Abstract
Background
The introduction of MALDI-TOF MS in the clinical microbiology laboratory has modified the approaches for the identification of fungi. Thanks to this tool, it is possible to identify cryptic species, which possess critical susceptibility patterns. Clinical strains were identified using the MicroScan and MALDI-TOF MS systems. Discrepant results from both methods were investigated using ITS rDNA barcoding. Finally, these isolates were also tested for in vitro susceptibility.
Results
The percentage of agreement between both methods to 498 yeast isolates was of 93.6% (32 discrepant isolates). The concordance of ITS sequencing with MALDI-TOF MS was higher (99%) than that of MicroScan (94%). Several of these discordant yeasts displayed high MICs for antifungal agents.
Conclusions
Our study highlights the need of the MS and molecular approaches such as MALDI-TOF MS and ITS rDNA barcoding for the correct identification of emerging or cryptic yeast species; besides, some of these could be multidrug resistant.
This work was the first experience in the implementation of the MALDI-TOF MS technology in Colombia. We found the first uncommon yeasts including Candida auris and we could identify Trichosporon faecalis. Our work highlights a clear necessity of an accurate yeast identification as a much more pertinent technique than the susceptibility profiles, because the most unusual yeasts exhibit resistance profiles to the few available antifungals.
Similar content being viewed by others
Background
In the last decades, the incidence of invasive fungal infections has progressively increased, especially in critically ill and immunocompromised patients [1, 2]. Yeast infections are mostly caused by Candida followed by Cryptococcus, and less frequently by Rhodotorula, Saccharomyces, Trichosporon and Pichia. These emerging species are also contributing to the epidemiological changes recorded in recent years, which significantly impact therapeutic regimens in patients as some yeasts can exhibit innate drug resistance [3,4,5,6].
Within the genus Candida, C. albicans is still the most common fungal pathogen worldwide. Despite being part of the mucocutaneous, gastrointestinal and genitourinary mycobiota in humans, C. albicans can be responsible for nearly 50% of candidaemia. In the 90’s the prevalence of candidaemia was between 64 and 48% but this percentage decreased to 38% during the period 2008–2011 due to increased prevalence of non C. albicans species [5, 7, 8]. For Cryptococcus, reports indicate a prevalence of 4%. Such a low percentage has not ever been described for emerging species [9,10,11].
The accurate identification of the fungal species that cause the infection is of paramount importance since the necessity to initiate the appropriate antifungal therapy. Furthermore, the time required for diagnosis/identification is also critical as any delay will affect the prognosis dramatically when dealing with invasive candidiasis as well as complicate a relevant stewardship [12, 13].
On South America mycological identification usually relies on phenotypic, biochemical, enzymatic and immunological approaches [14]. The MicroScan system is a rapid technology for microbial identification able to provide results after 4 h of inoculation [15]. Only available for yeast identification, this method relies on enzymatic reactions in a panel. The enzyme activities of each isolate are determined by colour changes in the chromogenic substrates (or a pH indicator). The biochemical reactions generate numerical profiles, which are then compared with a numerical database. In recent years, Mass Spectrometry (MS) using MALDI-TOF MS technology has been increasingly used as a tool for microbiological identification due to its high performance and less time required when compared with conventional methods [16].
Our aim was to compare the performance of MicroScan with that of MALDI-TOF MS for yeast identification. This study was conducted on a large collection of clinical isolates collected prospectively at the San Ignacio Hospital, Bogota, Colombia. Those isolates yielding discrepant results were further identified by the gold standard ITS and D1-D2 rDNA barcoding. Because of the few data on rare/emerging yeasts, susceptibility profiles were also determined.
Results
Performance of MicroScan in comparison with Bruker MALDI-TOF MS
With the MicroScan technology 497/498 (99.7%) strains were identified, belonging to 16 distinct species from 7 genera. When performing MALDI-TOF MS identification with the Bruker instrument 494 /498 (99%) strains were identified, belonging to 21 distinct species from 6 genera. The percentage of agreement between both methods was 93.5% (466 isolates) (Table 1). The remaining 32 isolates yielding discrepant results (n = 27) or being no-identified by one of the two methods were subjected to further investigations for molecular identification as the gold standard.
Resolution of the discrepancies by sequencing
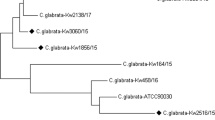
The 32 discordant identifications occurred mostly with rare and/or emergent species. Table 2, 3 presents the total and individual cases of discordance, and the species with which there was confusion. The vast majority of errors (n = 30) occurred with the MicroScan system while MALDI-TOF MS Bruker showed the highest agreement with ITS rDNA barcoding. Using the Bruker Biotyper instrument, the four isolates (0.8%) yielding “no identification” (score < 1.7) were finally identified by sequencing as Saprocheta suaveolens (n = 1) and Trichosporon faecale (n = 3). Notably, the MALDI-TOF MS Bruker could identify several cryptic species such as the multidrug resistant C. auris (n = 5), C. intermedia (n = 3), C. metapsilosis (n = 4), C. nivariensis (n = 1), C. orthopsilosis (n = 2), C. gattii (n = 2), C. neoformans var. grubii (n = 2), and G. candidum (n = 2) (Tables 2, 3).
In vitro antifungal susceptibility
We assessed the susceptibility profiles of isolates that showed discrepancies in their identification. Among Candida yeasts, C. auris presented the highest MIC values for FLU and AMB; C. metapatilosis for AMB and ITC. C. dubliniensis was the only species with a low MIC for VRC. C. intermedia and C. nivariensis were the only two species susceptible to all of the drugs evaluated (Table 4).
On the other hand, all the other yeasts (non Candida) showed higher MIC values for FLU and generally for azoles but were sensitive to AMB. All isolates had high MICs for echinocandins except P. kluyveri (Table 5).
Discussion
Proteomic analysis using the MALDI-TOF MS methodology offers a great opportunity for identifying microorganisms that are difficult to identify by biochemical methods. In this study, we compared two methodologies for the identification of clinically relevant yeasts: the Microscan system (biochemical approach) and the Bruker MS system (proteomic approach). This study also confirmed the wide diversity of species obtained from patients in the clinical setting, highlighting the importance of a correct identification at the species level for the determination of the appropriate therapy (since cryptic species may have non-predicted susceptibility profiles). The correct identification of clinical yeast isolates has become essential for optimal clinical management, as well as for detailed epidemiological studies and the prevention and containment of outbreaks. It is in our interest to analyse in depth the cases where the identification errors are important in the treatment, as it is in the case of the isolations of C. auris, as a misidentification yeast by conventional technology [17].
A correct identification by morphological and conventional testing with MicroScan was achieved for 94% of the isolates, a result similar (96% of n = 357 clinical isolates) to a previously reported one by other authors using the same system [15]. The automated system MicroScan handles 10% of colorimetric biochemistry and 90% of enzymatic biochemistry; the latter are reversible tests depending on the reading time. This is of great importance since if there is no identification in 4 h (time indicated by the commercial house) the biochemical reactions can revert themselves, leading to false negative results. In addition, the performances of this phenotypic system are intimately dependent on regular updates of the database. Since MicroScan is not capable of identifying Geotrichum spp. (at the species level), we recorded those isolate identifications as erroneous. In addition, cryptic or recently emerging species such as C. auris, C. intermedia C. metapsilosis, C. nivariensis, C. orthopsilosis, C. gattii and S. suaveolens, which are not yet present in the MicroScan database version 4.11.1020.1 cannot be readily identified.
MALDI-TOF MS systems have recently been developed and implemented in diagnostic microbiology laboratories for bacterial and fungal identification due to their efficacy, rapidity and minimum hands-on time. This technology, as highlighted here, provides valid results for most yeast species (84–99%). However, optimal results depend on the robustness of the system libraries [18, 19]. For those reasons, the accuracy of the MALDI-TOF MS System is modifying the way in which yeast identification is being performed, surpassing the conventional techniques. In this study, MALDI-TOF MS could correctly identify 98.9 of the strains analysed. The 5 errors were due to a misidentification of C. metapsilosis and the lack of identification of Trichosporon faecale and Saprochaete suaveolens even though they are present in the Biotyper database. We believe that the error was possibly due to the frequent difficulty in protein extraction for these species. In the case of Saprochaete suaveolens the database is not robust thus leading to erroneous results. A solution to the identification of these microorganisms is the creation or improvement of libraries [20].
Regarding susceptibility the change in the epidemiology of Candida species has occurred at the same time with the emergence of Candida strains resistant to antifungal drugs mainly to fluconazole, which is used as the first line pharmacological treatment [21, 22]. In this study, we did not handle cut-off points in the interpretation of susceptibility profiles since some of the yeasts do not have an established MIC [23]. It is important to mention that in all cases the non-albicans yeasts showed high MIC values, at least for one of the antifungals evaluated.
Successful treatment of fungal infections depends on the early identification of species and patterns of sensitivity to antifungal agents. The growth rate of resistance confirms the importance of monitoring changes in the distribution of pathogenic species. The sensitivity pattern of the Candida species revealed in this study shows that amphotericin B, voriconazole and caspofungin with the lowest MICs appear to be suitable drugs. Moreover, as expected from previous studies, for most of non-Candida species, amphotericin B was the antifungal displaying the highest in vitro activity.
In conclusion, yeast identification has advanced enabling the recognition of new “cryptic species”. These yeasts are yet not easily identifiable by traditional phenotypic methods commonly employed in clinical laboratories. In this study, we have shown the high performance of MALDI-TOF MS technology for the identification of clinically relevant yeasts. Moreover, continued increase on the number of susceptible patients and the selection pressures imposed using antifungal drugs continue to result in the emergence of new microorganisms that cannot be easily identified in laboratories. It is important that physicians take into account the identification of pathogenic yeasts down to the species level due to the diversity of antifungal sensitivity profiles.
Conclusions
Our study highlights the need of the MS and molecular approaches such as MALDI-TOF MS and ITS rDNA barcoding for the correct identification of emerging or cryptic yeast species; besides, some of these could be multidrug resistant.
Methods
Study design
The study was a single-center prospective analysis covering the period from November of 2013 to March of 2015. All the yeast isolates obtained from hospitalized immunocompromised patients of the San Ignacio Hospital were included in the present study. Yeasts were identified at the Laboratory of Microbiology of the San Ignacio Hospital using MicroScan (MicroScan WalkAway-96 Plus, Siemens, Deerfield, IL, USA) and at the Laboratory of Proteomics and Human Mycoses of the Faculty of Science of the Pontificia Universidad Javeriana using MALDI-TOF MS (Bruker Daltonics, Bremen, Germany).
A total of 498 clinical isolates were recovered from different samples such as respiratory secretions (n = 168), stool (n = 133), urine (n = 55), blood (n = 50), swabs (n = 44), biopsies (n = 24), medical devices (n = 10), cerebrospinal fluids (n = 10), abdominal fluids (n = 3) and bone marrow (n = 1). Species identification was performed using MicroScan in parallel with MALDI-TOF MS. Discrepant isolates were subjected to molecular identification as the gold standard.
MicroScan WalkAway system
The MicroScan system for yeast identification consists of a 96-well microdilution plate with 27 dehydrated chromogenic substrates. The enzyme activity of each isolate is determined by colour change. Isolates were previously grown on Sabouraud dextrose agar (SDA) at 30 °C for 24–36 h. Suspensions were prepared and calibrated against the MicroScan turbidity standard, and the panel was incubated aerobically for 4 h at 37 °C. Biochemical reactions generate numerical profiles that are compared with a database to identify organisms [15]. Urease assimilation and growth on canavanine-glycine-bromothymol blue agar confirmed the identification for Cryptococcus isolates.
MALDI-TOF MS
Bruker Biotyper
After incubation of clinical strains at 35 °C for 24–36 h on SDA, protein extraction was performed using the formic acid/ethanol method according to the Bruker Daltonics protocol. Briefly, two or three colonies were mixed with 300 μl of HPLC grade water until homogenization and then 600 μl of 100% ethanol (Sigma-Aldrich, St Louis, MO) were added. After centrifugation at 15,000 g for 2 min, the pellet was dried at 25 °C, reconstituted in equal volumes of 100% formic acid (Sigma- Aldrich) and acetonitrile (Sigma-Aldrich) (20 μL each), mixed thoroughly, and centrifuged at 15, 000 g for 2 min. One microliter of supernatant was spotted onto a 96-spot steel plate (Bruker Daltonik) and allowed to dry at room temperature before the addition of 1 mL of the HCCA matrix (provided by the supplier). Each sample was tested in duplicate. Only the spot returning the highest probability score of identification was considered [24]. The protein mass spectra was analysed using the Flex Control software and the MALDI Biotyper version 3.1 7311 reference spectra (main spectra) (Bruker Daltonics, Bremen, Germany). MALDI-TOF MS results were then compared and a score was obtained according to the manufacturer’s technical specifications, as follows: correct genus and species identification (≥2.0), correct genus identification (1.7–2.0), and no reliable identification (< 1.7) [25]. The 498 clinical isolates was a score above 2.
Molecular identification
Molecular identification of discrepant isolate results between both methods was subjected to molecular identification by amplification and sequencing of the ITS rDNA regions without prior DNA extraction step (colony PCR) [26]. Amplification of the ITS rDNA was achieved using the universal primers ITS1 (TCCGTAGGTGAACCTGCGG) and ITS4 (TCCTCCGCTTATTGATATGC) [27]. Nucleotide sequences were assembled using the Seq Scape software (Applied Biosystems, Foster City, CA, USA.) and compared with the GenBank database using the BLAST algorithm or with the MycoBank database. For some isolates, identification was further confirmed by amplification of the D1-D2 region of the 28S rDNA using the primers NL1 (GCATATCAATAAGCGGAGGAAAAG) and NL4 (GGTCCGTGTTTCAAGACGG) [27]. A similarity ≥98% between the unknown sequence and the closest matching sequence from the reference database was used as the criterion to identify an isolate to the species level. Additional primer pairs targeting the IGS1, namely IGS1-F (ATCCTTTGCAGACGACTTGA) and IGS1-R (GTGATCAGTGCATTGCATGA) for Cryptococcus, and 26SF (ATCCTTTGCAGACGACTTGA) and (AGCTTGACTTCGCAGATCGG) for Trichosporon were used to obtain a reliable identification at the species level [28, 29].
In vitro antifungal susceptibility testing
Each isolate displaying discrepant results was subjected to in vitro susceptibility testing using the E-test (Biomérieux) against triazoles (fluconazole, itraconazole and voriconazole), echinocandins (anidulafungin and caspofungin), and amphotericin B, according to the manufacturer’s instructions. The MIC was read as the drug concentration that leads to complete inhibition (100%) for amphotericin B, and 80% inhibition for azoles and echinocandins. Two control strains (C. parapsilosis ATCC 22019 and C. krusei ATCC 6258) were included in each set of experiments.
Abbreviations
- AMB:
-
Amphotericin B
- FLU:
-
Fluconazole
- ITC:
-
Itraconazole
- ITS:
-
Internal transcribed spacer
- MALDI-TOF MS:
-
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
- MIC:
-
Minimum inhibitory concentration
- VRC:
-
Voriconazole
References
Low C-Y, Rotstein C. Emerging fungal infections in immunocompromised patients. F1000 Med Rep. 2011;3:14.
Quindós G. Epidemiology of candidaemia and invasive candidiasis. A changing face. Rev Iberoam Micol. 2014;31(1):42–8.
Spampinato C, Leonardi D. Candida infections, causes, targets, and resistance mechanisms: traditional and alternative antifungal agents. Biomed Res Int. 2013;2013:204237.
Posteraro B, Ruggeri A, De Carolis E, Torelli R, Vella A, De Maio F, et al. Comparative evaluation of BD Phoenix and vitek 2 systems for species identification of common and uncommon pathogenic yeasts. J Clin Microbiol. 2013 Nov 1;51(11):3841–5.
Yapar N. Epidemiology and risk factors for invasive candidiasis. Ther Clin Risk Manag. 2014;10:95–105.
Leaw SN, Chang HC, Barton R, Bouchara J-P, Chang TC. Identification of medically important Candida and non-Candida yeast species by an oligonucleotide array. J Clin Microbiol. 2007;45(7):2220–9.
Puig-Asensio M, Ruiz-Camps I, Fernández-Ruiz M, Aguado JM, Muñoz P, Valerio M, et al. Epidemiology and outcome of candidaemia in patients with oncological and haematological malignancies: results from a population-based surveillance in Spain. Clin Microbiol Infect. 2015;21(5):491.e1–491.e10.
Kullberg BJ, Arendrup MC. Invasive Candidiasis. Campion EW, editor. N Engl J Med 2015;373(15):1445–1456.
D’Souza CA, Kronstad JW, Taylor G, Warren R, Yuen M, Hu G, et al. Genome Variation in Cryptococcus gattii, an Emerging Pathogen of Immunocompetent Hosts. MBio. 2011;2(1):e00342–10.
Desalermos A, Kourkoumpetis TK, Mylonakis E. Update on the epidemiology and management of cryptococcal meningitis. Expert Opin Pharmacother. 2012;13(6):783–9.
de APADF, Pedroso RDS, Borges AS, Moreira T de A, de ALB, Röder DVD de B. The epidemiology of cryptococcosis and the characterization of Cryptococcus neoformans isolated in a Brazilian University hospital. Rev Inst Med Trop Sao Paulo. 2017;59:e13.
Morrell M, Fraser VJ, Kollef MH. Delaying the empiric treatment of Candida bloodstream infection until positive blood culture results are obtained: a potential risk factor for hospital mortality. Antimicrob Agents Chemother. 2005;49(9):3640–5.
Hsueh P-R, Graybill JR, Playford EG, Watcharananan SP, Oh M-D, Ja’alam K, et al. Consensus statement on the management of invasive candidiasis in intensive care units in the Asia-Pacific region. Int J Antimicrob Agents. 2009;34(3):205–9.
Lohmann C, Sabou M, Moussaoui W, Prévost G, Delarbre J-M, Candolfi E, et al. Comparison between the Biflex III-Biotyper and the Axima-SARAMIS systems for yeast identification by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2013;51(4):1231–6.
St.-Germain G. G, Beauchesne D. Evaluation of the MicroScan rapid yeast identification panel. J Clin Microbiol. 1991;29(10):2296–9.
Putignani L, Del Chierico F, Onori M, Mancinelli L, Argentieri M, Bernaschi P, et al. MALDI-TOF mass spectrometry proteomic phenotyping of clinically relevant fungi. Mol BioSyst. 2011;7(3):620–9.
Parra-Giraldo CM, Valderrama SL, Cortes-Fraile G, Garzón JR, Ariza BE, Morio F, et al. First report of sporadic cases of Candida auris in Colombia. Int J Infect Dis. 2018;69:63.
Jamal WY, Ahmad S, Khan ZU, Rotimi VO. Comparative evaluation of two matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) systems for the identification of clinically significant yeasts. Int J Infect Dis. 2014;26:167–70.
Haas M, Grenouillet F, Loubersac S, Ariza B, Pepin-Puget L, Alvarez-Moreno CA, et al. Identification of cryptic Candida species by MALDI-TOF mass spectrometry, not all MALDI-TOF systems are the same: focus on the C. parapsilosis species complex. Diagn Microbiol Infect Dis. 2016;86(4):385–6.
Kolecka A, Khayhan K, Groenewald M, Theelen B, Arabatzis M, Velegraki A, et al. Identification of medically relevant species of arthroconidial yeasts by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2013;51(8):2491–500.
Wiederhold NP. Antifungal resistance: current trends and future strategies to combat. Infect Drug Resist. 2017;10:249–59.
Mnge P, Okeleye BI, Vasaikar SD, Apalata T. Species distribution and antifungal susceptibility patterns of Candida isolates from a public tertiary teaching hospital in the eastern Cape Province, South Africa. 2017;15:50.
Clinical and Laboratory Standards Institute (CLSI). Reference method for broth dilution antifungal susceptibility testing of yeasts; fourth informational supplement. Wayne: Clinical and Laboratory Standards Institute. 2012. (Document M27–S4).
Pinto A, Halliday C, Zahra M, van Hal S, Olma T, Maszewska K, et al. Matrix-assisted laser desorption ionization-time of flight mass spectrometry identification of yeasts is contingent on robust reference spectra. PLoS One. 2011;6(10):e25712.
Rodríguez-Leguizamón G, Fiori A, López LF, Gómez BL, Parra-Giraldo CM, Gómez-López A, et al. Characterising atypical Candida albicans clinical isolates from six third-level hospitals in Bogotá, Colombia. BMC Microbiol. 2015;15(1):199.
Luo G, Mitchell TG. Rapid identification of pathogenic fungi directly from cultures by using multiplex PCR. J Clin Microbiol. 2002;40(8):2860–5.
White TJ, Bruns S, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications; 1990.
Sugita T, Nakajima M, Ikeda R, Matsushima T, Shinoda T. Sequence analysis of the ribosomal DNA intergenic spacer 1 regions of Trichosporon species. J Clin Microbiol. 2002;40(5):1826–30.
Meyer W, Aanensen DM, Boekhout T, Cogliati M, Diaz MR, Esposto MC, et al. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med Mycol. 2009;47:561–70.
Acknowledgements
We would like to thank Sandra Constantino for linguistic assistance.
Funding
The research office of Hospital Universitario San Ignacio and vice rectory of research at the Pontificia Universidad Javeriana Bogotá, Colombia supported the research (grants no. 2014–52 and 7710) and ECOS Nord mobility (Colombia-France).
Availability of data and materials
The datasets corresponding to Mass spectrometry from MALDI TOF are available with the corresponding author CMPG, for sequencing data from molecular identification are supervised and storage by PLP. This can be requested by mail. We are interested to work in the construction of new libraries in MALDI TOF with different institution around the world.
Author information
Authors and Affiliations
Contributions
AC-G, GC, ELZ-C performance of experiments, MYL, BEA, and FM analyzed microbial data, SLV and JRG Analyzed the clinical data, CAA, CMP-G, PLP,: Conceived and designed the experiments. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The research and ethics committee of the Hospital Universitario San Ignacio (HUSI) approved this study (no. FM-CIE-8053-14). All patients are anonymized and only the code of isolates were transferred for this investigation. Therefore, no informed consent was required.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Ceballos-Garzón, A., Cortes, G., Morio, F. et al. Comparison between MALDI-TOF MS and MicroScan in the identification of emerging and multidrug resistant yeasts in a fourth-level hospital in Bogotá, Colombia. BMC Microbiol 19, 106 (2019). https://doi.org/10.1186/s12866-019-1482-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-019-1482-y