Abstract
Background
The Escherichia coli sequence type 131 (ST131) is a well established clone causing significant extraintestinal infections worldwide. However, no studies have been reported the phenotypic and molecular traits of ST131 isolates in comparison to other clones of E. coli from Iran. So, we determined the differences between 69 ST131 strains collected during a one year surveillance study and 84 non-ST131 isolates, including 56 clinical fluoroquinolone resistant and 28 broiler colibacillosis isolates in terms of clonality and genetic background.
Results
ST131 isolates were associated with phylogroup B2 (68 out of 69 isolates, 98.4%), while clinical non-ST131 and fluoroquinolone resistant broiler isolates mainly belonged to phylogroup A. The highest virulence score was observed in ST131 clone, while they showed less diversity in virulence profiles than other clinical isolates. Almost all of the ST131 isolates (95.6%) were ExPEC and had the highest virulence scores, but their resistance scores were less than clinical non-ST131 isolates. Broiler isolates showed higher prevalence of ExPEC-associated virulence genes and CTX-M-G1/G9 resistance determinants as compared to clinical non-ST131 isolates. While blaOXA-48/NDM carbapenemases were mostly found in ST131 clone, resistance rate against ertapenem was higher among clinical non-ST131 strains. According to ERIC-based fingerprinting, the ST131 strains were more genetically similar, followed by non-ST131 and broiler isolates.
Conclusions
ST131 isolates possess the ability to make a balance between clonality and extent of resistance/virulence genes content, so this phenomenon gives a fitness advantage over other E. coli clones. The broilers E. coli population poses a potential zoonotic risk which could be transmitted to the community through the food chain. A number of factors are involved in the dissemination of and infections due to ST131 clone.
Similar content being viewed by others
Background
Extraintestinal infections caused by Escherichia coli, including urinary tract infections, bloodstream infections and neonatal meningitis are common, costly and are major causes of morbidity and mortality. Emerging antimicrobial resistance complicates the management of these infections [1].
E. coli sequence type 131 (ST131) is defined by multilocus sequence typing (MLST) technique and is characterized by resistance to first line antibiotics such as fluoroquinolone and extended spectrum cephalosporins (ESCs). Two major subclones which were identified within the ST131 population are H30 and H30Rx. Except for rare strains, almost all H30 isolates are fluoroquinolone resistant, while H30Rx subclone has been associated with Extended Spectrum β-lactamases (ESBLs), namely CTX-M-15 [2, 3].
Fluoroquinolones are widely used in farm animals, mainly in broilers [4, 5] and fluoroquinolone resistant E. coli strains are frequently isolated from healthy and diseased birds [4]. It has been hypothesized that avian E. coli isolates are reservoirs of virulence and resistance determinants, and strains harboring these elements may be able to cause foodborne diseases in humans. Characterization of overlapping traits between avian pathogenic E. coli (APEC) and urinary pathogenic E. coli (UPEC), such as O-serogroups, virulence markers and assignments to phylogroups, encouraged this hypothesis and subsequent researches comparing APEC and human E. coli isolates including UPEC, neonatal meningitis E. coli (NMEC) and E. coli causing septicemia showed common molecular traits between human and avian strains [4, 6, 7]. While the presence of ST131 clone among E. coli isolates contaminating raw chicken meat wasn’t confirmed in the studies from Spain and Italy [8, 9], another survey from Spain found this specific clone in 7% of retail chicken samples [10]. Vincent et al. also had identified E. coli ST131 from retail chicken samples in Canada but at a significant lower prevalence (0.4%) than in Spain [11].
The presented reasons for the success of the ST131 pandemic clone are the acquisition of antimicrobial resistance and additional virulence factors as well as its predominance in the human gut. There are some reports which suggest that epidemiological differences exist between CTX-M-producing strains of ST131 and non-ST131 clones [12]. Our previous study from Iran reported important data on clinical E. coli ST131 isolates [13]. However, a comparative work describing fluoroquinolone resistant/ESBL producing ST131 E. coli and other non-ST131 isolates or isolates from avian sources has not been reported so far from Iran. Therefore, the objectives of this study were to compare the clonality, virulence characteristics and resistance genotypes of ST131 isolates with that of human non-ST131 and broiler colibacillosis strains in order to determine whether these genotypes were shared or not between these groups of isolates.
Results
Human-sourced E. coli isolates
Sixty-nine strains out of 338 collected human E. coli isolates belonged to ST131 clonal group. The O25b/O16 serogroups, H30/H30Rx subclones and virotypes of 63 isolates have been previously described [13]. The remaining six newly identified ST131 isolates belonged to phylogroup B2, O25b subgroup, virotype C and H30 subclone. Of the remaining 269 E. coli isolates, 56 fluoroquinolone resistant isolates were randomly selected. Thus, a total of 125 unique human E. coli isolates (isolated from 125 admitted patients) were subjected to the thorough investigation described below. Among the ST131 isolates, 39 (56.5%) were from female patients with a mean age of 65.49 years and 30 (43.5%) were from male patients, with a mean age of 73 years, whereas among non-ST131 isolates 39 (69.6%) were from female patients and 17 (30.4%) were from male patients with mean ages of 66.6 and 64.8 years, respectively. Overall, specimen types for ST131 and clinical non-ST131 isolates included blood (0 and 1.8%), wound (5.8% and 1.8%), respiratory (11.6% and 12.5%) and urine (82.6% and 83.9%). The prevalence of ST131 and clinical non-ST131 isolates were higher among women, however, sex distribution did not vary significantly between two groups of clinical isolates.
Prevalence of ESBL-producing E .coli and antimicrobial susceptibility
Fluoroquinolone resistance was detected among 63 ST131 (all belonged to subgroup O25b) and 18 (64.2%) broiler isolates. The highest frequency of resistance was obtained against cefotaxime (95.7%), cefotaxime (100%) and trimethoprim/sulfamethoxazole (71.4%) among ST131, non-ST131 and broiler isolates, respectively. Overall, except for carbapenems, a high proportion of ESBL producing isolates were non-susceptible to antibiotics tested and were categorized as MDR (98.4%, 121/123, P: 0.001).
The resistance rates against studied antibiotics among non-ST131 isolates were higher than ST131 and the differences were significant for trimethoprim/sulfamethoxazole, amoxicillin/clavulanate, piperacillin-tazobactam, ampicillin/sulbactam, fluoroquinolones and tobramycin (Table 1). The highest resistance score was obtained among non-ST-131 (median: 9, IQR: 3) as compared with ST131 (median: 8, IQR: 4, P: 0.01) and broiler isolates (median: 2, IQR: 2, P: 0.001).
Characterization of resistance genes
The predominant resistance gene detected among the three groups of isolates was CTX-M-G1, followed by aac6Ib-cr in ST131 (45, 65.2%), blaTEM- in non-ST131 (45, 80.4%) and blaCTX-M-15 in broiler (20, 71.4%) isolates. The prevalence of blaSHV-, blaCTX-M-15, CTX-M-G1, CTX-M-G8, CTX-M-G9, CTX-M-G25 and blaOXA-48 was higher among ST131 as compared with clinical non-ST131 isolates; however the differences were only significant for the last three resistance determinants. The broiler isolates showed higher prevalence of CTX-M-G1 and CTX-M-G9 than both of two clinical groups (Table 1). The main gene contributing to the ESBL phenotype among the ST131 strains was CTX-M-G1 (60/66, 90.9%), followed by blaCTX-M-15 (54/66, 81.8%), while in non-ST131 isolates, it was CTX-M-G1 (47/56, 83.9%) and blaTEM- (45/56, 80.4%). The blaCTX-M-15 was more frequently found among ST131 (57/69, 82.6%) than non-ST131 (38/56, 67.9%) and broiler (20/28, 71.4%) isolates, however, the differences were not significant. Among the clinical isolates carrying blaCTX-M-15, resistance rates were higher against gentamicin (57.9%, P: 0.01), amoxicillin-clavulanate (73.7%, P, 0.02), cefepime (69.5%, P: 0.005), amikacin (15.8%, P: 0.02) and tobramycin (65.3%, P: 0.001). The blaOXA-48 and blaOXA-48/NDM combination were detected in 23 (33.3%) and three (4.3%) of the ST131 isolates respectively, of those 19 isolates including three blaNDM carrying strains were found susceptible to carbapenems. Two out of the four (7.1%) blaOXA-48 carrying non-ST131 isolates were also categorized as susceptible to carbapenems.
Phylogrouping
All the 153 strains formed seven distinct phylogroups (A, B1, B2, C, D, E and F). Table 2 shows the phylogroup distribution for the E. coli isolates stratified by source and by ciprofloxacin susceptibility pattern. Among the total population, group B2 was the most prevalent phylogroup, corresponding to 55.6% (85/153) of the isolates, followed in prevalence by the other groups: A (17%), E (13.1%), F (5.2%), B1 and C (each 3.9%), and D (1.3%). Almost all ST131 isolates were from group B2 (98.6%, 68/69), whereas the non-ST131 and broiler isolates exhibited diverse phylogroups, with group A being predominant in these two groups. Considering the clinical non-ST131 and broiler isolates, fluoroquinolone resistant strains found mainly belonged to phylogroup A, although the differences were not significant with respect to phylogroup distribution of resistant isolates. None of broiler isolates were identified as ST131 clonal group. Multiplex ST-PCR detected one urine cultured non-ST131 isolate as ST95 (phylogroup A), and broiler isolates did not belong to the investigated five major ST.
Virulence genotyping
Based on the molecular definition of ExPEC, all ST131 isolates except one O16 and two O25b strains were attributed with the status of ExPEC, in contrast to only 42.9% (24/56, P: 0.001) and 75% (21/28, P: 0.006) of clinical non-ST131 and broiler isolates, respectively. Considering the 84 non-ST131 isolates, including both of the clinical and broiler strains, fluoroquinolone susceptible isolates showed higher virulence scores as compared with fluoroquinolone resistant strains (median; 11 vs. 6.50, P: 0.045). The conserved pattern of virulence genes (i.e., gene prevalence > 90%) of the ST131 isolates included fyuA, F10 papA, iutA, chuA, sat, malX, usp and traT. Eighty-five different virulence profiles were found (named 1–85). Twenty of them grouped more than one isolate; profile 26 was the most frequently identified (n = 28), while profile 11 showed the highest virulence score (one ST131 isolate, virotype B, virulence score: 16). Clinical non-ST131 and broiler isolates were spread among 42 and 21 different virulence profiles, respectively, whereas ST131 isolates showed 25 different profiles, with the virulence profiles number 26 (n = 26) and 6 (n = 15) containing the majority of isolates (Fig. 1). A single pattern (pattern 84; fimH30+) was detected between human (non-ST131) and broiler isolates.
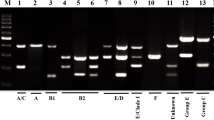
ERIC-PCR based dendogram of the 152 E.coli isolates and virulence gene profile. Cluster analysis of the Dice similarity indices based on the unweighted pair group method using average linkages (UPGMA) was done to generate a dendogram describing the relationship among the ERIC profiles. (One non-ST131 isolate didn’t amplify in ERIC-PCR, so it is not included in the dendogram image). P, poultry (broilers); ST, ST131; NST, non-ST131; VP, virulence profile number. Filed squares indicate the presence of the gene
Among ST131 isolates, the prevalence of K5 and afaDraBC was associated with O25b (fluoroquinolone resistant) and O16 (fluoroquinolone susceptible) subgroups, respectively. The presence of ompT, hlyF, iss, ibeA, iroN and kpsMT II genes was associated with broiler isolates. The median virulence score and the virulence score range values decreased progressively from a high of 11 (IQR: 3, range 7–16) for ST131 isolates to a low of 8.50 (IQR: 8, range, 0–14) for broiler isolates, to a score of 6 (IQR: 5, range, 1–14) for non-ST131 isolates (P < 0.001) (Table 3).
Regardless of ST131 status, H30 positive isolates were overwhelmingly fluoroquinolone resistant (105/106, 99.1%) (P: 0.001). H30Rx sublineage was mainly detected among ST131 isolates (27/69, 39.1%), whereas one and four strains of broiler and non-ST131 isolates belonged to H30Rx sublineage, respectively (P: 0.001). Resistant phenotypes which were associated with H30Rx subclone were against cefotaxime, amikacin, tobramycin, gentamycin, fluoroquinolones, amoxicillin-clavulanate and aztreonam. Almost all (31/32, 96.8%) H30Rx isolates were qualified as ExPEC and were associated with CTX-M-G1, CTX-M-G9 (P: 0.01 for both) and aac6-Ib/ aac6Ib-cr (P: 0.001, for both). H30Rx isolates showed higher prevalence of CTX-M-15 gene as compared with H30-non-Rx subgroup, but the difference was not significant (87.5% vs. 71.9%). H30Rx sublineage of ST131 clone was significantly associated with the virulence genes of cnf1, hlyA, k5, kpsMT II, papA, papC and papEF.
Genotyping
The highest similarity rates were observed among ST131 isolates, followed by the non-ST131 strains and then the broiler isolates (Fig. 1). Two out the three blaNDM carrying isolates exhibited > 80% similarity in ERIC-PCR based fingerprinting. Several dissimilar isolates belonging to all the three groups harboring blaCTX-M-15 and/or blaOXA-48 were scattered throughout the dendogram, suggesting possible horizontal gene transfer.
Discussion
Alarming increase in the isolation of CTX-M-15 producing E. coli isolates have been reported from different countries and this phenomenon has been linked to the clonal expansion of ST131 [14, 15]. The association of ST131 with higher ExPEC fraction and multidrug resistance phenotype, together with their ability for widespread dissemination, has led to the worldwide predominance of ST131 among other E. coli populations. Past studies have reported the association of selected factors with ST131 and non-ST131 isolates [16]. However, comprehensive analyses to study the association of several determinants with clinical ST131, non-ST131 and E. coli isolates from broiler subjects, particularly from developing countries like Iran, remain scarce.
To our knowledge, this is the first study of phenotypic and molecular traits of ST131 and its main subclones, including H30 and H30Rx and non-H30 strains within a population of systematically collected E. coli clinical isolates from Iran. We found that among fluoroquinolone resistant isolates, ST131 clone shows priority over non-ST131 isolates including more virulence genes profiles, higher proportion of ExPEC and high similarity for phylogroup B2 background (98.4% vs. 23.2%).
We found a predominance of blaCTX-M-15 (82.6%) among ESBL producing ST131 E. coli isolates. Notably, approximately 60% of non-ST131 and broiler isolates also harbored blaCTX-M-15, providing evidence of the extended prevalence of blaCTX-M-15 and a significant association of ST131 with blaCTX-M-15. Moreover, the blaCTX-M-15 was significantly associated with resistance to first line antibiotics, notably amoxicillin/clavulanate, aminoglycosides and cefepime, and also among clinical non-ST131 isolates with greater resistance scores. This event would help the rising expansion of blaCTX-M-15 carrying strains over other ESBL producing E. coli isolates in the presence of selection pressure due to the corresponding antibiotics [17].
Studies have reported a core set of virulence factors among ST131 strains including (fimH, fyuA, iutA, traT, malX), with occasional variation [14, 18]. In our study, these core virulence determinants were detected in > 89% of ST131 isolates screened, along with F10 papA, chuA and usp. Because the present ST131 strains were significantly more likely to be from group B2 than the non-ST131 and broiler E. coli isolates, they may have fitness priority over other E. coli isolates.
The proportions of fimH, fyuA, iutA, traT, and PAI genes were similar to those in previous reports [19, 20], although the proportions of hlyA/cnf1 and ompT/sat were different from those reported in previous Korean/Indian and U. S studies [20,21,22]. This result suggests that differences in virulence profiles among ST131 strains may depend on their geographical origin, whether the study was hospital/ “long-term care facility” or community based, and the source of isolates.
The high diversity of virulence gene profiles that we describe in our ESBL producers has been previously described [23], but we found that ST131 has less profile diversity, which implies to the selective distribution of certain sublineages within ST131. Furthermore, the similarity of broiler isolates with human pathogenic E. coli based on virulence markers was confirmed in this study, since a total of 75% of broiler isolates belonged to the ExPEC fraction. As also suggested by other studies, certain APEC subgroups, especially a large fraction of phylogroup A isolates may be considered potential zoonotic agents [24].
In this study, evaluation of the potential zoonotic risk of the E. coli isolates from broiler relies on the phylogroup distribution associated with ciprofloxacin resistance. Regarding the phylogroups, overall, the proportion of group A strains of clinical non-ST131 isolates was greater than the other phylogroups. Considering that half of the fluoroquinolone resistant broiler E. coli isolates we tested belonged to group A and that chicken meat has previously been found prevailed by A and B1 isolates [25], this finding suggests the hypothesis that avian might be a source for at least some human ciprofloxacin resistant group A strains. The change in phylogroup background of isolates from group B2 in drug-resistant ExPEC has been previously observed [26]; however, the appearance and expansion of the ciprofloxacin-resistant B2 ST131 isolates which was also dominant among this clone seems to disagree with these observations [27].
Interestingly, we didn’t find any document indicative for the carriage of ST131 isolates by broiler species which is in agreement with previous reports of rare isolation of ST131 positive cases among avian sources [27]. However, the main limitation of our study was the relatively small number of broiler isolates which restricted statistical power. Based on our results, the ST131 clone is associated with human microbiota and also predominates among fluoroquinolone-resistant strains, while it can be present among susceptible isolates, even in a low number.
Conclusion
The present study confirms that the H30 allele is prevalent among fluoroquinolone resistant isolates. Our data suggest that ST131 clone possess more virulence traits but less multidrug resistance patterns than other clinical fluoroquinolone resistant/ESBL-producing isolates. Among ESBL-positive isolates, ST131 isolates—mostly representing the H30-Rx subclone— have higher virulence scores than non-ST131 isolates, implying higher virulence power and thereby the possible reason for their high prevalence. Non-ST131 isolates including both of the clinical and broiler strains showed an inverse relationship between the prevalence of virulence markers and the resistance phenotype in which higher resistance is accompanied by lower frequency of virulence traits. Further work is needed to be done to understand the significance of avian E. coli isolates contributing to human disease.
Methods
Isolate collection
In this one year cross-sectional study (March 2015–March 2016), a total of 338 E. coli isolates were cultured from patients with extra-intestinal infections admitted to Kosar university hospital of Semnan, Iran. The clinical samples were taken as part of standard care for the patients. Sixty-three out of the 338 isolates had been previously described [13]. Moreover, a total of 28 tissue swabs of culled-animals affected with colibacillosis arrived to the laboratory at the same time period. Samples were taken from chickens already sacrificed for diagnostic purposes. The colibacillosis samples were collected from cases submitted to the Diagnostic Service of the Veterinary School of the Semnan University (Semnan Province). Swabs were taken from two broiler farms located in Semnan Province. The samples were plated onto MacConkey agar and incubated overnight at 37 °C [28].
Antibiotic susceptibility testing and phenotypic detection of ESBL producers
Antibiotic susceptibility patterns for collected E. coli isolates were determined by using the standard disc diffusion. The results of disc diffusion were interpreted based on the Clinical Laboratory Standard Institute (CLSI) recommendations [29]. We considered intermediate susceptibility patterns as resistant. Isolates with an inhibition zone of < 23 mm to any of the imipenem, meropenem, or ertapenem were categorized as carbapenem-resistant [29]. The resistance score was the number of antibiotics to which an isolate was resistant. Multidrug-resistant (MDR) was defined according to resistance to at least one representatives of ≥3 antimicrobial classes [30]. Phenotypic combined disc test based on the CLSI recommendations was used to detect the Extended Spectrum β-lactamase (ESBL) producing isolates [29].
Phylogenetic analysis and sequence typing PCR
The CTAB method was used for extraction of DNA templates for PCR [31]. Major E. coli phylogroups (A, B1, B2, C, D, E and F) were determined by quadruplex PCR [32]. For detection of major ExPEC STs (ST131, ST95, ST73, ST69 and ST127), multiplex PCR was used as described previously [33].
Screening of ST131clonal group
Single nucleotide polymorphism (SNP) in mdh and gyrB genes associated with ST131 clone was detected by using of the gyrB/mdh duplex PCR for all isolates belonged to phylogroups B2, D and F. The H30 and H30-Rx subclones of ST131 isolates were detected, and the two most common O genotypes, O25b and O16 were screened by PCR among study ST131 isolates [13].
Virulence genotyping
The presence of 24 putative virulence genes was assessed by multiplex PCR [34]. Isolates which were positive for ≥2 of the five markers, including afaDrBC, papAH, and/or papC, sfa, KpsM II, or iutA were considered as ExPEC, and the virulence score was calculated as described previously [35]. The genes described previously by Johnson et al as the minimal predictors of APEC virulence; iroN, ompT, hlyF, iutA and iss were detected by a multiplex PCR [36].
Detection of resistance genes
Isolates carrying resistance markers, including carbapenemases (blaNDM, blaOXA-48, blaIMP-, blaVIM- and blaKPC) [37], (β-lactamases) (blaTEM-, blaSHV-,blaCTX-M groups 1, 2, 8, 9, 25) [38, 39] and plasmid mediated quinolone resistance (PMQR) (qnrA, qnrB, qnrS and aac6Ib-cr) were detected by multiplex PCR according to previously published methods [40]. The presence of the blaCTX-M-15 was detected using a single PCR [41]. Moreover, aminoglycoside resistance determinants (ARD), including aac6-Ib/aac3-IIa and 16S rRNA methylase genes (armA, rmtB, rmtC) were screened by PCR [42, 43].
ERIC-PCR analysis
Clonal relatedness was established by Enterobacterial repetitive intergenic consensus sequence polymerase chain reaction (ERIC-PCR). BioNumerics software, version 6.1, (Applied Maths, Sint-Martens-Laten, Belgium) was used to analyz the ERIC patterns. The similarities in amplicon profiles were compared using a Dice coefficient at 1% tolerance and 0.5% optimization, and by using the unweighted-pair group method with arithmetic mean clustering (UPGMA) method, with a cut-off of 80% similarity a dendrogram was constructed [44].
Statistical analysis
Continuous variables were described as means and standard deviations or as medians and interquartile ranges (IQRs). Dichotomous variables were described using frequencies and percentages, and they were compared using chi-square test, as appropriate. The criterion for statistical significance was P < 0.05. Data were analyzed with software SPSS version 16.
Abbreviations
- APEC:
-
Avian pathogenic E.coli
- E. coli :
-
Escherichia coli
- ECOR:
-
E. coli reference collection
- ERIC-PCR:
-
Enterobacterial repetitive intergenic consensus sequence polymerase chain reaction
- ESBL:
-
Extended Spectrum β-lactamase
- ExPEC:
-
Extraintestinal Pathogenic E.coli
- MDR:
-
Multidrug-resistant
- MLST:
-
Multilocus sequence typing
- NMEC:
-
Neonatal meningitis E. coli
- ST:
-
Sequence type
- UPEC:
-
Urinary pathogenic E. coli
- UPGMA:
-
Unweighted-pair group method with arithmetic mean clustering
References
Talan DA, Takhar SS, Krishnadasan A. Fluoroquinolone-resistant and extended-spectrum β-lactamase-producing Escherichia coli infections in patients with pyelonephritis, United States. Emerg Inf Dis. 2016;22:1594–603.
Nicolas-Chanoine MH, Bertrand X, Madec JY. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev. 2014;27:543–74.
Price LB, Johnson JR, Aziz M. The epidemic of extended spectrum-b-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. MBio. 2013;4:e00377–13.
Jakobsen L, Spangholm DJ, Pedersen K, Pedersen K, Jensen LB, Emborg HD, Agersø Y, et al. Broiler chickens, broiler chicken meat, pigs and pork as sources of ExPEC related virulence genes and resistance in Escherichia coli isolates from community dwelling humans and UTI patients. Int J Food Microbiol. 2010;142:264–72.
Ansari-Lari M, Shekarforoush S, Mehrshad S, Safari H. Prevalence and risk factors for Salmonella spp. colonization in broiler flocks in shiraz, southern Iran. Vet Res Forum. 2014;5:65–8.
Ewers C, Li G, Wilking H, Kieling S, Alt K, Antao EM. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int J Med Microbiol. 2007;297:163–76.
Moulin-Schouleur M, Répérant M, Laurent S, Brée A, Mignon-Grasteau S, Germon P. Extraintestinal pathogenic Escherichia coli strains of avian and human origin: link between phylogenetic relationships and common virulence patterns. J Clin Microbiol. 2007;45:3366–7336.
Egea P, Lopez-Cerero L, Torres E, Gomez-Sanchez M, del C, Serrano L, et al. Increased raw poultry meat colonization by extended-spectrum β-lactamase-producing Escherichia coli in the south of Spain. Int J Food Microbiol. 2012;159:69–73.
Caruso G, Giammanco A, Cardamone C, Oliveri G, Mascarella C, Capra G. Extra-intestinal fluoroquinolone resistant Escherichia coli strains isolated from meat. Biomed Res Int. 2019;18:8714975.
Mora A, Herrera A, Mamani R, Lopez C, Alonso MP, et al. Recent emergence of clonal group O25b:K1:H4-B2-ST131 ibeA strains among Escherichia coli poultry isolates, including CTX-M-9-producing strains, and comparison with clinical human isolates. Appl Environ Microbiol. 2010;76:6991–7.
Vincent C, Boerlin P, Daignault D, Dozois CM, Dutil L, et al. Food reservoir for Escherichia coli causing urinary tract infections. Emerg Infect Dis. 2010;16:88–95.
Nicolas-Chanoine MH, Robert J, Vigan M, Laouénan C, Brisse S, Mentré F. Different factors associated with CTX-M-producing ST131 and non ST131 E. coli clinical isolates. PLoS One. 2013;8:e72191.
Hojabri Z, Mirmohammadkhani M, Kamali F, Ghassemi K, Taghavipour S, Pajand O. Molecular epidemiology of Escherichia coli sequence type 131 and its H30/H30-Rx subclones recovered from extra-intestinal infections: first report of OXA-48 producing ST131 clone from Iran. Eur J Clin Microbiol Infect Dis. 2017;36:1859–66.
Clark G, Paszkiewicz K, Hale J, Weston V, Constantinidou C, Penn C. Genomic analysis uncovers a phenotypically diverse but genetically homogeneous Escherichia coli ST131 clone circulating in unrelated urinary tract infections. J Antimicrob Chemother. 2012;67:868–77.
Fasciana T, Giordano G, DI CARLO P, Colomba C, Mascarella C, Tricoli M (2017) Virulence factors and antimicrobial resistance of ESCHERICHIA COLI ST131 in community-onset healthcare-associated infections in SICILY, Italy. Pharmacol Online 1: 12–21.
Chung HC, Lai CH, Lin JN, Huang CK, Liang SH, Chen WF. Bacteremia caused by extended-spectrum-beta lactamase- producing Escherichia coli sequence type ST131 and non- ST131 clones: comparison of demographic data, clinical features, and mortality. Antimicrob Agents Chemother. 2012;56:618–22.
Johnson JR, Urban C, Weissman SJ, Jorgensen JH, Lewis JS 2nd, Hansen G. Molecular epidemiological analysis of Escherichia coli sequence type ST131 (O25:H4) and bla CTX-M-15 among extended-Spectrum-β- lactamase-Producing E. coli from the United States, 2000 to 2009. Antimicrob Agents Chemother. 2012;56:2364–70.
Calhau V, Ribeiro G, Mendonca N, Da Silva GJ. Prevalent combination of virulence and plasmidic-encoded resistance in ST 131 Escherichia coli strains. Virulence. 2013;4:726–9.
Dahbi G, Mora A, Lopez C, Alonso MP, Mamani R, Marzoa J. Emergence of new variants of ST131 clonal group among extraintestinal pathogenic Escherichia coli producing extended-spectrum beta-lactamases. Int J Antimicrob Agents. 2013;42:347–51.
Cha MK, Kang CI, Kim SH, Cho SY, Ha YE, Wi YM. Comparison of the microbiological characteristics and virulence factors of ST131 and non-ST131 clones among extended-spectrum β-lactamase–producing Escherichia coli causing bacteremia. Diagn Microbiol Infect Dis. 2016;84:102–4.
Hussain A, Ranjan A, Nandanwar N, Babbar A, Jadhav S, Ahmed N. Genotypic and phenotypic profiles of Escherichia coli isolates belonging to clinical sequence type 131 (ST131), clinical non-ST131, and fecal non-ST131 lineages from India. Antimicrob Agents Chemother. 2014;58:7240–9.
Han JH, Garrigan C, Johnston B, Nachamkin I, Clabots C, Bilker WB. Epidemiology and characteristics of Escherichia coli sequence type 131 (ST131) from long-term care facility residents colonized intestinally with fluoroquinolone-resistant Escherichia coli. Diagn Microbiol Infect Dis. 2017;87:275–80.
Merino I, Shaw E, Horcajada JP, Cercenado E, Mirelis B, Pallarés MA. CTX-M-15-H30Rx-ST131 subclone is one of the main causes of healthcare-associated ESBL-producing Escherichia coli bacteraemia of urinary origin in Spain. J Antimicrob Chemother. 2016;71:2125–30.
Solà-Ginés M, Cameron-Veas K, Badiola I, Dolz R, Majó N, Dahbi G. Diversity of multi-drug resistant avian pathogenic Escherichia coli (APEC) causing outbreaks of Colibacillosis in broilers during 2012 in Spain. PLoS One. 2015;10:e0143191.
Jakobsen L, Kurbasic A, Skjøt-Rasmussen L. Escherichia coli isolates from broiler chicken meat, broiler chickens, pork, and pigs share phylogroups and antimicrobial resistance with community-dwelling humans and patients with urinary tract infection. Foodborne Pathog Dis. 2010;7:537–47.
Moreno E, Prats G, Sabate´ M. Quinolone, fluoroquinolone and trimethoprim/sulfamethoxazole resistance in relation to virulence determinants and phylogenetic background among uropathogenic Escherichia coli. J Antimicrob Chemother. 2006;57:204–11.
Giufrè M, Graziani C, Accogli M, Luzzi I, Busani L, Cerquetti M. Escherichia coli of human and avian origin: detection of clonal groups associated with fluoroquinolone and multidrug resistance in Italy. J Antimicrob Chemother. 2012;67:860–7.
Heininger A, Binder M, Schmidt S, Unertl K, Botzenhart K, Doring G. PCR and blood culture for detection of Escherichia coli bacteremia in rats. J Clin Microbiol. 1999;37:2479–82.
Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; twentieth informational supplement M100S-S26. Wayne, PA: CLSI; 2016.
Johnson JR, Johnson B, Clabots C, Kuskowski MA, Pendyala S, DebRoy C. Escherichia coli sequence type ST131 as an emerging fluoroquinolone-resistant uropathogen among renal transplant recipients. Antimicrob Agents Chemother. 2010;54:546–5450.
Ghotaslou R, Aghazadeh M, Ahangarzadeh Rezaee M, Moshafi MH, Forootanfar H, Hojabri Z. The prevalence of aminoglycoside-modifying enzymes among coagulase negative staphylococci in Iranian pediatric patients. J Infect Chemother. 2014;20:569–73.
Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep. 2013;5:58–65.
Hojabri Z, Mirmohammadkhani M, Darabi N, Arab M, Pajand O. Characterization of antibiotic-susceptibility patterns and virulence genes of five major sequence types of Escherichia coli isolates cultured from extraintestinal specimens: a 1-year surveillance study from Iran. Infect and Drug Resist. 2019;12:893–903.
Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with Urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000;181:261–72.
Johnson JR, Porter S, Thuras P, Castanheira M. The pandemic H30 subclone of sequence type 131 (ST131) as the leading cause of multidrug-resistant Escherichia coli infections in the United States (2011-2012). Open Forum Infect Dis. 2017;4:ofx089.
Johnson TJ, Wannemuehler Y, Doetkott C, Johnson SJ, Rosenberger SC, Nolan LK. Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. J Clin Microbiol. 2008;46:3987–96.
Hojabri Z, Arab M, Darabi N, Kia NS, Lopes BS, Pajand O. Evaluation of the commercial combined disk test and minimum inhibitory concentration (MIC) determination for detection of carbapenemase producers among gram-negative bacilli isolated in a region with high prevalence of blaOXA-48 and blaNDM. Int Microbiol. 2019;22:81–9.
Hojabri Z, Pajand O, Bonura C, Aleo A, Giammanco A, Mammina C. Molecular epidemiology of Acinetobacter baumannii in Iran: endemic and epidemic spread of multiresistant isolates. J Antimicrob Chemother. 2014;69:2383–7.
Woodford N, Fagan EJ, Ellington MJ. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum (beta)-lactamases. J Antimicrob Chemother. 2006;57:154–5.
Ciesielczuk H, Hornsey M, Choi V, Woodford N, Wareham DW. Development and evaluation of a multiplex PCR for eight plasmid-mediated quinolone-resistance determinants. J Med Microbiol. 2013;62:1823–7.
Blanco J, Mora A, Mamani R, López C, Blanco M, Dahbi G, et al. Four Main Virotypes among extended-Spectrum-β-lactamase- producing isolates of Escherichia coli O25b:H4-B2-ST131: bacterial, epidemiological, and clinical characteristics, vol. 51; 2013. p. 3358–67.
Haldorsen BC, Simonsen GS, Sundsfjord A, Samuelsen O, Norwegian Study Group on Aminoglycoside Resistance. Increased prevalence of aminoglycoside resistance in clinical isolates of Escherichia coli and Klebsiella spp. in Norway is associated with the acquisition of AAC (3)-II and AAC (6′) Ib. Diagn Microbiol Infect Dis. 2014;78:66–9.
Pajand O, Hojabri Z, Nahaei MR, Hajibonabi F, Pirzadeh T, Aghazadeh M. In vitro activities of tetracyclines against different clones of multidrug-resistant Acinetobacter baumannii isolates from two Iranian hospitals. Int J Antimicrob Agents. 2014;43:476–8.
McLellan SL, Daniels AD, Salmore AK. Genetic characterization of Escherichia coli populations from host sources of fecal pollution by using DNA fingerprinting. Appl Environ Microbiol. 2003;69:2587–94.
Acknowledgements
None.
Funding
This work was supported fully by Semnan University of Medical Sciences (grant No. 1180). This is a report of a database from research projects entitled; “Detection of Virulence Factors Encoding Genes among sequence type 131 of Escherichia coli isolates recovered from hospitalized patients in Kowsar hospital, Iran”, registered in Semnan University of Medical Sciences, Semnan, Iran.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
All authors read and approved the final manuscript, ZH: analyzed the dendogram, MA & ND: did laboratory works, OP: wrote the manuscript, FS: analyzed the SPSS.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Hojabri, Z., Darabi, N., Arab, M. et al. Clonal diversity, virulence genes content and subclone status of Escherichia coli sequence type 131: comparative analysis of E. coli ST131 and non-ST131 isolates from Iran. BMC Microbiol 19, 117 (2019). https://doi.org/10.1186/s12866-019-1493-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-019-1493-8