Abstract
Background
Tsetse flies are cyclical vectors of African trypanosomiasis (AT). The flies have established symbiotic associations with different bacteria that influence certain aspects of their physiology. Vector competence of tsetse flies for different trypanosome species is highly variable and is suggested to be affected by bacterial endosymbionts amongst other factors. Symbiotic interactions may provide an avenue for AT control. The current study provided prevalence of three tsetse symbionts in Glossina species from Cameroon, Chad and Nigeria.
Results
Tsetse flies were collected and dissected from five different locations. DNA was extracted and polymerase chain reaction used to detect presence of Sodalis glossinidius, Spiroplasma species and Wolbachia endosymbionts, using species specific primers. A total of 848 tsetse samples were analysed: Glossina morsitans submorsitans (47.52%), Glossina palpalis palpalis (37.26%), Glossina fuscipes fuscipes (9.08%) and Glossina tachinoides (6.13%). Only 95 (11.20%) were infected with at least one of the three symbionts. Among infected flies, six (6.31%) had Wolbachia and Spiroplasma mixed infection. The overall symbiont prevalence was 0.88, 3.66 and 11.00% respectively, for Sodalis glossinidius, Spiroplasma species and Wolbachia endosymbionts. Prevalence varied between countries and tsetse fly species. Neither Spiroplasma species nor S. glossinidius were detected in samples from Cameroon and Nigeria respectively.
Conclusion
The present study revealed, for the first time, presence of Spiroplasma species infections in tsetse fly populations in Chad and Nigeria. These findings provide useful information on repertoire of bacterial flora of tsetse flies and incite more investigations to understand their implication in the vector competence of tsetse flies.
Similar content being viewed by others
Background
Trypanosomiasis disease is caused by trypanosomes, extracellular flagellated protozoan parasites of the genus Trypanosoma, and is one of major endemic diseases in sub-Saharan Africa. The disease exists as Human African Trypanosomiasis (HAT) (sleeping sickness) and the animal African trypanosomiasis (AAT) (nagana) forms. In humans, the disease is caused by Trypanosoma brucei rhodesiense, responsible for acute form of the disease in eastern and southern Africa, and Trypanosoma brucei gambiense, responsible for chronic form of the disease in western and central Africa [1]. Approximately 56 million people are estimated to be at different levels of risk of contracting HAT and more than 1.18 million Km2 are still at risk of T. b. gambiense infection [2]. In 2019, 992 cases recorded in Africa [1]. The AAT is caused by several species and subspecies of trypanosomes that include Trypanosoma congolense, Trypanosoma vivax, Trypanosoma simiae, Trypanosoma uniforme, Trypanosoma godfreyi, Trypanosoma brucei brucei and Trypanosoma grayi, and is a major constraint to Agricultural development on the continent, causing pathogenic infections in cattle, sheep, goats, pigs, dogs, camels and horses [3,4,5].
Trypanosomes are cyclically transmitted between different vertebrate hosts by tsetse flies (Diptera: Glossinidae). Thirty-one species and subspecies of tsetse flies have been described. The flies are grouped into three groups or subgenera based on common characteristics and morphology due to bio-ecological and genetic similarities; the riverine Palpalis, the savannah Morsitans and the forest Fusca [6, 7]. The flies acquire trypanosomes when feeding on an infected vertebrate host, which (trypanosomes) undergo a series of transformations and multiplication in their gut, giving rise to infective forms which inoculated into a new host during feeding [8].
Tsetse fly gut harbours diverse bacteria acquired from environment or maternally [9]. Previous studies have shown that these bacterial populations vary considerably depending both on tsetse fly species or sub-species and geographic origin of the flies [10]. The microbial community influences several aspects of tsetse fly’s physiology, including nutrition, fecundity development and maturation of innate immune system and vector competence [11, 12].
Tsetse flies have established long-term associations with four vertically transmitted endosymbiotic bacteria including Wigglesworthia glossinidia, Sodalis glossinidius, Wolbachia species and Spiroplasma species that was recently established as the fourth tsetse symbiont in Glossina fuscipes fuscipes, Glossina tachinoides, and Glossina palpalis palpalis [13]. They show different types of relation with their host.
All tsetse flies house W. glossinidia as primary and obligate endosymbiont. The W. glossinidia resides intracellularly within bacteriome in anterior midgut and extracellularly in milk gland of the fly [11]. The W. glossinidia provides dietary supplements absent in the tsetse fly vertebrate blood-meal restricted diet, supports larval development and contributes to maturation of adult immune system [12, 14]. Wolbachia endosymbionts are obligatory intracellular bacteria belonging to the Order Rickettsiales, infecting broad range of arthropod and filarial nematode species and probably most prevalent endosymbiont in insect germlines [11]. Within Wolbachia genus, 17 supergroups (A - Q) are currently recognized based on sequences of the five (fbpA, coxA, ftsZ, gatB, coxA and hcpA) conserved genes and the amino acid sequences of the four hypervariable regions of the Wsp protein [11, 15]. Majority of insect infections fall into supergroups A and B [16]. Phylogenetic analysis based on a concatenated dataset of MLST loci and wsp gene of Wolbachia strains detected in tsetse flies collected in ten African countries revealed that the Wolbachia strains infecting G. m. morsitans, G. m. centralis, G. brevipalpis, G. pallidipes and G. austeni belong to supergroup A, while the Wolbachia strain infecting G. p. gambiensis fell into supergroup B [17].
In tsetse fly, Wolbachia mainly resides in reproductive tissues and is maternally transmitted from generation to generation through trans-ovarian transmission. Wolbachia is also transferred horizontally among arthropods. Wolbachia infection in the tsetse fly host results in a variety of reproductive abnormalities such as parthenogenesis, male killing, feminization and cytoplasmic incompatibility [16,17,18]. Cytoplasmic incompatibility (CI) results in embryonic mortality in progeny derived from mating between insects with different Wolbachia infection status, where infected male mates with an uninfected female or infected female with a different strain of the bacterium [16]. The presence of this symbiont in the Glossina morsitans morsitans tsetse fly is associated with the induction of CI [18], which confers indirect reproductive advantages to infected females and is considered as a potential vector control alternative [11, 18]. Some studies have shown that Wolbachia infections limit mosquito-transmitted pathogens including dengue virus, chikungunya virus, Plasmodium parasites, yellow fever virus, Zika virus and filarial nematodes [19,20,21,22].
The tsetse fly’s secondary and facultative symbiont is the commensal S. glossinidius. It is a gram-negative organism belonging to Enterobacteriaceae family. The S. glossinidius is widely spread in numerous tissues of tsetse fly (midgut, fat body, milk gland, salivary glands and reproductive system), present intracellularly and extracellularly [23]. The S. glossinidius genome consists of one circular chromosome of 4.17 Mbp, three extrachromosomal plasmids designated pSG1, pSG2, and pSG3, as well as a phage, ФSG1. However, its genome sequence shows reduced coding capacity with a large number of pseudogenes [24]. The S. glossinidius can be transmitted maternally via haemolymph, milk gland secretions, and horizontally during mating [11, 25]. In tsetse flies, specific role of this symbiont is still not clear. However, it has been shown to affect host longevity and has been suspected to play a role in potentiating susceptibility to trypanosome infection in tsetse by influencing efficacy of tsetse immune system possibly through lectin-inhibitory activity [26].
The Spiroplasma genus belongs to Mollicutes class, and Tenericutes phylum. Spiroplasma species are abundant in insects gut or haemolymph where they have large variety of commensal, pathogenic or mutualist interactions with host [13]. Spiroplasma confers protection against pathogens such as fruit fly Drosophila neotestacea from nematode [27], pea aphid Acyrthosiphon pisum against fungi [28] and fruit fly Drosophila hydei against a parasitoid wasp Leptopilina heterotoma [29]. However, reproductive alterations such as CI, male-killing and sex determination are related to numerous species of Spiroplasma [13].Spiroplasma recently, been established as a new class of tsetse fly symbiont in G. f. fuscipes, G. tachinoides, and G. p. palpalis. The interactions between Spiroplasma and tsetse flies seem to be beneficial because of its ability to extend lifespan and reduce t vector competence for Trypanosoma [11, 30].
Current control measures against trypanosomiasis are mainly based on chemotherapy. In absence of effective vaccine and to address limitations associated with chemotherapy, disruption of trypanosomes transmission through vector control is crucial. Transmission of pathogens by vector depends on its vector competence, which can be affected by several factors, including vector endosymbionts [31]. Due to their importance, interactions between the symbionts and their hosts are being harnessed toward development of novel approaches for vector and disease control [32,33,34].
The present study assessed presence of Sodalis glossinidius, Spiroplasma species and Wolbachia in wild populations of tsetse flies in Cameroon, Nigeria and Chad.
Results
For this study, 848 tsetse fly samples from Cameroon, Chad and Nigeria were used. The flies belonged Glossina morsitans submorsitans: 403 (47.52%); G. p. palpalis: 316 (37.26%); G. f. fuscipes: 77 (9.08%); G. tachinoides: 52 (6.13%) species. Distributions of prevalence of different symbiotic bacteria are presented in Table 1.
Out of 848 flies, 95 (11.20%) were infected with at least one of the endosymbionts. Among infected flies, six (6.31%) had Wolbachia-Spiroplasma mixed infection.
Sodalis glossinidius infection prevalence
Presence of S. glossinidius was investigated in 678 field-collected tsetse flies from Cameroon (149), Chad (259) and Nigeria (270) using a Sodalis Hemolysin gene-based PCR. The S. glossinidius infection rates were 2.01%, 1.16% and 0.0% in Cameroon Chad Nigeria respectively (Table 1). Prevalence of S. glossinidius infection were similar (p > 0.05) between Cameroon and Chad. The sequence of our amplicons had > 99% similarity to Sodalis’s Hemolysin partial gene sequences (OQ458712.1, MH192369.1, LN854557.1, AP008232.1) in NCBI database. Maximum Likelihood phylogenetic tree (Fig. 1) revealed amplicons from Cameroon and Chad with different clades.
Phylogenetic tree of detected. Sodalis glossinidius’ hemolysin partial gene and its closed relatives. The evolutionary history conducted in MEGA X, was inferred by using the Maximum Likelihood method and Hasegawa-Kishino-Yano model. This analysis involved 10 nucleotide sequences and a total of 596 positions in the final dataset. Our isolates are marked by a black circle. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches
Spiroplasma infection prevalence
A total of 847 fly samples from Cameroon (154), Chad (423) and Nigeria (270) were used for molecular detection of Spiroplasma using a 16 S rRNA-based PCR approach with wspecF/wspecR primers. Spiroplasma was detected in 31 (3.66%) of the samples, with. prevalence varying significantly (p < 0.001) between countries (Table 1). Spiroplasma was detected in 0.0%, 0.95% and 10.00% samples from Cameroon, Chad and Nigeria respectively. The sequence of our amplicons had > 99% similarity to Spiroplasma 16 S rRNA partial gene sequences (KX159383.1, NR 104720.1, OQ448933.1, CP018022.1) in NCBI database.
Maximum Likelihood phylogenetic tree (Fig. 2) revealed amplicons from Nigeria and Chad with different clades.
Phylogenetic tree of detected Spiroplasma’s 16S rRNA partial gene and its closed relatives. The evolutionary history conducted in MEGA X, was inferred by using the Maximum Likelihood method and Kimura 2-parameter model. This analysis involved 20 nucleotide sequences and a total of 383 positions in the final dataset. Our isolates are marked by a black circle. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches
Wolbachia infection prevalence
A total of 582 tsetse samples (gut or abdomen) were screened for presence of Wolbachia using 16 S rRNA-based PCR approach with wspecF/wspecR set of primers. The samples were collected in Cameroon: 155, Chad: 157 and Nigeria: 270. The results of the screening (Table 1) revealed that prevalence of Wolbachia infections varied, but not significantly (p = 0.06) between the three countries. The Wolbachia prevalence in Cameroon, Chad and Nigeria was 9.68%, 7.00% and 14.07% respectively. The sequence of our amplicons had > 99% similarity to Wolbachia 16 S rRNA gene sequences (MK277440.1, JQ726770.1, JF494910.1, KM517499.1) in NCBI database. Maximum Likelihood phylogenetic tree (Fig. 3) revealed amplicons from Nigeria and Cameroon on same clades, and separate from that of chad.
Phylogenetic tree of detected Wolbachia’s partial 16 S rRNA gene and its closed relatives. The evolutionary history conducted in MEGA X, was inferred by using the Maximum Likelihood method and Kimura 2-parameter model. A discrete Gamma distribution was used to model evolutionary rate differences among sites. This analysis involved 18 nucleotide sequences and a total of 377 positions in the final dataset. Our isolates are marked by a black circle. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches
All 16 S rRNA and hemolysin gene sequences generated in this study are deposited into GenBank under accession numbers OQ448931 to OQ448937 and OQ458709 to OQ458712 (See additional file 1).
Prevalence according to the fly species
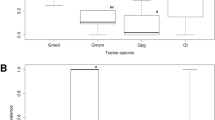
All the four tsetse fly species were infected at different rates by all the endosymbionts, except G. tachinoides flies that was devoid of S. glossinidius infections (Table 2). There was no significant difference (p = 0.28) between the prevalence of S. glossinidius among the four species. The prevalence of Spiroplasma was significantly higher (p = 0.005) in G. p. palpalis (6.35%) and G. tachinoides (5.77%) compared to G. m. submorsitans (1.49) and G. f. fuscipes (2.60%). For Wolbachia, the prevalence was significantly lower (p = 0.005) in G. m. submorsitans (4.79%) compared to other fly species.
When the collection site is considered (prevalence compared between different fly species living in the same environment), symbiont prevalence among the three tsetse species in Lac Iro were similar. However, in Yankari collection site, the Wolbachia infection was significantly higher (p = 0.017) in G. tachinoides.
Discussion
In this study, tsetse symbiotic bacteria (S. glossinidius, Spiroplasma and Wolbachia) were detected in 11.20% of tsetse samples examined with varying prevalence within countries, collection sites and tsetse species. This overall symbiont infection rate is lower compared to that of other previous studies [35,36,37]. Varying levels of infection rates may be attributed to environmental factors (vegetation, humidity, temperature) encountered in different ecological settings and to the intrinsic characters of each tsetse species.
The overall S. glossinidius infection rate of 0.88% obtained in the present study is much lower than the 12.69% global prevalence of Sodalis in tsetse samples from 15 African countries [35]. However, in the same study, similar prevalence was obtained in Burkina Faso, Mali, Ghana and Senegal. The Sodalis prevalence in Chad (1.16%) was lower than the 9.0% previously reported in the same area [38]. The prevalence in Cameroon was also lower than the 37.2% previously reported in the neighbouring area [36]. The variation may be due to PCR screening approach. The primers used in the previous works (pSG2 primers) were targeting a portion of the extrachromosomal plasmid (pSG2) while in this present work, the primers (Hem primers) were targeting the nuclear hemolysin gene. Comparing different sets of primers, it has been reported that the use of the hemolysin gene provided a more reliable assessment of prevalence than the pSG2 that was giving higher but non-consistent prevalence [39]. Moreover, tsetse flies screened in Cameroon for in the present study were all G. p. palpalis while the previous study screened G. tachinoides and G. m. submorsitans. There is no published data on S. glossinidius prevalence in tsetse flies in Nigeria. The presence of S. glossinidius has been suspected to be involved in vector competence of tsetse fly. But the confirmation is still under debate. Numerous reports showed a positive association between Sodalis and trypanosome establishment in the midgut possibly through lectin-inhibitory activity involving the production of N-acetyl glucosamine [26, 38, 40]. However, other studies reported absence of association between the presence of Sodalis and that of Trypanosome [35,36,37].
The Spiroplasma general infection rate of 3.66% obtained in present study is much lower than 17.17% and 44.5% reported respectively in Burkina Faso for G. tachinoides and in Uganda for G. f. fuscipes [30, 41]. The study in Uganda showed a high (0–62% variation of prevalence across the 26 sampling sites ranging and was correlated with geographic origin and season of collection of G. f. fuscipes [30]. The prevalence in Nigeria (10.0%) was significantly higher (p = 0.00) compared to that of Cameroon (0.0%) and Chad (= 0.95%). Few investigations have been done on the prevalence of Spiroplasma in tsetse flies in Africa. There is no data on tsetse Spiroplasma prevalence in the three countries where our samples were collected. Spiroplasma is known to induce reproductive abnormalities and pathogen protective phenotypes in various arthropods hosts [11]. A negative correlation of Spiroplasma with trypanosome infection found in G. f. fuscipes, indicate that Spiroplasma infections may have an important effect in tsetse fly resistance to infection with trypanosomes [30, 42]. Experimental study by Son et al. [42] on G. f. fuscipes established that Spiroplasma infection induced changes in sex–biased gene expression in the reproductive tissues, a depletion in the availability of nutrients in pregnant females resulting in delayed larval development, and compromised sperm fitness. These findings indicate that Spiroplasma could be exploited for reducing tsetse population size and therefore, the disease transmission [42].
The Wolbachia global prevalence of 11.00% and countries prevalence obtained in the present study are in the range of prevalence reported by Doudoumis et al. [17]. All these values were lower than 80.5% and 78.9% reported in Zambia for G. m. morsitans and G. pallidipes respectively [37]. In Cameroon and Chad, the prevalence previously obtained in the same and neighbouring study sites (67.6% and 14.5% respectively) were much higher than those of the present study [36, 38]. The primers used in this study were targeting the Wolbachia specific 16 S rRNA while the previous studies used primers targeting the Wolbachia surface protein (wsp) gene. The comparison of results generated by 16 S rRNA and wsp primers by [43] did not find significant difference between the two primers, even though the prevalence with 16 S rRNA primers (65%) was higher than the prevalence with wsp primers (60.5%). The tsetse species were globally different amongst the two studies. The prevalence in Nigeria (14.07%) was relatively higher than those of the two other countries (Cameroon: 9.68% and Chad: 7.00%). There is no data on Wolbachia infection in tsetse flies in Nigeria.
Wolbachia has received increased interest in recent years because of its high rates of distribution in a wide range of arthropods and nematodes, its unique effects on host physiology and its potential in disease control. Concerning trypanosomiasis, there is no confirmed implication of Wolbachia in tsetse vector competence. Investigations on the tripartite association between tsetse fly, Wolbachia and trypanosomes reported contrasting results. When in G. f. fuscipes there was a negative association between Wolbachia and trypanosome [44] suggesting prevention of trypanosome infections by the presence of Wolbachia; some studies did not find association between presence of Wolbachia and ability of tsetse flies to harbor trypanosomes [36, 37, 43].
Spiroplasma infected the four tsetse fly species, but with a low infection rate on G. m. submorsitans and G. f. fuscipes compared to G. tachinoides and G. p. palpalis. On the contrary, the investigations by Doudoumis et al. [13] found Spiroplasma infection only in palpalis group and none in the morsitans and fusca groups. Attributing this result to their frequent infection with Wolbachia which may have led to development of competitive exclusion with Spiroplasma. Wolbachia was detected in all the four tsetse fly species with varying prevalence within species. This variation agrees with findings from similar studies on tsetse fly symbionts. In the present study, G. tachinoides was found to be more infected with Wolbachia than the three other tsetse fly species. The S. glossinidius was not detected in G. tachinoides, but has been detected in G. tachinoides in other studies [35, 36].
Conclusion
The present study revealed for the first time, the presence of infection by Spiroplasma in tsetse flies in Chad and Nigeria. The infection rates of S. glossinidius, Spiroplasma species and Wolbachia varied between countries and collection sites probably due to environmental factors. Few tsetse flies harboured symbiont co-infections. These findings provide useful information to repertoire of bacterial flora of tsetse flies and call for more investigations to understand the implication of these symbiotic bacteria in the vector competence of tsetse flies. More data could help development of environmentally friendly methods for tsetse and trypanosomiasis control.
Methods
Study areas
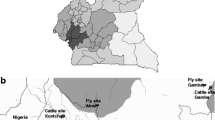
Tsetse flies used in this study were collected from Cameroon, Chad, Nigeria (Fig. 4). In Cameroon flies were trapped in Dodeo (Latitude 7° 27.994’ N and Longitude 12° 04.101′ E) located in the “Faro et Déo” division of the Adamawa region. The village is close to the Cameroon-Nigeria Border. The vegetation is predominantly characterized by gallery forest along rivers and the climate type is of Sudano-Sahelian with rainy season from May to October and a dry season from November to April. This area has a dense hydrographic network and offers pastures for livestock [5, 36].
In Chad, flies were collected in Maro and Lake Iro areas, all situated in the Moyen-Chari province in southern Chad. The area has a climate of Sudano-Sahelian type with A rainy season from May to October and a dry season from November to April of equal duration. Lac Iro is situated between Latitude 9°59’ N and Longitude 19°26′ E, along Salamat River. The vegetation is dominated by floodplains and dense forests containing shrubs. The area is considered as a buffer zone of Zakouma National Park where domestic and wild animals meet [38]. Maro (8°24.807’ N and 18°46.139’ E) is located at Chad-Central African Republic border. The area is crossed by many rivers and their multiple tributaries; the most important is the Chari River and its confluent the Grande Sido [45]. Maro is close to the Bamingui-Bangouran National Parc.
In Nigeria, flies were collected in Yankari and in Ija-Gwari. Yankari Game Reserve (9°45.240’N and 10°30.448E) is situated in the south-central part of Bauchi State in the North-East of the country. It covers an area of 2244 km2 and has a Sudan savannah vegetation. It has well-developed patches of woodland with scattered shrubs and trees. This national park has a rich wildlife diversity. Ija-Gwari is located at 9° 18.860’N and 7° 26.814’E in Tafa Local Government Area of Niger State. The vegetation is of riverine fringing forest forming a dense two-storey canopy. The collection site is an open area where human activities (agriculture and cattle grazing) are regular [46].
Sampling of tsetse
Biconical traps, very effective in catching riverine tsetse of the palpalis group [47] and widely used throughout Africa were used for this study, set up in various tsetse fly-favourable biotopes at least 100 m intervals. Tsetse flies were collected once a day and transported in cool boxes to a base camp. The collected flies were identified using morphological identification keys [48, 49], numbered and dissected on the same day.
Dissection
Flies were dissected in a drop of sterile saline solution as described by Ngomtcho et al. and Shaida et al., [5, 46]. The wings were removed first, followed by legs, proboscis, salivary glands and gut from each fly. After each fly, dissection tools (forceps, pins and slide) were decontaminated by immersion in 5% sodium hypochlorite for approximately ten minutes, followed by immersion in 70% ethanol and final immersion in sterile normal saline. Wings were stored dry for geometric morphometrics. Legs, proboscis and salivary glands were preserved in nucleic acid preservation agent (NAPA: 25 mM sodium citrate, 10 mM EDTA, 70 g ammonium sulphate/100 mL solution, pH 7.5) in 1.5 mL cryotubes. Gut tissue was homogenised in 200 µL of 50 mM Tris-Cl, pH 9.0, using four 2.38 mm metal beads (MoBio Laboratories, Carlsbad, CA, USA). Fifty microliters of the homogenate were added to 500 µL of NAPA. Most of the flies from Chad were dead. For this reason, after the removal of wings, legs and proboscis, the entire abdomen was transferred in ethanol.
DNA extraction
DNA was extracted from homogenised gut tissue in NAPA with the DNeasy Blood and Tissue Kit (Qiagen, Germany) according to the manufacturer’s instructions with slight adjustments (100 µL of homogenate were used for purification and 100 µL elution buffer were used at elution step).
For the dead flies from Chad, DNA was extracted from the abdomen using 5% Chelex-100 Resin (BIO-RAD, Hercules, California, USA). Briefly, the abdomen was transferred from ethanol to a new 1.5 mL centrifuge tube and allowed to dry. The abdomen was then crushed using the micro-pipette tips. Thereafter, 100 µL of chelex 5% solution were added. The mixture was vortexed and incubated at 56° C for 30 min in a Thermomixer. After this incubation, the mixture was vortexed and incubated for an additional 5 min at 95° C. The mixture was then mixed thoroughly before brief centrifugation at 7000 rpm for 1 min and stored at -20 °C.
Molecular detection of Wolbachia
The detection of Wolbachia was carried out by amplifying a 438 bp fragment of the 16 S rRNA gene with the 16 S W-Spec primers (Table 3) designed by Werren & Windsor [50]. PCR amplifications were performed in 20 µL reaction mixture containing 1 X DreamTaq buffer (10X), 150 µM dNTPs, 0.2 µM of each primer, 0.5 U of DreamTaq polymerase and 2 µl of template DNA. The cycling condition was as described by Doudoumis et al. [17]. It started by 95 °C for 5 min followed by 35 cycles of 95 °C for 30 s, 30 s at 54 °C, 1 min at 72 °C and a final extension step of 72 °C for 10 min. Wolbachia positive control sample was the genomic DNA from Glossina morsitans morsitans infected with Wolbachia, donated by the laboratory of Alvaro Acosta Serrano at the Liverpool School of Tropical Medicine. For negative control, ddH2O was used instead of fly DNA template. After amplification, the PCR products were analyzed by electrophoresis on a 1.5% agarose gel containing Stain-G and visualized under UV light. PCR products of one Wolbachia positive sample was randomly selected in each country and sequenced.
Molecular detection of S. glossinidius
The presence of S. glossinidius was determined by PCR with Hem primers (Table 3) that target the gene encoding the haemolysin protein of the bacterium [51]. The primers targeted a 650 bp fragment of the hemolysin gene. The PCR reaction was performed in a final volume of 20 µL containing 1X Dream taq buffer, 150 µM dNTPs, 0.2 µM of each primer, 0.5 U of Dream Taq polymerase and 2 µL of DNA template. PCR cycles were: 95 °C for 5 min; 35 cycles of 95 °C for 30 s, 54 °C for 30 s and 72 °C for 60 s; and a final elongation at 72 °C for 10 min. S. glossinidius positive control sample was the genomic DNA from Glossina morsitans morsitans infected with S. glossinidius, donated by the laboratory of Alvaro Acosta Serrano at the Liverpool School of Tropical Medicine. For negative control, ddH2O was used instead of fly DNA template.
After amplification, the PCR products were analyzed by electrophoresis on a 1.5% agarose gel containing Stain-G and visualized under UV light. PCR products of two S. glossinidius positive samples were randomly selected in each country and sequenced.
Molecular detection of Spiroplasma
Screening for Spiroplasma was carried out by amplifying 455 bp fragment of 16 S rRNA as described by Doudoumis et al. [13] with specific primers (Table 3). PCR reactions were performed in 20 µl reaction mixture containing 1X DreamTaq buffer, 150 µM dNTPs, 0.65 µM of each primer, 0.5 U of DreamTaq polymerase and 2 µl of template DNA. The PCR temperature profile was 95 °C for 5 min followed by 35 cycles of 95 °C for 30 s, 30 s at 59 °C, 1 min at 72 °C and a final extension step of 72 °C for 10 min.
To obtain Spiroplasma positive control, a PCR with randomly selected samples was ran. The sample with the band at the target position was re-amplified and its PCR product was purified and sequenced for confirmation. For Negative controls, ddH2O was used instead of the DNA template.
After amplification, the PCR products were analyzed by electrophoresis on a 1.5% agarose gel containing Stain-G and visualized under UV light. PCR products of two Spiroplasma positive samples were randomly selected in each country and sequenced.
Sequencing
Selected positive samples, were re-amplified in PCR reaction volume of 50 µL (maintaining the same concentration of reactants). Five microliters were visualised on 1.5% agarose gel for confirmation of the amplification. The remaining 45 µL PCR products were separated on 2% agarose gel. The amplicons bands were extracted and purified using GeneJET Gel Extraction Kit (Thermo Scientific) according to manufacturer instructions. The purified amplicons were sent for sanger sequencing to a commercial company (SeqLab, Göttingen, Germany).
Phylogenetic analysis
Obtained sequences were manually checked and edited using Geneious Pro version 5.5.9 software. The Basic Local Alignment Search Tool (BLASTn) [52] from National Center for Biotechnology Information (NCBI) was used to confirmed identity of amplicons and to determine closest related sequences in the GenBank. Sample sequences and reference sequences obtained from NCBI were aligned using MUSCLE [53] alignment tool with its default setting implemented in MEGAX [54] which was also used to infer phylogenetic relationships. Maximum likelihood method was performed with the Kimura-2 model [55] for Wolbachia and Spiroplasma, then Hasegawa-Kishino-Yano model [56] for Sodalis as determined by the MEGA model finder tool with 1000 bootstraps replicates.
Statistics data analysis
Endosymbionts hosted by tsetse flies were expressed in percentage as symbiont prevalence. The Pearson’s chi-square test (χ2) was used to compare symbiont prevalence between countries and collection sites. The differences were considered significant when the p-values were lower than 0.05. Statistical tests were performed using SPSS for Windows, version 20.
Data availability
The datasets generated and analysed during this study are included in this publication. The 16 S rRNA and hemolysin partial gene sequences generated have been deposited into GenBank under accession numbers OQ448931 to OQ448937 and OQ458709 to OQ458712.
Abbreviations
- BLAST:
-
Basic Local Alignment Search Tool
- NAPA:
-
Nucleic Acid Preservation Agent
- NCBI:
-
National Center for Biotechnology Information
References
WHO. Trypanosomiasis, human African (sleeping sickness) [Internet]. 2021 [cited 2021 Aug 9]. Available from: https://www.who.int/news-room/fact-sheets/detail/trypanosomiasis-human-african-(sleeping-sickness).
Simarro PP, Cecchi G, Franco JR, Paone M, Diarra A, Priotto G et al. Monitoring the Progress towards the Elimination of Gambiense Human African Trypanosomiasis. Matovu E, editor. PLoS Negl Trop Dis. 2015;9(6):e0003785.
Gibson W. Resolution of the species problem in African trypanosomes. Int J Parasitol. 2007;37(8–9):829–38.
Adams ER, Hamilton PB, Gibson WC. African trypanosomes: celebrating diversity. Trends Parasitol. 2010;26(7):324–8.
Ngomtcho SCH, Weber JS, Ngo Bum E, Gbem TT, Kelm S, Achukwi MD. Molecular screening of tsetse flies and cattle reveal different Trypanosoma species including T. grayi and T. theileri in northern Cameroon. Parasit Vectors. 2017;10(1):631.
Leak SGA. Tsetse biology and ecology: their role in the epidemiology and control of trypanosomosis. New York: CABI Publishing in association with International Livestock Research Institute;: Oxon; 1999. p. 568.
WHO. Control and surveillance of human African trypanosomiasis: report of a WHO expert committee [Internet]. Geneva: World Health Organization. ; 2013 [cited 2022 Feb 20]. (WHO technical report series; no. 984). Available from: https://apps.who.int/iris/handle/10665/95732.
Uilenberg G, Boyt WP. A field guide for the diagnosis, treatment and prevention of african animal trypanosomosis. New ed. Rome: Food and Agriculture Organization of the United Nations; 1998. p. 158.
Griffith B. Analysis Of The Gut-Specific Microbiome Of Field-Captured Tsetse Flies, And Its Potential Relevance To Host Trypanosome Vector Competence [Internet] [Masters of Public Health]. [Department of Epidemiology of Microbial Diseases, Yale School of Public Health, New Haven]: Yale University; 2014. Available from: https://elischolar.library.yale.edu/ysphtdl/1115.
Geiger A, Fardeau ML, Njiokou F, Ollivier B. Glossina spp. gut bacterial flora and their putative role in fly-hosted trypanosome development. Front Cell Infect Microbiol [Internet]. 2013 [cited 2021 Aug 1];3. Available from: http://journal.frontiersin.org/article/https://doi.org/10.3389/fcimb.2013.00034/abstract.
Attardo GM, Scolari F, Malacrida A. Bacterial Symbionts of Tsetse Flies: Relationships and Functional Interactions Between Tsetse Flies and Their Symbionts. In: Kloc M, editor. Symbiosis: Cellular, Molecular, Medical and Evolutionary Aspects [Internet]. Cham: Springer International Publishing; 2020 [cited 2021 Nov 4]. p. 497–536. (Results and Problems in Cell Differentiation; vol. 69). Available from: https://link.springer.com/https://doi.org/10.1007/978-3-030-51849-3_19.
Weiss BL, Maltz M, Aksoy S. Obligate symbionts activate immune system development in the tsetse fly. J Immunol Baltim Md 1950. 2012;188(7):3395–403.
Doudoumis V, Blow F, Saridaki A, Augustinos A, Dyer NA, Goodhead I, et al. Challenging the Wigglesworthia, Sodalis, Wolbachia symbiosis dogma in tsetse flies: Spiroplasma is present in both laboratory and natural populations. Sci Rep. 2017;7(1):4699.
Akman L, Yamashita A, Watanabe H, Oshima K, Shiba T, Hattori M, et al. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat Genet. 2002;32(3):402–7.
Baldo L, Dunning Hotopp JC, Jolley KA, Bordenstein SR, Biber SA, Choudhury RR, et al. Multilocus sequence typing system for the Endosymbiont Wolbachia pipientis. Appl Environ Microbiol. 2006;72(11):7098–110.
Hughes GL, Rasgon JL. Wolbachia Infections in Arthropod Hosts. In: Insect Pathology [Internet]. Elsevier; 2012 [cited 2021 Aug 7]. p. 351–66. Available from: https://linkinghub.elsevier.com/retrieve/pii/B9780123849847000099.
Doudoumis V, Tsiamis G, Wamwiri F, Brelsfoard C, Alam U, Aksoy E, et al. Detection and characterization of Wolbachia infections in laboratory and natural populations of different species of tsetse flies (genus glossina). BMC Microbiol. 2012;12(1):3.
Alam U, Medlock J, Brelsfoard C, Pais R, Lohs C, Balmand S et al. Wolbachia Symbiont Infections Induce Strong Cytoplasmic Incompatibility in the Tsetse Fly Glossina morsitans. Schneider DS, editor. PLoS Pathog. 2011;7(12):e1002415.
Chouin-Carneiro T, Ant TH, Herd C, Louis F, Failloux AB, Sinkins SP. Wolbachia strain wAlbA blocks Zika virus transmission in Aedes aegypti. Med Vet Entomol. 2020;34(1):116–9.
Kambris Z, Cook PE, Phuc HK, Sinkins SP. Immune activation by life-shortening Wolbachia and reduced filarial competence in mosquitoes. Science. 2009;326(5949):134–6.
Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, Lu G, Pyke AT, Hedges LM, et al. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell. 2009;139(7):1268–78.
van den Hurk AF, Hall-Mendelin S, Pyke AT, Frentiu FD, McElroy K, Day A, et al. Impact of Wolbachia on infection with chikungunya and yellow fever viruses in the mosquito vector aedes aegypti. PLoS Negl Trop Dis. 2012;6(11):e1892.
Wang J, Weiss BL, Aksoy S. Tsetse fly microbiota: form and function. Front Cell Infect Microbiol [Internet]. 2013 [cited 2021 Aug 2];3. Available from: http://journal.frontiersin.org/article/https://doi.org/10.3389/fcimb.2013.00069/abstract.
Toh H, Weiss BL, Perkin SAH, Yamashita A, Oshima K, Hattori M, et al. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 2006;16(2):149–56.
Channumsin M, Ciosi M, Masiga D, Turner CMR, Mable BK. Sodalis glossinidius presence in wild tsetse is only associated with presence of trypanosomes in complex interactions with other tsetse-specific factors. BMC Microbiol. 2018;18(1):163.
Dale C, Welburn SC. The endosymbionts of tsetse flies: manipulating host-parasite interactions. Int J Parasitol. 2001;31(5–6):628–31.
Jaenike J, Unckless R, Cockburn SN, Boelio LM, Perlman SJ. Adaptation via symbiosis: recent spread of a Drosophila Defensive Symbiont. Science. 2010;329(5988):212–5.
Lukasik P, Guo H, van Asch M, Ferrari J, Godfray HCJ. Protection against a fungal pathogen conferred by the aphid facultative endosymbionts Rickettsia and Spiroplasma is expressed in multiple host genotypes and species and is not influenced by co-infection with another symbiont. J Evol Biol. 2013;26(12):2654–61.
Xie J, Vilchez I, Mateos M. Spiroplasma bacteria enhance survival of Drosophila hydei attacked by the parasitic Wasp Leptopilina heterotoma. PLoS ONE. 2010;5(8):e12149.
Schneider DI, Saarman N, Onyango MG, Hyseni C, Opiro R, Echodu R, et al. Spatio-temporal distribution of Spiroplasma infections in the tsetse fly (Glossina fuscipes fuscipes) in northern Uganda. PLoS Negl Trop Dis. 2019;13(8):e0007340.
Kariithi HM, Meki IK, Schneider DI, De Vooght L, Khamis FM, Geiger A, et al. Enhancing vector refractoriness to trypanosome infection: achievements, challenges and perspectives. BMC Microbiol. 2018;18(S1):179.
De Vooght L, Van Keer S, Van Den Abbeele J. Towards improving tsetse fly paratransgenesis: stable colonization of Glossina morsitans morsitans with genetically modified Sodalis. BMC Microbiol. 2018;18(1):165.
Weiss B, Vigneron A, Aksoy E, Aksoy S. Paratransgenic manipulation of microRNA-275 expression in the tsetse fly midgut, and the downstream impact on trypanosome infection outcomes. In ESA; 2018 [cited 2021 Aug 2]. Available from: https://esa.confex.com/esa/2018/meetingapp.cgi/Paper/138889.
Yang L, Weiss BL, Williams AE, Aksoy E, Orfano A, de Son S. Paratransgenic manipulation of a tsetse microRNA alters the physiological homeostasis of the fly’s midgut environment. PLOS Pathog. 2021;17(6):e1009475.
Dieng MM, Dera K, sida M, Moyaba P, Ouedraogo GMS, Demirbas-Uzel G, Gstöttenmayer F, et al. Prevalence of Trypanosoma and Sodalis in wild populations of tsetse flies and their impact on sterile insect technique programmes for tsetse eradication. Sci Rep. 2022;12(1):3322.
Kame-Ngasse GI, Njiokou F, Melachio-Tanekou TT, Farikou O, Simo G, Geiger A. Prevalence of symbionts and trypanosome infections in tsetse flies of two villages of the Faro and Déo division of the Adamawa region of Cameroon. BMC Microbiol. 2018;18(1):159.
Mulenga GM, Namangala B, Gummow B. Prevalence of trypanosomes and selected symbionts in tsetse species of eastern Zambia. Parasitology. 2022;149(11):1406–10.
Djoukzoumka S, Mahamat Hassane H, Khan Payne V, Ibrahim MAM, Tagueu Kanté S, Mfopit YM, et al. Sodalis glossinidius and Wolbachia infections in wild population of Glossina morsitans submorsitans caught in the area of Lake Iro in the south of Chad. J Invertebr Pathol. 2022;195:107835.
Wongserepipatana M. Prevalence and associations of Trypanosoma spp. and Sodalis glossinidius with intrinsic factors of tsetse flies [Internet] [PhD]. University of Glasgow; 2016 [cited 2023 Feb 11]. Available from: https://eleanor.lib.gla.ac.uk/record=b3173282.
Farikou O, Njiokou F, Mbida Mbida JA, Njitchouang GR, Djeunga HN, Asonganyi T, et al. Tripartite interactions between tsetse flies, Sodalis glossinidius and trypanosomes—An epidemiological approach in two historical human african trypanosomiasis foci in Cameroon. Infect Genet Evol. 2010;10(1):115–21.
Khamlichi SE, Maurady A, Asimakis E, Stathopoulou P, Sedqui A, Tsiamis G. Detection and Characterization of Spiroplasma and Wolbachia in a Natural Population of Glossina Tachinoides. 2020 Dec 7 [cited 2022 Feb 26]; Available from: https://easychair.org/publications/preprint/jnLt.
Son JH, Weiss BL, Schneider DI, Dera K, sida M, Gstöttenmayer F, Opiro R, et al. Infection with endosymbiotic Spiroplasma disrupts tsetse (Glossina fuscipes fuscipes) metabolic and reproductive homeostasis. PLoS Pathog. 2021;17(9):e1009539.
Kanté ST, Melachio T, Ofon E, Njiokou F, Simo G. Detection of Wolbachia and different trypanosome species in Glossina palpalis palpalis populations from three sleeping sickness foci of southern Cameroon. Parasit Vectors. 2018;11(1):630.
Alam U, Hyseni C, Symula RE, Brelsfoard C, Wu Y, Kruglov O, et al. Implications of microfauna-host interactions for Trypanosome Transmission Dynamics in Glossina fuscipes fuscipes in Uganda. Appl Environ Microbiol. 2012;78(13):4627–37.
Ibrahim MAM, Weber JS, Ngomtcho SCH, Signaboubo D, Berger P, Hassane HM, et al. Diversity of trypanosomes in humans and cattle in the HAT foci Mandoul and Maro, Southern Chad—A matter of concern for zoonotic potential? Phillips MA, editor. PLoS Negl Trop Dis. 2021;15(6):e0009323.
Shaida SS, Weber JS, Gbem TT, Ngomtcho SCH, Musa UB, Achukwi MD, et al. Diversity and phylogenetic relationships of Glossina populations in Nigeria and the cameroonian border region. BMC Microbiol. 2018;18(1):180.
Challier A, Laveissière C. Un nouveau piège pour la capture des glossines (Glossina: Diptera, Muscidae) : description et essais sur le terrain. Cah ORSTOMSérie Entomol Médicale Parasitol. 1973;11:251–62.
Pollock JN. Training manual for Tsetse control personnel Vol. 1: Tsetse biology, systematics and distribution, techniques [Internet]. Rome, Italy: FAO; 1982 [cited 2022 Mar 20]. 280 p. Available from: https://www.fao.org/documents/card/fr/c/4f831550-7aa2-57f8-8a54-b5714b0abb2b/.
Leak SGA, Ejigu D, Vreysen MJB. Collection of entomological baseline data for tsetse area-wide integrated pest management programmes. Rome: FAO; 2008. (FAO animal production and health).
Werren JH, Windsor DM. Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc R Soc Lond B Biol Sci. 2000;267(1450):1277–85.
Pais R, Lohs C, Wu Y, Wang J, Aksoy S. The obligate mutualist Wigglesworthia glossinidia influences reproduction, digestion, and immunity processes of its host, the tsetse fly. Appl Environ Microbiol. 2008;74(19):5965–74.
Sayers EW, Bolton EE, Brister JR, Canese K, Chan J, Comeau DC, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2021;50(D1):D20–6.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018;35(6):1547–9.
Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–20.
Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22(2):160–74.
Acknowledgements
The authors would like to thank the Institute of Agricultural Research for Development: IRAD in Cameroon, the “Institut de Recherche en Elevage pour le Développement: IRED”, and the “Programme National de Lutte contre la Trypanosomiase Humaine Africaine : PNLTHA ” in Chad, The Nigerian Institute for Trypanosomiasis Research: NITR in Nigeria for their assistance and contributions during fieldwork. We are grateful to the African Centre of Excellence for Neglected Tropical Diseases and Forensic Biotechnology: ACENTDFB, The TOZARD Research Laboratory and the Centre for Biomolecular Interactions Bremen: CBIB, AG Kelm for the laboratory facilities. Special thanks go to Petra Berger for the technical assistance. The laboratory of Alvaro Acosta Serrano at the Liverpool School of Tropical Medicine for the donation of Genomic DNA of Glossina morsitans morsitans infected with S. glossinidius and Wolbachia that was used as positive control. We are grateful to the administrative authorities, traditional leaders, rangers and security personnel for their collaboration and field survey supports.
Funding
Financial support for the project came from WANIDA (West African Network of Infectious Diseases ACEs) doctoral scholarship (WAN100629X), scholarship package funded by the Agence Française de Développement (AFD), Institut de Recherche pour le Développement (IRD) and World Bank under the African Centers of Excellence (ACE) Partner Project, from Deutsche Forschungsgemeinschaft (DFG; project grants to SK, Ke428/8–1, Ke428/10–1, Ke428/13 − 1), and from Africa Centre of Excellence for Neglected Tropical Diseases and Forensic Biotechnology (ACE-NTDFB). EOB is a recipient of Emerging Global Leader (K43) Award and supported by the Fogarty International Center of the National Institutes of Health under Award Number K43TW012015. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
YMM, JK and SK conceived the study; YMM, DMA, MM, EOB, MNS, JK and SK designed the study protocol; YMM, JSE, GDD, MAMI, DS, DMA, SK collected samples; DMA, MM and SK supervised the fieldwork; MM, EOB, MNS, JK and SK supervised laboratory experiments; YMM, JSE, GDC, MAMI and DS carried out the experiments; YMM drafted the manuscript; All authors read, revised and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Additional file 1:
Sequence accession numbers. Table indicating which accession number corresponds to each sequence.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Mfopit, Y.M., Engel, J.S., Chechet, G.D. et al. Molecular detection of Sodalis glossinidius, Spiroplasma species and Wolbachia endosymbionts in wild population of tsetse flies collected in Cameroon, Chad and Nigeria. BMC Microbiol 23, 260 (2023). https://doi.org/10.1186/s12866-023-03005-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-023-03005-6