Abstract
Background
Helicobacter pylori (H. pylori) infects nearly half of the world’s population with a variation in incidence among different geographic regions. Genetic variants in the promoter regions of the IL1B gene can affect cytokine expression and creates a condition of hypoacidity which favors the survival and colonization of H. pylori. Therefore, the aim of this study was to characterize the polymorphic sites in the 5′- region [−687_ + 297] of IL1B in H. pylori infection using in silico tools.
Results
A total of five nucleotide variations were detected in the 5′-regulatory region [−687_ + 297] of IL1B which led to the addition or alteration of transcription factor binding sites (TFBSs) or composite regulatory elements (CEs). Genotyping of IL1B − 31 C > T revealed a significant association between -31 T and susceptibility to H. pylori infection in the studied population (P = 0.0363). Comparative analysis showed conservation rates of IL1B upstream [−368_ + 10] region above 70% in chimpanzee, rhesus monkey, a domesticated dog, cow and rat.
Conclusions
In H. pylori-infected patients, three detected SNPs (− 338, − 155 and − 31) located in the IL1B promoter were predicted to alter TFBSs and CE, which might affect the gene expression. These in silico predictions provide insight for further experimental in vitro and in vivo studies of the regulation of IL1B expression and its relationship to H. pylori infection. However, the recognition of regulatory motifs by computer algorithms is fundamental for understanding gene expression patterns.
Similar content being viewed by others
Background
Helicobacter pylori (H. pylori) is a Gram-negative, spiral-shaped and microaerophilic bacterium that infects nearly half of the world’s population with a variation in incidence among different geographic regions [1, 2]. Epidemiological studies have indicated that the highest prevalence of H. pylori was found in Africa (79.1%), and the lowest prevalence was found in Northern America (37.1%) and Oceania (24.4%) with an overall global H. pylori prevalence of 44.3%, ranging from 50.8% in developing countries to 34.7% in developed countries [3,4,5,6]. The global annual recurrence rate of H. pylori was (4.3%) and it was found to be related to the human development index and prevalence of infection [3]. However, the clinical aspects of chronic infection with H. pylori vary from gastroduodenal inflammation and peptic ulceration to the most dangerous aspects, gastric carcinoma and mucosa-associated lymphoid tissue (MALT) lymphoma [7, 8]. Also, H. pylori may be implicated in several extra-gastric diseases such as iron deficiency anemia, idiopathic thrombocytopenic purpura, several dermatological disorders, hepatic encephalopathy, diabetes, and pulmonary and cardiovascular diseases [7, 9]. Indeed, the susceptibility to H. pylori infection and its diverse clinical presentation is determined by multiple factors, including heterogeneity of H. pylori strains and their virulence factors, environmental factors, and the host genetic background, especially those regarding polymorphisms in certain cytokines, gene regulation and their receptor antagonist genes [10,11,12,13]. One of these cytokines is the interleukin 1-beta (IL1B) gene.
IL-1 family genes, spanning ~ 430 kb, cluster on chromosome 2q13–21 and consist of IL-1A, IL-1B, and IL-1RN genes which encode the pro-inflammatory cytokines IL-1α and IL-1β and the endogenous receptor antagonist IL-1ra, respectively [14]. IL-1β, the crucial cytokine in the gastrointestinal tract [15], has a variety of biological activities on a wide range of tissues and plays an important role in inflammatory, metabolic, physiologic, hematopoietic, and immunological processes (for a review, see references (16) and [17]). Because of the ability of IL-1β to inhibit gastric acid secretion, it may have a profound effect on the natural history of H. pylori infection by allowing expansion of H. pylori colonization from the gastric antrum to the corpus [15, 18,19,20]. On a molar basis, IL-1β is 100-fold more potent than proton pump inhibitors (PPIs) and 6000-fold more potent than H2 receptor antagonists [21].
IL-1β is expressed at high levels in myeloid cell lineages in response to tissue injury and microbial invasion [22, 23]. Also, many different types of cells, e.g. B cells, T cells, NK cells, dendritic cells, fibroblasts and epithelial cells, express this protein in response to a broad range of stimuli and under inflammatory conditions but at much lower level [24,25,26]. LPS-inducible IL1β expression is regulated by two regions: a proximal promoter that contains a TATA box and an upstream LPS-responsive enhancer (located between − 3757 and − 2729), which is also known as the upstream inducible sequence [27, 28]. In monocyte, this promoter is packaged into a highly accessible chromatin structure which is characterized by the constitutive association of PU.1 and C/ EBPβ, but the inducible association of RNA polymerase II [24, 29, 30]. The following multiple transcription factors that constitutively and inducibly associate with IL-1β regulatory regions have been identified: Spi-1/PU.1 (Spi1), NF-κB, C/EBPβ, AP-1, TBP, SSRP, or c-Jun and c-fos [29].
Genetic variations in the promoter region of genes encoding cytokines were shown to correlate with individual differences in the expression of respective cytokines which may influence the intensity of the inflammatory response and susceptibility to many diseases [31,32,33,34,35]. IL-1B gene has two allelic variants (CT; dbSNP: rs16944) and (TC; dbSNP: rs1143627) which are located at positions − 511 and − 31, respectively, in the promoter region. These SNPs have been proposed to be associated with the susceptibility to H. pylori infection; and H. pylori-related gastric cancer and peptic ulcer disease [36], but it is still a contradictory topic of debate. Many studies have been published analyzing the contribution of IL1B promoter polymorphisms to H. pylori susceptibility with conflicting results explained, in part, by ethnic differences [19, 32, 36]. In the present study, genomic DNA Sanger sequencing was applied to detect SNPs in the region [−687_ + 297] of IL1B in H. pylori-infected patients; and bioinformatics analyses were used to study whether these mutations would alter transcription factor binding sites (TFBSs). Further computational analysis was also made to investigate other potential regulatory elements in this region. Finally, comparative profiling was conducted to assess the conservation of these genetic variations in 11 species. However, to our knowledge, the association between promoter polymorphisms of the IL1B gene and H. pylori infection in the Sudanese population has not been studied. It is imaginable that individual differences in H. pylori susceptibility or individual differences in H. pylori-related disease severity are linked to genetically determined differences in IL1B production. Therefore, studying the regulation of IL1B gene expression is of great significance.
Results
Nucleotide variations in the 5′-regulatory region of the IL1B gene
In Sudanese H. pylori infected patients, a total of five nucleotide variations were detected in the 5′-regulatory region. Among which, three are bimodally mutated heterozygous SNPs, and they were newly discovered; the positions of these three SNPs are − 338, − 155 and + 38. The other two SNPs were rs16944 and rs1143627, see Table 1 for more illustration. The nucleotide sequences of the IL1B 5′- region [−687_ + 297] were deposited in the GenBank database under the following accession numbers: from MT767762 to MT767775.
In silico prediction of the IL1B promoter regions
Five types of promoter prediction programs were employed to predict the promoter regions of the IL1B 5′- region [−687_ + 297] and the results are presented in Table 2. The Promoter 2.0 Prediction Server predicted no promoter region. Neural Network Promoter Prediction (NNPP version 2.2) predicted three promoter regions, located at -328 bp, − 124 bp and + 1 bp relative to the IL1B translational start codon (transcript NM_000576.3), whose prediction scores were 0.97, 0.60 and 0.96, respectively. While Fprom, TSSG and TSSW programs predicted one promoter region, + 1 bp, which is the only region predicted by all used prediction programs.
In silico analysis of predicted IL1B promoter regions
ENCODE data showed a high level of DNase I hypersensitivity, promoter associated histone modifications and transcription factor occupancy patterns at − 124 and + 1 bp promoter regions. While no Nuclease hypersensitivity around − 328 bp region. However, no CpG islands were detected in the predicted promoter regions (Fig. 1). Also, ENCODE data confirmed no presence of the CpG island in the predicted promoters.
Conservancy of the predicted promoter region [− 368_ + 10]
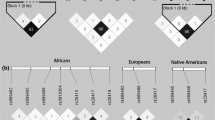
The ECR Browser revealed mammalian conservation for the [− 368_ + 10] region in chimpanzee (Pan troglodytes - pan-Tro2) (99.2%), rhesus monkey (Macaca mulatta - rheMac2) (93.4%), domesticated dog (Canis lupus familiaris - canFam2) (73%), cow (Bos Taurus - bosTau3) (71.1%), rat (Rattus norvegicus - rn4) (70.5%) and mouse (Mus musculus - mm9) (69.3%). But the region was not conserved in opossum (Monodelphis domestica), chicken (Gallus gallus), frog (Xenopus laevis), zebrafish (Danio rerio), fugu pufferfish (Takifugu rubripes), and spotted green pufferfish (Tetraodon nigrovoridis), see Fig. 2 for more illustration.
Shows conservation of the IL1B 5′-upstream region compared to the human sequence in region [−687_ + 297] (hg19 chr2:113594193–113,594,801). The height of the conservation plot at each position represents the number of nucleotides conserved in a window of 100 nucleotides centered on that position. The pink rectangles at the top of the plot represent the evolutionary conserved regions, which are defined as regions of 100 nucleotides with at least 70% identity. Blue boxes represent the IL1B exons, while yellow boxes indicate the IL1B 5′-UTR. Intragenic positions are highlighted in red, or in green when corresponding to transposable elements and simple repeats. Overview from the ECR Browser [37]
Prediction of TFBSs
In this study, five programs were used to predict TFBSs and to insure proper analysis we only selected factors that are predicted by three out of the five programs or the factors predicted by two programs but verified in the literature. The five prediction programs reported multiple putative TFBSs within the [−368_ + 10] region, see Table 3 and Figs. 3, 4 and 5. However, screening of this region, by using the NCBI SNP databases (dbSNPs), revealed the presence of 9 SNPs upstream of the IL1B core promoter region which are shown in Table 4. The ECR Browser and NCBI BLASTn showed the conservation of these SNPs in chimpanzee, rhesus monkey, cow and dog. Mulan revealed multiple TFBSs to be located at rs749558279, rs140623868 and -338A > T. The overview of conserved TFBSs predicted by Mulan to be conserved (100%) between human, chimpanzee, rhesus monkey, cow and domesticated dog is summarized in Table 4.
a) Illustrates PCR-CTPP products analyzed on a 2% agarose gel stained with ethidium bromide. Three genotypes can be seen. 1and 3 show 240 bp, 155 bp and 122 bp which indicate a heterozygous genotype. 2, 5 and 6 show 240 bp and 122 bp which indicate a homozygous T genotype. While 4 and 7 show 240 bp and 155 bp which indicate a homozygous C genotype. b) shows the sequencing result of the polymorphism by using Finch TV software. c) Illustrates the conservation of (TC; dbSNP: rs1143627) at position − 31 among different species
Prediction of CEs
MatrixCatch was used to find known regulatory elements (both single sites and pairs) which were verified experimentally. Also, it found novel regulatory elements by computational comparison but without experimental verification on functionality. These elements were found by using similarity to known ones in a library of CE models [38]. The summary of predicted CEs by MatrixCatch is presented in Table 5.
Allele and genotype frequencies of IL1B-31 and susceptibility to H. pylori infection
Sixty-one patients and 61 uninfected controls were successfully genotyped for the IL1B-31 C/T polymorphism (Fig. 4). The frequency of allele T and IL1B-31 T/C + T/T genotypes were significantly higher in H. pylori-infected individuals compared to uninfected controls (36.89% versus 23.77%, P = 0.0363 and 54.1% versus 34.43%, P = 0.0445, resp.). The allelic and genotype distributions of IL1B-31 C/T polymorphism followed those expected in Hardy-Weinberg equilibrium (HWE) for control population (P = 0.1366) (Table 6).
Discussion
Genetic variants in the promoter region of IL1B gene can affect cytokine expression and create a condition of hypoacidity which favors the survival and colonization of H. pylori [15, 36]. In the present study, we functionally analyzed SNPs in the IL1B 5′-region [−687_ + 297] of Sudanese patients infected with H. pylori and developed divergent clinical outcomes. We observed three novel mutations (− 338, −155 and + 38) and interestingly, two of them (− 338 and − 155) were located at in silico-predicted promoter regions. Thus, these mutations might play a role in regulating the expression of IL1B. In this study, the computational analysis predicted three promoter regions at − 328, − 124 and + 1, but two of them (− 328 and − 124) were only predicted by the NNPP algorithm that uses neural networks (NNs). NNs have been applied for promoter prediction since 1991 [39]. The study conducted by Liu and States et al. compared different available prediction techniques during the development of their own technique, and showed that although NNPP2.2 is competitive with several other freely available techniques, the technique suffers from a high level of false positives [40]. However, many studies have used this technique for promoter predictions such as [41,42,43,44]. Clearly, the result obtained by this technique or other in silico tools cannot substitute for the experimental proofs but it can provide a direction or guidance for such experiments to validate computational predictions.
Nuclease hypersensitivity and histone modifications are characteristic for cis-regulatory regions such as promoters. The ENCODE data shows these hallmarks to be present in the putative promoter region at the + 1 bp region. The upstream region around − 124 bp showed some of these characteristics, although to a lesser degree, while the region around − 328 bp showed only histone marks [45,46,47]. Also, no CpG islands were detected in predicted promoter regions, however, most promoters with a TATA box do not have high GC content [48]. In silico comparative analysis showed the [−368_ + 10] region to be mammalian conserved, with conservation rates above 70% in chimpanzee, rhesus monkey, a domesticated dog, cow and rat. This conservation might indicate a possible regulatory role for this region (Fig. 2). But the region was not conserved in opossum, chicken, frog, zebrafish, fugu pufferfish, and spotted green pufferfish; it is possible that the regulation of IL1B in these species is controlled by a different mechanism or pathway.
Regulation of gene transcription depends on the interaction between TFs and TFBSs. Any changes in these sites may develop significant effects on the binding of TFs to regulatory sequences and then the expression products of genes [44, 49, 50]. In this study, an in silico-based prediction analysis using different algorithms indicated that the transcription factors NF, C/EBP, Spi-1/PU.1, NF-kappaB, AP-1, TBP, IRFs and STAT, c-Myb and GATA-1 are involved in the regulation of IL1B gene expression and have the potential to bind in the polymorphic regions (Table 3). This indication is in agreement with the results of previous studies [26, 51, 52]. The two novel SNPs located in the in silico-predicted promoter region led to the addition or alteration of the TFBSs. As illustrated in Table 7, − 338 (A > T) polymorphism resulted in the alteration of GR to PU.1 and the − 155(G > C) polymorphism led to an addition of a C/EBPbeta. T allele in position − 31 instead of C allele resulted in an addition of RSRFC4 protein. This finding is partially in agreement with experiments that assessed allele-specific oligonucleotides for − 31. The experiments reported that there were one or more TFs resulting in a fivefold increase in DNA binding on the IL1B-31 T oligonucleotide after LPS stimulation. These TFs may be unable to interact with the C-bearing IL1B-31 allele to form the transcription initiation complex [53].
The extensively studied SNPs in relation to H. pylori infection (−31 and − 511) were also detected in our patients. We observed a significant association between -31 T and susceptibility to H. pylori infection in the studied population (P = 0.0363). This result is in concordance with a number of studies conducted in different ethnic groups that showed an association between IL1B-31 T and H. pylori infection [53,54,55,56,57,58]. Also, there are some studies that found a negative association [33, 58, 59]. This variation could be due to differences in genetic backgrounds of the studied population, the method of genotyping and sample size [36, 60]. Interestingly, we found that the T-511C SNP was not located in the in silico-predicted promoter regions, hence it could not affect the expression of IL1B. While − 31 which involves a TATA-box could directly affect the induction of IL1B. These findings are in agreement with the result obtained by Al-Omer et al. that the − 31 polymorphism was markedly affecting DNA-protein interactions in vitro while − 511 does not alter in vitro protein secretion and its effect may be mediated by linkage disequilibrium (LD) with − 31 [53]. Also, R Kimura et al. found that the expression of the -31 T allele was 2.2 times of the -31C allele and this higher transcription efficiency may correspond to the fact that C-31 T is located in a TATA box [61]. In contrast, other observations of IL-1β production have suggested that there was no significant association between the known allelisms in the IL-1B gene and IL-1β induction in vitro and that the -31C was the higher expressing allele in vivo [61,62,63]. However, the production of IL1β is affected by several factors besides gene polymorphisms such as epigenetic conditions and other genetic backgrounds. To exclude the influence of trans-acting factors which are able to confound the effects of the polymorphisms, the allele-specific transcript quantification coupled with haplotype analysis [61, 64] is recommended to identify the cis-acting effect of T-511C polymorphism and our novel detected polymorphisms (− 338 and − 155) on the IL1B transcription and susceptibility to multifactorial diseases including H. pylori infection.
However, recognition of regulatory motifs by computer algorithms is fundamental for understanding gene expression patterns, as well as, cell specificity and development [49]. Identifying SNPs that might be a genetic modifiers in IL1B gene may be valuable in preventive, diagnostic, and therapeutic strategies against the incidence and progression of H. pylori infection. This study revealed three nucleotide variations in the IL1B 5′-region which possibly lead to modification of transcriptional regulation in H. pylori infection, however, this conclusion requires further in vitro and in vivo validation in subsequent studies.
Conclusions
In H. pylori-infected patients, three detected SNPs located in the IL1B promoter were predicted to alter CEs and TFBSs, which might affect the gene expression. This computational analysis provide insight for further experimental in vitro and in vivo studies of the regulation of IL1B expression and its relationship to H. pylori infection. However, recognition of regulatory motifs by computer algorithms is fundamental for understanding gene expression patterns.
Methods
Study methodology
In this study, genomic DNA Sanger sequencing was used to detect SNPs in the region [−687_ + 297] of IL1B in 14 H. pylori-infected patients. Then, computational analyses of the IL-1B promoter region [−687_ + 297] were applied in two steps: 1) in silico prediction of the promoter region and 2) in silico analysis of the predicted promoter region [−368_ + 10]. Furthermore, genotyping of IL1B-31 C > T polymorphism was performed using PCR-CTPP in 122 participants to study its association with the susceptibility to H. pylori infection in the Sudanese population. The methodology followed in this study is described in Fig. 6.
Study setting and study population
This study was carried out at public and private hospitals in Khartoum state. The hospitals included Ibin Sina specialized hospital, Soba teaching hospital, Modern Medical Centre and Al Faisal Specialized Hospital. Sample size was calculated using Epi Info software version 7 [65, 66]. The matched case-control formula was selected assuming 95% confidence level, 80% power of study, 1 ratio of control to case, 15% of controls exposed, 3.36 odds ratio and 37.2% of cases exposed. Based on the sample size calculation, a total of 122 individuals were recruited for this study.
The 122 participants had been referred for endoscopy. Out of that, 15 had gastric cancer, 27 had peptic ulceration, 61 had gastroduodenal inflammation, 10 had esophageal diseases, while nine showed normal upper gastroduodenoscopic features. The diagnosis of gastroduodenal diseases had been made by an experienced gastroenterologist during the upper gastrointestinal (GI) endoscopy procedure. While gastric cancer was diagnosed based on histology. Participants’ demographic and clinical data were obtained by a structured questionnaire, personal interviews, and a review of case records. The selection criteria included the Sudanese population from both sexes, no antibiotic or non-steroidal anti-inflammatory drugs (NSAIDs) uses. All the participants were informed with the objectives and purposes of the study and the written informed consents were taken. The demographic characteristics of participants is presented in Table 8.
DNA extraction
Gastric biopsies were collected in 400 μl phosphate buffer saline (PBS). For histological examination, the biopsies were transported in formalin. DNA extraction was carried out by using innuPREP DNA Mini Kit (analytikjena AG, Germany) according to the protocol given by the manufacturer, as previously described in [67].
PCR amplification of specific 16S rRNA of H. pylori
The specific 16S rRNA gene of H. pylori was amplified by using the following primers (primers: F:5′-GCGCAATCAGCGTCAGGTAATG-3′) (R:5′-GCTAAGAGAGCAGCCTATGTCC-3′) [68]. The PCR condition was previously described [69].
PCR amplification and sequencing of the IL1B promoter region
The IL-1B-511 and − 31, promoter polymorphisms, were amplified using the following primers: F:5′- CATCCATGAGATTGGCTAG-3′ and R:5′- AGCACCTAGTTGTAAGGAAG-3′ [70]. The cycling conditions were an initial denaturation at 94 °C for 5 min, followed by 35 cycles of 94 °C for 1 min, 60 °C for 1 min and 72 °C for 1 min, with a final extension at 72 °C for 7 min. The amplified PCR product is 800 bp and was located between − 687 bp upstream and + 297 bp downstream of the IL-1B gene.
Out of 14 PCR products of H. pylori-infected subjects, which have the clearest bands, were sent for DNA purification and Sanger dideoxy sequencing. Both DNA strands were sequenced commercially by Macrogen Inc., Korea.
Sequence analysis and SNPs detection
The sequencing results, two chromatograms for each patient (forward and reverse), were visualized, checked for quality, and analyzed using the Finch TV program version 1.4.0 [71]. The nucleotide Basic Local Alignment Search Tool (BLASTn; https://blast.ncbi.nlm.nih.gov/) was used to assess nucleotide sequence similarities [72].
To determine the SNPs in the IL-1B promoter region, multiple sequence alignment (MSA) for tested sequences with a reference sequence (NG_008851) were performed by using BioEdit software [73].
Bioinformatics analysis of the IL-1B promoter region in H. pylori-infected subjects
In silico prediction of the promoter
The crucial element for initiating and regulating messenger RNA transcription is the promoter sequence which is generally located in the 5′ upstream region of a structural gene [44]. Promoters have complex and specific architecture, and contain multiple TFs involved in specific regulation of transcription [74]. Different features of a promoter region may have different power for promoter identification [49], therefore, we applied a variety of programs for prediction of promoter regions in order to obtain accurate results for subsequent experimental proof. These programs include: (1) Promoter 2.0 Prediction Server (http://www.cbs.dtu.dk/) which takes advantage of a combination of elements similar to neural networks and genetic algorithms to recognize a set of discrete sub-patterns with variable separation as one pattern: a promoter [75]; (2) Neural Network Promoter Prediction (NNPP2.2) (http://www.fruitfly.org/) which applying multiple hidden layers and time-delay neural networks (TDNNs) for promoter annotation [76]; (3) TSSW (http://softberry.com/) that uses functional motifs from the Wingender et al. database [77] and linear discriminant function combining characteristics describing function motifs and oligonucleotide composition of these sites [78]; (4) TSSG program (http://softberry.com/) program that uses the same approach of TSSW but the TFD database of functional motifs [79]; (5) Fprom program (http://softberry.com/) which is TSSG variant with different learning set of promoter sequences [49].
In silico analysis of the predicted promoter region
Assessment for the presence of promoter associated features
In silico predicted promoter region was additionally assessed for the presence of promoter associated features, including promoter-associated histone marks, broad chromatin state segmentation, transcription factor ChIP-seq, and DNase I hypersensitivity clusters, using the ENCODE data (https://epd.epfl.ch/cgi-bin/get_doc?db=hgEpdNew&format=genome&entry=IL1B_1) [45,46,47].
Prediction of CpG Islands
A CpG island is often regarded as a marker for the initiation of gene expression. It is a segment of DNA with high GC and CpG dinucleotide contents which is located in the 5′ UTR (untranslated regions) of genes. In this study, MethPrimer [44, 80] and GpC finder software (http://www.softberry.com/berry.phtml?topic=cpgfinder&group=programs&subgroup=promoter) were employed to predict CpG islands in the promoter. CpG finder is intended to search for CpG islands in sequences, while MethPrimer is developed to design PCR primers for methylation mapping and primers are picked around the predicted CpG islands. CpG islands are predicted by using a simple sliding window algorithm to examine the GC content and the ratio observed/expected (Obs/Exp) across the sequence. The search parameter values for the software were CpG island length > 200 bp, CG% > 50%, and Obs/Exp > 0.6.
Prediction of transcription factor binding sites (TFBSs)
One of the important steps in the chain of promoter analytical events is the prediction of the potentially functional TFBSs. Protein binding sites in a promoter represent the most important elements and the corresponding proteins are called transcription factors (TFs). In this step, the promoter region was analyzed for possible TFBSs using five prediction software. (1) Alggen Promo (http://alggen.lsi.upc.es/cgi-bin/promo_v3/) in which positional weight matrices (PWM) are constructed from known binding sites extracted from TRANSFAC [38] and used for the identification of potential binding sites in sequences [81, 82]. (2) AliBaBa2 (http://www.gene-regulation.com/) which works based on the assumption that each binding site has an unknown context that determines its sequence and this leads to a construct of specific matrices for each sequence we are analyzing. And to do so a context-specific process starting at a dataset of known binding sites and ending with the identification of a potential new binding site [83]. (3) Gene Promoter Miner (GPMiner) (http://GPMiner.mbc.nctu.edu.tw/) which is an integrated system that identifies promoter regions, regulatory elements and DNA stability by incorporating the support vector machine (SVM) with nucleotide composition features, over-represented hexamer nucleotides, and DNA stability. For predicting TFBSs, MATCH tool [84] was utilized to scan TFBSs in an input sequence using the TF binding profiles from TRANSFAC public release version 7.0 [85] and JASPAR [86, 87]. (4) TF-Bind (http://tfbind.hgc.jp/) which uses positional weight matrices (PWMs) and Bucher’s calculating method [88] to calculate the matching score between an input sequence and a set of known TF binding sites. To estimate TF binding sites, a robust cut-off value determining algorithm was proposed using the background rate estimated on non-promoters sequences [89]. (5) Tfsitescan (http://www.ifti.org) which is an object-oriented transcription factors database (ooTFD)-retrieval tool that is used for transcription factors sites analyses. It constructs an image-map in association with sequence analysis results which is linked to individual sites entries [90].
Prediction of composite regulatory elements (CEs)
CE is the minimal functional unit, which can provide combinatorial transcriptional regulation of gene expression. Structurally, a CE consists of two closely located DNA binding sites (BSs) for distinct transcription factors. But its regulatory function is qualitatively different from regulation effects of either individual DNA binding sites. In this study, we identified the composite regulatory elements in our region by using MatrixCatch algorithm (http://gnaweb.helmholtz-hzi.de/cgi-bin/MCatch/MatrixCatch.pl). The basic idea of MatrixCatch is to recruit data collected for respective binding sites separately from each other in order to complement the lack of knowledge on sequence variation of each DNA BS in CEs, and such information is compiled in position weight matrices (PWMs). The CE model consists of two PWMs, as well as their minimal scores, relative orientation and distance. Moreover, MatrixCatch is supplied with a library of 265 matrix models used for recognition which represents the widest scope of known CEs available to date [91].
Comparative analysis
Promoter region was analyzed for possible conservation using the ECR Browser (http://ecrbrowser.dcode.org) [37], NCBI BLASTn (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and ClustalW (https://www.genome.jp/tools-bin/clustalw). Conservation was assessed in 11 species: chimpanzee (Pan troglodytes), rhesus monkey (Macacamulatta), mouse (Mus musculus), rat (Rattusnorvegicus), dog (Canisfamiliaris), cow (Bostaurus), opossum (Monodelphisdomestica), chicken (Gallus gallus), frog (Xenopuslaevis), zebrafish (Danio rerio), fugu pufferfish (Takifugurubripes), and spotted green pufferfish (Tetraodon nigrovoridis).
Also, conservation of SNPs was evaluated and the possible conservation of TFBSs at these SNP locations was screened with Multiple-sequence local alignment and visualization (Mulan) search engine (https://mulan.dcode.org/) [92].
Detection of the IL-1B-31 C/T polymorphism using PCR with confronting two-pair primer (PCR-CTPP)
For detection of the IL-1B-31 polymorphism, PCR-CTPP was applied. The primers for the C allele were (F:5′-ACT TCT GCT TTT GAA GGC C-3′) and (R:5′-TAG CAC CTA GTT GTA AGG A-3′); and those for the T allele were (F:5′-AGA AGC TTC CAC CAA TAC T-3′) and (R:5′-CTC CCT CGC TGT TTT TAT A-3′) [93]. One μl of extracted DNA was used in a 25 μl reaction mixture with a prepared Maxime PCR PreMix Kit (i-Taq) (iNtRON BIOTECHNOLOGY, Seongnam, Korea), 23 μl of de-ionized sterile water, 0.25 μl of each primer. PCR conditions were as follow: 5 min of initial denaturation at 94 °C, followed by 25 cycles of 1 min at 94 °C, 1 min at 54 °C, and 1 min at 72 °C, and a 5 min final incubation at 72 °C. The PCR products were visualized by electrophoresis on a 2% agarose gel stained with ethidium bromide. Genotyping was performed as follows; 240, 155 bp for CC genotype, 240, 155, 122 bp for CT genotype, and 240, 122 bp for TT genotype [93].
Statistical analysis
Deviations from Hardy-Weinberg equilibrium in control were examined by χ2 test. According to prevalence of H. pylori infection, differences in distribution by age were assessed by Mann-Whitney test, while differences in distribution by categorical variables were examined by χ2 test or Fisher’s test. Odds ratios (ORs) were calculated and reported within the 95% confidence intervals (CIs). P < 0.05 was considered to be statistically significant. The statistical analyses were performed using the GraphPad Prism 5.
Availability of data and materials
The data regarding IL1B-31C > T genotypes and alleles distributions among participants and the in silico results of software used to support the findings of this study are available from the corresponding author on easonable request.
Abbreviations
- 16S rRNA :
-
16 Svedberg ribosomal RNA
- AP-1:
-
Activator protein 1
- BDGP:
-
Berkeley Drosophila Genome Project
- BLASTn:
-
The nucleotide Basic Local Alignment Search Tool
- bosTau3:
-
Bos Taurus (cow) full genome as provided by UCSC (Jan 2008)
- bp:
-
base pair
- C/ EBPβ:
-
CCAAT-enhancer binding protein beta
- C/EBPα:
-
CCAAT-enhancer binding protein alpha
- canFam2:
-
Canis lupus familiaris (domesticated dog) full genome as provided by UCSC (Jan 2008)
- CE:
-
Composite regulatory element
- ChIP-seq:
-
Chromatin Immunoprecipitation Sequencing
- CI:
-
Confidence interval
- CS:
-
Composite score
- CTPP:
-
Confronting two-pair primer
- DR1:
-
Down-regulator of transcription 1
- ECR:
-
Evolutionary conserved region
- ENCODE:
-
Encyclopedia of DNA elements
- FPROM:
-
Human promoter prediction
- GATA-1:
-
Globin transcription factor 1
- GR:
-
Glucocorticoid receptor
- H. pylori :
-
Helicobacter pylori
- hg19:
-
Homo sapiens (Human) full genome as provided by UCSC (Feb 2009)
- HNF4:
-
Hepatocyte nuclear factor 4
- HSF:
-
Heat-shock factor
- HSTF:
-
Heat shock transcription factor
- HWE:
-
Hardy-Weinberg equilibrium
- IL-1 :
-
interleukin 1
- IL1A :
-
interleukin 1-alpha
- IL1B :
-
interleukin 1-beta
- IL-1RN :
-
interleukin 1 receptor antagonist
- IRF:
-
Interferon-regulatory factor
- LD:
-
Linkage disequilibrium
- LDF:
-
Linear discriminant factor
- LPS:
-
Lipopolysaccharide
- MALT:
-
Mucosa-associated lymphoid tissue lymphoma
- MAZ:
-
Myc-associated zinc finger protein
- mm9:
-
Mus musculus (mouse) full genome as provided by UCSC (Jan 2008)
- MSA:
-
Multiple sequence alignment
- Mulan:
-
Multiple-sequence local alignment and visualization
- NCBI:
-
National Center for Biotechnology Information
- NF-AT:
-
Nuclear factor of activated T cells
- NF-Y:
-
Nuclear transcription factor Y
- NF-κB:
-
Nuclear factor Kappa β
- NK:
-
Natural killer cell
- NNPP:
-
Neural Network Promoter Prediction
- NSAIDS:
-
Non-steroidal anti-inflammatory drugs
- Oct-1:
-
Octamer transcription factor
- OR:
-
Odds ratio
- pan-Tro2:
-
Pan troglodytes (chimpanzee) full genome as provided by UCSC (Jan 2008)
- PBS:
-
Phosphate buffer saline
- PCR:
-
Polymerase chain reaction
- PPAR:
-
Peroxisome proliferator-activated receptor
- PWM:
-
Position weight matrices
- rheMac2:
-
Macaca mulatta (rhesus monkey) full genome as provided by UCSC (Jan 2008)
- rn4:
-
Rattus norvegicus (rat) full genome as provided by UCSC (Jan 2008)
- SNP:
-
Single nucleotide polymorphism
- Spi-1:
-
SV40 promoter-1
- SRF:
-
Serum response factor
- SSRP:
-
Structure specific recognition protein 1
- STAT1:
-
Signal transducer and activator of transcription 1
- Std:
-
Standard
- T cell:
-
T lymphocyte cell
- T:
-
Thymidine
- TBP:
-
TATA- binding protein
- TF:
-
Transcription factor
- TFBS:
-
Transcription factor binding site
- TSS:
-
Transcription start site
- TSSG:
-
Recognition of human PolII promoter region and start of transcription
- TSSW:
-
Recognition of human PolII promoter region and start of transcription
- UTR:
-
Unstranslated region
- ZTA:
-
The Epstein-Barr virus bZIP transcription factor
References
Moodley Y, Linz B, Bond RP, Nieuwoudt M, Soodyall H, Schlebusch CM, et al. Age of the association between Helicobacter pylori and man. PLoS Pathog. 2012;8(5):e1002693.
Suerbaum S, Michetti P. Helicobacter pylori infection. New England J Med. 2002;347:1175–86.
Sjomina O, Pavlova J, Niv Y, Leja M. Epidemiology of helicobacter pylori infection. Helicobacter. 2018;23(S1):e12514.
Zamani M, Ebrahimtabar F, Zamani V, Miller WH, Alizadeh-Navaei R, Shokri-Shirvani J, et al. Systematic review with meta-analysis: the worldwide prevalence of helicobacter pylori infection. Aliment Pharmacol Ther. 2018;47(7):868–76.
Goh K-L, Chan W-K, Shiota S, Yamaoka Y. Epidemiology of Helicobacter pylori infection and public health implications. Helicobacter. 2011;16 Suppl 1(1):1–9.
Fock KM, Ang TL. Epidemiology of helicobacter pylori infection and gastric cancer in Asia. J Gastroenterol Hepatol. 2010;25(3):479–86.
Chmiela M, Karwowska Z, Gonciarz W, Allushi B, Staczek P. Host pathogen interactions in helicobacter pylori related gastric cancer. World J Gastroenterol. 2017;23(9):1521–40.
Kodaman N, Pazos A, Schneider BG, Piazuelo MB, Mera R, Sobota RS, et al. Human and helicobacter pylori coevolution shapes the risk of gastric disease. Proc Natl Acad Sci U S A. 2014;111(4):1455–60.
Testerman TL, Morris J. Beyond the stomach: An updated view of helicobacter pylori pathogenesis, diagnosis, and treatment. World J Gastroenterol. 2014;20(36):12781–808.
Cadamuro ACT, Rossi AFT, Maniezzo NM, Silva AE. Helicobacter pylori infection: host immune response, implications on gene expression and microRNAs. World J Gastroenterol. 2014;20(6):1424–37.
Forman D, Burley VJ. Gastric cancer: global pattern of the disease and an overview of environmental risk factors. Best Pract Res Clin Gastroenterol. 2006;20(4):633–49.
de Sablet T, Piazuelo MB, Shaffer CL, Schneider BG, Asim M, Chaturvedi R, et al. Phylogeographic origin of helicobacter pylori is a determinant of gastric cancer risk. Gut. 2011;60(9):1189–95.
Chang W-L, Yeh Y-C, Sheu B-S. The impacts of H. pylori virulence factors on the development of gastroduodenal diseases. J Biomed Sci. 2018;25(1):68.
Nicklin MJH, Weith A, Duff GW. A physical map of the region encompassing the human interleukin-1α, interleukin-1β, and Interleukin-1 receptor antagonist genes. Genomics. 1994;19(2):382–4.
El-Omar EM. The importance of interleukin 1beta in helicobacter pylori associated disease. Gut. 2001;48(6):743–7.
Dinarello CA. Interleukin-1 and interleukin-1 antagonism. Blood. 1991;77(8):1627–52.
Ren K, Torres R. Role of interleukin-1beta during pain and inflammation. Brain Res Rev. 2009;60(1):57–64.
Rad R, Dossumbekova A, Neu B, Lang R, Bauer S, Saur D, et al. Cytokine gene polymorphisms influence mucosal cytokine expression, gastric inflammation, and host specific colonisation during helicobacter pylori infection. Gut. 2004;53(8):1082–9.
Sun X, Cai H, Li Z, Li S, Yin W, Dong G, et al. Association between IL-1b polymorphisms and gastritis risk. Medicines. 2017;96:5.
Motamedi Rad N, Rezaeishahmirzadi M, Shakeri S, Abbaszadegan MR, Shekari M. Association of IL-1B+3954 and IL-1RN polymorphisms in chronic gastritis and peptic ulcer. Iran J Public Health. 2018;47(9):1364–70.
Robert A, Olafsson AS, Lancaster C, Zhang WR. Interleukin-1 is cytoprotective, antisecretory, stimulates PGE2 synthesis by the stomach, and retards gastric emptying. Life Sci. 1991;48(2):123–34.
Pulugulla SH, Packard TA, Galloway NLK, Grimmett ZW, Doitsh G, Adamik J, et al. Distinct mechanisms regulate IL1B gene transcription in lymphoid CD4 T cells and monocytes. Cytokine. 2018;111:373–81.
Adamik J, Wang KZQ, Unlu S, Su A-JA, Tannahill GM, Galson DL, et al. Distinct mechanisms for induction and tolerance regulate the immediate early genes encoding interleukin 1β and tumor necrosis factor α. PloS One. 2013;8(8):e70622.
Marecki S, Riendeau CJ, Liang MD, Fenton MJ. PU.1 and multiple IFN regulatory factor proteins synergize to mediate transcriptional activation of the human IL-1β gene. J Immunol. 2001;166(11):6829–38.
Dinarello C. Biologic basis for interleukin-1 in disease. Blood. 1996;87(6):2095–147.
Kominato Y, Galson D, Waterman WR, Webb AC, Auron PE. Monocyte expression of the human prointerleukin 1 beta gene (IL1B) is dependent on promoter sequences which bind the hematopoietic transcription factor Spi-1/PU.1. Mol Cell Biol. 1995;15(1):59–68.
Auron PE, Webb AC. Interleukin-1: a gene expression system regulated at multiple levels. Eur Cytokine Netw. 1994;5(6):573–92.
Listman JA, Race JE, Walker-Kopp N, Unlu S, Auron PE. Inhibition of IL-1beta transcription by peptides derived from the hCMV IE2 transactivator. Mol Immunol. 2008;45(9):2667–77.
Zhang Y, Saccani S, Shin H, Nikolajczyk BS. Dynamic protein associations define two phases of IL-1beta transcriptional activation. J Immunol. 2008;181(1):503–12.
Shirakawa F, Saito K, Bonagura CA, Galson DL, Fenton MJ, Webb AC, et al. The human prointerleukin 1 beta gene requires DNA sequences both proximal and distal to the transcription start site for tissue-specific induction. Mol Cell Biol. 1993;13(3):1332–44.
Ahmed AB, Zidi S, Sghaier I, Ghazouani E, Mezlini A, Almawi W, et al. Common variants in IL-1RN, IL-1beta and TNF-alpha and the risk of ovarian cancer: a case control study. Cent Eur J Immunol. 2017;42(2):150–5.
Figueiredo CA, Marques CR, Costa Rdos S, da Silva HB, Alcantara-Neves NM. Cytokines, cytokine gene polymorphisms and helicobacter pylori infection: friend or foe? World J Gastroenterol. 2014;20(18):5235–43.
Lu W, Pan K, Zhang L, Lin D, Miao X, You W. Genetic polymorphisms of interleukin (IL)-1B, IL-1RN, IL-8, IL-10 and tumor necrosis factor {alpha} and risk of gastric cancer in a Chinese population. Carcinogenesis. 2005;26(3):631–6.
Singh H, Samani D, Nema V, Ghate MV, Gangakhedkar RR. IL-1RN and IL-1beta polymorphism and ARV-associated hepatotoxicity. Mediat Inflamm. 2018;2018:4398150.
Shehjar F, Afroze D, Misgar RA, Malik SA, Laway BA. Association of polymorphic variants of IL-1beta and IL-1RN genes in the development of Graves’ disease in Kashmiri population (North India). Hum Immunol. 2018;79(4):228–32.
Ma J, Wu D, Hu X, Li J, Cao M, Dong W. Associations between cytokine gene polymorphisms and susceptibility to helicobacter pylori infection and helicobacter pylori related gastric cancer, peptic ulcer disease: a meta-analysis. PLoS One. 2017;12(4):e0176463.
Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32:W280–W6.
Kel-Margoulis OV, Kel AE, Reuter I, Deineko IV, Wingender E. TRANSCompel®: a database on composite regulatory elements in eukaryotic genes. Nucleic Acids Res. 2002;30(2):332–4.
Demeler B, Zhou GW. Neural network optimization for E. coli promoter prediction. Nucleic Acids Res. 1991;19(7):1593–9.
Liu R, States DJ. Consensus promoter identification in the human genome utilizing expressed gene markers and gene modeling. Genome Res. 2002;12(3):462–9.
Jennes I, Zuntini M, Mees K, Palagani A, Pedrini E, De Cock G, et al. Identification and functional characterization of the human EXT1 promoter region. Gene. 2012;492(1):148–59.
Song H, Wang M, Wang Z, Liu J, Qi J, Zhang Q. Characterization of kiss2 and kissr2 genes and the regulation of kisspeptin on the HPG axis in Cynoglossus semilaevis. Fish Physiol Biochem. 2017;43(3):731–53.
Võ Anh Khoa Đ, Ponsuksili S, Murani E, Wimmers K. Polymorphic sites in the 5′-region of the porcine C8A gene. Arch Anim Breeding. 2011;54:430–8.
Xin S, Wang X, Dai G, Zhang J, An T, Zou W, et al. Bioinformatics analysis of SNPs in IL-6 gene promoter of Jinghai yellow chickens. Genes. 2018;9:446.
ENCODE Project Consortium. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 2011;9(4):e1001046.
Sloan CA, Chan ET, Davidson JM, Malladi VS, Strattan JS, Hitz BC, et al. ENCODE data at the ENCODE portal. Nucleic Acids Res. 2016;44(D1):D726–D32.
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74.
Yang C, Bolotin E, Jiang T, Sladek FM, Martinez E. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene. 2007;389(1):52–65.
Solovyev VV, Shahmuradov IA, Salamov AA. Identification of promoter regions and regulatory sites. Methods Mol Biol. 2010;674:57–83.
Wang J, Hannenhalli S. A mammalian promoter model links cis elements to genetic networks. Biochem Biophys Res Commun. 2006;347(1):166–77.
Zhang G, Zhou B, Li S, Yue J, Yang H, Wen Y, et al. Allele-Specific Induction of IL-1β Expression by C/EBPβ and PU.1 Contributes to Increased Tuberculosis Susceptibility. PLOS Pathog. 2014;10(10):e1004426.
Lind H, Haugen A, Zienolddiny S. Differential binding of proteins to the IL1B −31 T/C polymorphism in lung epithelial cells. Cytokine. 2007;38(1):43–8.
El-Omar EM, Carrington M, Chow W-H, McColl KEL, Bream JH, Young HA, et al. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404(6776):398–402.
Al-Moundhri MS, Al-Nabhani M, Al-Bahrani B, Burney IA, Al-Madhani A, Ganguly SS, et al. Interleukin-1beta gene (IL-1B) and interleukin 1 receptor antagonist gene (IL-1RN) polymorphisms and gastric cancer risk in an Omani Arab population. Gastric Cancer. 2006;9(4):284–90.
Hartland S, Newton JL, Griffin SM, Donaldson PT. A functional polymorphism in the interleukin-1 receptor-1 gene is associated with increased risk of helicobacter pylori infection but not with gastric cancer. Dig Dis Sci. 2004;49(9):1545–50.
Zhao Y, Wang JW, Tanaka T, Hosono A, Ando R, Tokudome S, et al. Association between TNF-alpha and IL-1beta genotypes vs helicobacter pylori infection in Indonesia. World J Gastroenterol. 2013;19(46):8758–63.
Drici Ael M, Moulessehoul S, Tifrit A, Diaf M, Turki DK, Bachir M, et al. Effect of IL-1beta and IL-1RN polymorphisms in carcinogenesis of the gastric mucosa in patients infected with helicobacter pylori in Algeria. Libyan J Med. 2016;11:31576.
Kumar S, Kumar A, Dixit VK. Evidences showing association of interleukin-1B polymorphisms with increased risk of gastric cancer in an Indian population. Biochem Biophys Res Commun. 2009;387(3):456–60.
Ruzzo A, Graziano F, Pizzagalli F, Santini D, Battistelli V, Panunzi S, et al. Interleukin 1B gene (IL-1B) and interleukin 1 receptor antagonist gene (IL-1RN) polymorphisms in helicobacter pylori-negative gastric cancer of intestinal and diffuse histotype. Ann Oncol. 2005;16(6):887–92.
Xue H, Lin B, Ni P, Xu H, Huang G. Interleukin-1B and interleukin-1 RN polymorphisms and gastric carcinoma risk: a meta-analysis. J Gastroenterol Hepatol. 2010;25(10):1604–17.
Kimura R, Nishioka T, Soemantri A, Ishida T. Cis-acting effect of the IL1B C−31T polymorphism on IL-1β mRNA expression. Genes Immun. 2004;5(7):572–5.
Santtila S, Savinainen K, Hurme M. Presence of the IL-1RA allele 2 (IL1RN*2) is associated with enhanced IL-1beta production in vitro. Scand J Immunol. 1998;47(3):195–8.
Hwang I-R, Kodama T, Kikuchi S, Sakai K, Peterson LE, Graham DY, et al. Effect of interleukin 1 polymorphisms on gastric mucosal interleukin 1β production in helicobacter pylori infection. Gastroenterology. 2002;123(6):1793–803.
Kaijzel EL, Bayley JP, van Krugten MV, Smith L, van de Linde P, Bakker AM, et al. Allele-specific quantification of tumor necrosis factor α (TNF) transcription and the role of promoter polymorphisms in rheumatoid arthritis patients and healthy individuals. Genes Immun. 2001;2(3):135–44.
Harbage B, Dean AG. Distribution of Epi info software: an evaluation using the internet. Am J Prev Med. 1999;16(4):314–7.
Su Y, Yoon SS. Epi info - present and future. AMIA Annu Symp Proc. 2003;2003:1023.
Idris AB, Ataelmanan AE, Eltaher SM, Idris EB, Osman Arbab BM, Ibn Idris A, et al. Independently Carriage of IL-1RN*2 Allele associated with increased risk of gastric cancer in the Sudanese Population. medRxiv. 2019:19013573. https://www.medrxiv.org/content/10.1101/19013573v1.
Carbone M, Maugeri TL, Gugliandolo C, La Camera E, Biondo C, Fera MT. Occurrence of helicobacter pylori DNA in the coastal environment of southern Italy (straits of Messina). J Appl Microbiol. 2005;98(3):768–74.
Idris AB, Hassan HG, Ali MAS, Eltaher SM, Idris LB, Altayb HN, et al. Molecular phylogenetic analysis of 16S rRNA sequences identified two lineages of helicobacter pylori strains detected from different regions in Sudan suggestive of differential evolution. Int J Microbiol. 2020;2020:8825718.
Furuta T, El-Omar EM, Xiao F, Shirai N, Takashima M, Sugimura H. Interleukin 1beta polymorphisms increase risk of hypochlorhydria and atrophic gastritis and reduce risk of duodenal ulcer recurrence in Japan. Gastroenterology. 2002;123(1):92–105.
FinchTV. 1.4.0 ed. USA: Geospiza, Inc.; Seattle, 2012.
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST. A new generation of protein database search programmes. Nucleic Acids Res. 1997;25(17):3389–402.
Hall TA. BioEdit: a user-friendly biologicalsequence alignment editor and analysis programfor windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–8.
Umarov R. Novel computational methods for promoter identification and analysis. Thuwal, Kingdom of Saudi Arabia: King Abdullah University of Science and Technology; 2020.
Knudsen S. Promoter2.0: for the recognition of PolII promoter sequences. Bioinformatics (Oxford, England). 1999;15(5):356–61.
Reese MG. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem. 2001;26(1):51–6.
Wingender E, Dietze P, Karas H, Knüppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24(1):238–41.
Solovyev V, Kosarev P, Seledsov I, Vorobyev D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006;7 Suppl 1(Suppl 1):S10.1–12.
Ghosh D. A relational database of transcription factors. Nucleic Acids Res. 1990;18(7):1749–56.
Li L-C, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics (Oxford, England). 2002;18(11):1427–31.
Farré D, Roset R, Huerta M, Adsuara JE, Roselló L, Albà MM, et al. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003;31(13):3651–3.
Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics (Oxford, England). 2002;18(2):333–4.
Grabe N. AliBaba2: context specific identification of transcription factor binding sites. In Silico Biol. 2002;2(1):S1–15.
Kel AE, Gößling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCHTM: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31(13):3576–9.
Fu Y, Weng Z. Improvement of TRANSFAC matrices using multiple local alignment of transcription factor binding site sequences. Genome Inform. 2005;16(1):68–72.
Lee T-Y, Chang W-C, Hsu J, Chang T-H, Shien D-M. GPMiner: An integrated system for mining combinatorial cis-regulatory elements in mammalian gene group. BMC Genomics. 2012;13 Suppl 1:S3.
Sandelin A, Alkema W, Engström P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucl Acids Res. 2004;32(suppl_1):D91–D4.
Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J Mol Biol. 1990;212(4):563–78.
Tsunoda T, Takagi T. Estimating transcription factor bindability on DNA. Bioinformatics (Oxford, England). 1999;15(7–8):622–30.
Ghosh D. Object-oriented transcription factors database (ooTFD). Nucleic Acids Res. 2000;28(1):308–10.
Deyneko IV, Kel AE, Kel-Margoulis OV, Deineko EV, Wingender E, Weiss S. MatrixCatch - a novel tool for the recognition of composite regulatory elements in promoters. BMC Bioinformatics. 2013;14(1):241.
Ovcharenko I, Loots GG, Giardine BM, Hou M, Ma J, Hardison RC, et al. Mulan: multiple-sequence local alignment and visualization for studying function and evolution. Genome Res. 2005;15(1):184–94.
Hamajima N, Saito T, Matsuo K, Kozaki K, Takahashi T, Tajima K. Polymerase chain reaction with confronting two-pair primers for polymorphism genotyping. Japanese J Cancer Res. 2000;91(9):865–8.
Acknowledgements
We gratefully acknowledge the participants and the staff of the gastroscopic unit in Ibin Sina specialized hospital, Soba teaching hospital, Modern Medical Centre, Al-Shorta hospital, and Al Faisal Specialized Hospital. We would also like to show our gratitude to the Department of Medical Microbiology, Faculty of Medical Laboratory Sciences, University of Khartoum for their cooperation.
Funding
The authors received no specific funding for this work.
Author information
Authors and Affiliations
Contributions
MAH and EMI supervised the methodology. ABI, AEA, AEAM and BMOA collected the samples. ABI, EBI and AEA extracted the DNA. ABI amplified the 16S rRNA and IL1B genes. ABI and EBI analyzed the data. ABI performed the bioinformatics and statistical analysis for the data. ABI wrote the manuscript. AEAM and MAH edited and revised the manuscript. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Khartoum Ministry of Health research department, University of Khartoum, Faculty of Medical Laboratory Sciences review board, and Research Ethics Committees of hospitals. Written informed consent was taken from participants before they enrolled.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
The structured questionnaire.
Additional file 2.
Original, full-length gel and blot images.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Idris, A.B., Idris, E.B., Ataelmanan, A.E. et al. First insights into the molecular basis association between promoter polymorphisms of the IL1B gene and Helicobacter pylori infection in the Sudanese population: computational approach. BMC Microbiol 21, 16 (2021). https://doi.org/10.1186/s12866-020-02072-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-020-02072-3