought-stress-inducible histone gene in Arabidopsis thaliana is a member of a distinct class of plant linker histone variants. Plant Mol Biol. 1997;34(4):629–41.
Titiz O, Tambasco-Studart M, Warzych E, Apel K, Amrhein N, Laloi C, et al. PDX1 is essential for vitamin B6 biosynthesis, development and stress tolerance in Arabidopsis. Plant J. 2006;48(6):933–46.
Vanderbeld B, Snedden WA. Developmental and stimulus-induced expression patterns of Arabidopsis calmodulin-like genes CML37, CML38 and CML39. Plant Mol Biol. 2007;64:683–97.
Lidder P, Gutiérrez RA, Salomé PA, McClung CR, Green PJ. Circadian control of messenger RNA stability. Association with a sequence‐specific messenger RNA decay pathway. Plant Physiol. 2005;138:2374–85.
Jiang Y, Yang B, Harris NS, Deyholos MK. Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J Exp Bot. 2007;58(13):3591–607.
de Bruxelles GL, Peacock WJ, Dennis ES, Dolferus R. Abscisic acid induces the alcohol dehydrogenase gene in Arabidopsis. Plant Physiol. 1996;111(2):381–91.
Libault M, Wan J, Czechowski T, Udvardi M, Stacey G. Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol Plant-Microbe Interact. 2007;20(8):900–11.
Pfalz M, Mikkelsen MD, Bednarek P, Olsen CE, Halkier BA, Kroymann J. Metabolic engineering in Nicotiana benthamiana reveals key enzyme functions in Arabidopsis indole glucosinolate modification. Plant Cell. 2011;23(2):716–29.
Sato M, Mitra RM, Coller J, Wang D, Spivey NW, Dewdney J, et al. A high-performance, small-scale microarray for expression profiling of many samples in Arabidopsis-pathogen studies. Plant J. 2006;49(3):565–77.
Devoto A, Turner JG. Regulation of jasmonate‐mediated plant responses in Arabidopsis. Ann Bot. 2003;92(3):329–37.
Weber APM, Oesterhelt C, Gross W, Bräutigam A, Imboden L, Krassovskaya I. EST-analysis of the thermo-acidophilic red microalga Galdieria sulphuraria reveals potential for lipid A biosynthesis and unveils the pathway of carbon export from rhodoplasts. Plant Mol Biol. 2004;55:17–32.
Zhang K, Bhuiya M-W, Pazo JR, Miao Y, Ralph J, Liu CJ. An engineered monolignol 4-O-methyltransferase depresses lignin biosynthesis and confers metabolic capability in Arabidopsis. J Biol Chem. 2012;285:277–85.
Chanroj S, Lu Y, Padmanaban S, Nanatani K, Uozumi N, Rao R, et al. Plant-specific cation/H+ exchanger 17 and its homologs are endomembrane K+ transporters with roles in protein sorting. J Biol Chem. 2011;286:33931–41.
Wu A, Allu AD, Garapati P, Siddiqui H, Dortay H, Zanor MI, et al. JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell. 2012;24(2):482–506.
Kim HG, Kwon SJ, Jang YJ, Nam MH, Chung JH, Na YC, et al. GDSL LIPASE1 modulates plant immunity through feedback regulation of ethylene signaling. Plant Physiol. 2013;163:1776–91.
Güimil S, Chang HS, Zhu T, Sesma A, Osbourn A, Roux C, et al. Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc Natl Acad Sci U S A. 2005;102:8066–70.
Campos-Soriano L, Segundo BS. New insights into the signaling pathways controlling defense gene expression in rice roots during the arbuscular mycorrhizal symbiosis. Plant Signal Behav. 2011;6:553–7.
Sherameti I, Venus Y, Drzewiecki C, Tripathi S, Dan VM, Nitz I, et al. PYK10, a beta-glucosidase located in the endoplasmatic reticulum, is crucial for the beneficial interaction between Arabidopsis thaliana and the endophytic fungus Piriformospora indica. Plant J. 2008;54:428–39.
Campos-Soriano L, García-Garrido JM, San Segundo B. Activation of basal defense mechanisms of rice plants by Glomus intraradices does not affect the arbuscular mycorrhizal symbiosis. New Phytol. 2010;188:597–614.
Jacobs S, Zechmann B, Molitor A, Trujillo M, Petutschnig E, Lipka V, et al. Broad-spectrum suppression of innate immunity is required for colonization of Arabidopsis roots by the fungus Piriformospora indica. Plant Physiol. 2011;156:726–40.
Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–97.
Hill TW, Kaefer E. Improved protocols for Aspergillus medium: trace elements and minimum media salt stock solutions. Fungal Genet New. 2001;48:20–1.
Johnson JM, Sherameti I, Ludwig A, Nongbri PL, Sun C, Lou B, et al. Protocols for Arabidopsis thaliana and Piriformospora indica co-cultivation - A model system to study plant beneficial traits. J Endocyt Cell Res. 2011;21:101–13.
Pfaffl MW. A new mathematical model for relative quantification in realtime RT-PCR. Nucleic Acids Res. 2001;29:e45.
Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, et al. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–39.
Usadel B, Nagel A, Thimm A, Redestig H, Bläsing OE, Palacios-Rojas N, et al. Extension of the visualisation tool MapMan to allow statistical analysis of arrays, display of co-responding genes and comparison with known responses. Plant Physiol. 2005;138:1195–204.
Vahabi K, Johnson JM, Drzewiecki C, Oelmüller R. Fungal staining tools to study the interaction between the beneficial endophyte Piriformospora indica with Arabidopsis thaliana roots. J Endocyt Cell Res. 2011;21:77–88.
Daudi A, Cheng Z, O'Brien JA, Mammarella N, Khan S, Ausubel FM, et al. The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell. 2012;24(1):275–87.
Gallyas F, Gorcs T, Merchenthaler I. High-grade intensification of the end-product of the diaminobenzidine reaction for peroxidase histochemistry. J Histochem Cytochem. 1982;30:183–4.
Vidossich P, Alfonso-Prieto M, Rovira C. Catalases versus peroxidases: DFT investigation of H2O2 oxidation in models systems and implications for heme protein engineering. J Inorg Biochem. 2012;117:292–7.
Greenfield L, Starkenburg S, Shallice M, Nyhus J and Leong L: Stable compositions comprising chromogenic compounds and methods of use. Patent 2010, United States 0120077211, WO2010077870 A2
Kramell R, Schmidt J, Schneider G, Sembdner G, Schreiber K. Synthesis of N-(jasmonyl) amino acid conjugates. Tetrahedron. 1988;44:5791–807.
Acknowledgements
We thank Sarah Muβbach and Claudia Röppischer for their excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
KV designed and carried out most of the experiments. IS, MB, AM and AL helped in transcriptome analysis. MR did the phytohormone analysis. KV, IS and RO wrote the article. RO supervised the research. All authors read and approved the final manuscript.
Additional file
Additional file 1: Figure S1.
Experimental design for the co-cultivation assays. Figure S2. Localization of P. indica mycelium and spores in and around Arabidopsis roots. Table S1. MAPMAN analysis of the genes which are regulated at least 3-fold in the roots of seedlings exposed to P. indica for two or six days. Table S2. Regulated genes in A. thaliana roots after two days interaction with P. indica grown on normal or split Petri dishes. Table S3. List of primers for RT-PCR used in this study.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Vahabi, K., Sherameti, I., Bakshi, M. et al. The interaction of Arabidopsis with Piriformospora indica shifts from initial transient stress induced by fungus-released chemical mediators to a mutualistic interaction after physical contact of the two symbionts. BMC Plant Biol 15, 58 (2015). https://doi.org/10.1186/s12870-015-0419-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-015-0419-3