Abstract
Background
Pasteurella multocida (P. multocida) is an important veterinary pathogen that can cause severe diseases in a wide range of mammals and birds. The global regulator crp gene has been found to regulate the virulence of some bacteria, and crp mutants have been demonstrated to be effective attenuated vaccines against Salmonella enterica and Yersinia enterocolitica. Here, we first characterized the crp gene in P. multocida, and we report the effects of a crp deletion.
Results
The P. multocida crp mutant exhibited a similar lipopolysaccharide and outer membrane protein profile but displayed defective growth and serum complement resistance in vitro compared with the parent strain. Furthermore, crp deletion decreased virulence but did not result in full attenuation. The 50 % lethal dose (LD50) of the Δcrp mutant was 85-fold higher than that of the parent strain for intranasal infection. Transcriptome sequencing analysis showed that 92 genes were up-regulated and 94 genes were down-regulated in the absence of the crp gene. Finally, we found that intranasal immunization with the Δcrp mutant triggered both systematic and mucosal antibody responses and conferred 60 % protection against virulent P. multocida challenge in ducks.
Conclusion
The deletion of the crp gene has an inhibitory effect on bacterial growth and bacterial resistance to serum complement in vitro. The P. multocida crp mutant was attenuated and conferred moderate protection in ducks. This work affords a platform for analyzing the function of crp and aiding the formulation of a novel vaccine against P. multocida.
Similar content being viewed by others
Background
Pasteurella multocida (P. multocida) is a non-motile, capsulated, Gram-negative facultative anaerobic bacterium and is recognized as an important veterinary pathogen [1]. P. multocida is classified into five serogroups (A to F) based on its capsular antigens and into 16 serovars based on its somatic antigens [2]. Certain strains of P. multocida cause fowl cholera, a disease of poultry and wild birds resulting in high mortality rates with economic significance [3]. Some control is achieved with adjuvanted bacterins, which provide some degree of protective immunity and limit the incidence and severity of clinical disease, but this type of vaccine lacks the ability to induce long-term immunity and cross-protection against heterologous serotypes, resulting in immunized animals that continue to suffer disease outbreaks [4]. In an attempt to mimic natural infection and elicit long-term humoral and cellular immunity, empirically derived, live, avirulent strains have been developed. However, the basis for attenuation is not known, and reversion to virulence occurs [4]. Thus, new vaccines, particularly well-defined live vaccines, are required, and a significant amount of current research is directed toward achieving this goal.
The design of such vaccines is based on a wealth of new information on the pathogenesis of this bacterium. Global regulators play a vital role in the adaption of bacteria to the environmental changes that are encountered during infection, such as the PhoP/PhoQ regulators, which induce modifications of lipopolysaccharides (LPS) or outer membrane proteins (OMPs) to deal with environmental extremes and promote bacterial survival [5]. Many of these regulators are closely related to virulence [6–11] and are proven mutation targets for vaccine development [12, 13]. Crp (cAMP receptor protein) was the first prokaryotic transcription factor to be purified [14] and crystallized [15] from Escherichia coli (E. coli) and is the best characterized. Crp plays a vital role in the transcription of a series of genes for the utilization of carbon sources other than glucose [16]. This protein regulates the expression of numerous genes in response to variations in the intracellular concentration of cAMP [17], which is synthesized by membrane-bound adenylate cyclase. The cya gene, which encodes adenylate cyclase, is activated in the absence of glucose [18]. Upon binding to cAMP, the cAMP-Crp complex undergoes a conformational change that allows it to bind to promoters containing the consensus TGTGAN6TCACA sequence [19]. After binding to promoter DNA, Crp recruits RNA polymerase, resulting in the formation of specific protein-protein interactions that lead to the transcription of target genes. At some promoters, Crp also inhibits transcription via several mechanisms, such as promoter occlusion [20, 21]. A total of 254 target promoters have been identified in E. coli [22]. Crp has been shown to control the expression of essential virulence factors, and crp mutants attenuate the virulence of many Gram-negative bacteria, including Salmonella enterica [23], Mycobacterium tuberculosis [24], Vibrio cholerae [25] and pathogenic Yersinia species [26, 27]. Furthermore, Salmonella enterica strains with mutations in crp either alone or in combination with other genes have served as effective vaccine candidates against salmonellosis [28, 29].
The sequence and functions of the crp gene in P. multocida have not yet been identified. In this study, P. multocida 0818 was selected to investigate the putative crp gene. The bacterium was isolated from livers of ducks with a typical clinical representation of pasteurellosis from a duck farm suffering a pasteurellosis outbreak in southwest China. P. multocida 0818 was typed as capsular serotype A:1, nontoxinogenic, and was highly virulent, with a 50 % muscular lethal dose of <100 CFU being observed for 3-day-old ducklings (unpublished data). Here, the crp gene was first characterized from the virulent P. multocida 0818 strain. Then, the non-polar Δcrp mutant of P. multocida 0818 was constructed, and its phenotype, including its virulence, resistance to serum complement and bacterial growth, lipopolysaccharide (LPS) and outer membrane protein (OMP) profiles, were systematically investigated. crp-regulated genes were also identified through transcriptome sequencing. Moreover, the immunogenicity and protective efficacy of the Δcrp mutant were determined.
Methods
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in this study are described in Table 1. P. multocida was grown at 37 °C in brain heart infusion (BHI) broth or on BHI agar (BD Bioscience, USA), and Salmonella enterica serovar Typhimurium (S. Typhimurium) and E. coli were grown in Luria-Bertani (LB) broth or on LB agar. When required, antibiotics and diaminopimelic acid (DAP) were added to the medium at the following concentrations: kanamycin, 50 μg/ml; ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml for S. Typhimurium and E. coli or 2.5 μg/ml for P. multocida; and DAP, 50 μg/ml [30]. The transformation of S. Typhimurium and P. multocida was performed via electroporation. Transformants were selected on LB or BHI agar plates containing appropriate antibiotics, and Asd+ plasmids were selected on LB agar plates.
Molecular and genetic procedures
Restriction digests and ligations were performed using enzymes purchased from New England Biolabs (NEB, Beverley, MA, USA) and TAKARA (Takara Bio Inc., Shiga, Japan), respectively, according to the manufacturer’s instructions. Plasmid DNA was extracted from bacteria using the TIANprep Mini Plasmid Kit (Tiangen Biotech Co., Ltd., Beijing, China), whereas genomic DNA was prepared using the cetyltrimethylammonium bromide method [31]. The DNA was amplified via PCR using PrimeSTAR Max DNA polymerase (Takara Bio Inc., Shiga, Japan) or Taq DNA polymerase (Tiangen Biotech) and purified using a DNA Purification Kit (Tiangen Biotech). The primers employed in this study were designed according to the published genome sequence of P. multocida strain Pm70 (GenBank, AE004439.1) [32] and are listed in Table 2. The DNA sequences were commercially determined by BGI Tech (BGI Tech Solutions Co., Ltd., Shenzhen, China), and sequence alignments were constructed using the Basic Local Alignment Search Tool (BLAST).
Plasmids and mutant strain construction
BLAST was applied to search for potential P. multocida crp gene (PM1157) via alignment of the amino acid sequences of S. Typhimurium Crp (Protein ID, NP_462369.1) and the genome of P. multocida Pm70. To clarify the potential gene in P. multocida, the PM1157 gene sequence was amplified from the P. multocida 0818 strain with the primers Ccrp-F1/Ccrp-R1. The amplified DNA fragment was inserted into pQK663 derived from pYA3332 [33] between the NcoI and BamHI digestion sites to generate pQK163, which was then transformed into the S. Typhimurium Δasd Δcrp strain for a maltose fermentation assay. For expression of the Crp protein, the complete P. multocida crp sequence was amplified from P. multocida 0818 chromosomal DNA using the primers pET-crp-F and pET-crp-R. The PCR fragment was then purified and digested with kpnI-HF and BamHI-HF (NEB) and subsequently ligated to the pET-32a expression vector (Novagen Inc., Madison, WI, USA) between the kpnI and BamHI sites to generate pET-32a-crp. To complement the crp mutant in P. multocida, the complete crp gene was amplified from P. multocida 0818 genomic DNA using the primers Ccrp-F2/Ccrp-R2, and the amplified fragment was then digested and inserted into the NotI and BamHI sites of a shuttle vector pMC-Express [34] (kindly donated by Paul R Langford from Imperial College London) to generate pQK176. The plasmids pQK163 and pQK176 were transformed into the crp mutant strains S411 (S. Typhimurium Δasd Δcrp) and S416 (P. multocida Δcrp), respectively, generating the corresponding complementary strains S411 (pQK163) and S416 (pQK176).
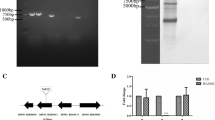
The P. multocida Δcrp mutant was constructed by allelic exchange using the suicide T-vector pYA4278 [35] as previously described [36]. Briefly, a 410-bp upstream segment and a 416-bp downstream segment of the crp gene were amplified with the primers Dcrp-1 F/Dcrp-1R and Dcrp-2 F/Dcrp-2R, respectively. The two segments were then linked via PCR using the primers Dcrp-1 F/Dcrp-2R. The PCR product was then ligated to AhdI-digested pYA4278 to generate the plasmid pQK174. Next, the kanamycin resistance (kanR) cassette amplified from pYA4372 with the primers kan-F/kan-R was inserted into the NotI and SbfI sites of pQK174 to generate the plasmid pQK175. This plasmid was subsequently introduced into P. multocida 0818 from E. coli χ7213 [37] via conjugation, and the Δcrp mutant designated S416 was selected on BHI agar containing kanamycin. Subsequently, the candidate mutant clones were verified by PCR screening using primers 1, 2, 3, 4, 5 and 6, which were designed based on the genomic sequence and the kanR cassette, as depicted in Fig. 2A. As a positive control, the 16S ribosomal RNA gene was amplified with the primers 16sRNA-F/16sRNA-R. Moreover, Crp expression was also measured in P. multocida 0818, S416 (P. multocida Δcrp) and S416 (pQK176) via western blotting with a 1:160-diluted polyclonal rabbit anti-Crp antibody.
Purification of Crp protein and preparation of a polyclonal rabbit anti-Crp antibody
The plasmid pET-32a-crp was transformed to E. coli BL21 (DE3) cells (Tiangen Biotech) with ampicillin selection. Recombinants were harvested after 6 h of induction with 1 mM IPTG (isopropyl b-D-1-Thiogalactopyranoside). The Crp protein was purified using 6× His/Ni-NTA affinity chromatography. To prepare the polyclonal anti-Crp antibody, two female New Zealand white rabbits were subcutaneously immunized with Crp protein (1 mg) adjuvanted with Freund’s complete/incomplete adjuvant (Sigma-Aldrich, St. Louis, MO, USA) four times at 14-day intervals. The titers of the antisera were then analyzed using the immune agar diffusion test. When the titers of the two rabbit antisera reached at least 1:32, blood samples were collected within approximately 14 days after the last immunization to obtain the anti-Crp antibody.
Maltose utilization test
To detect maltose utilization, S184 (S. Typhimurium Δasd) harboring the control plasmid pQK663, S411 (S184 Δcrp) harboring pQK663 and S411 harboring the complementary plasmid pQK163 were grown at 37 °C for 18 h on MacConkey indicator plates containing 1 % maltose, and the colony color was observed [38].
Phenotype determination
The growth curve of P. multocida strains was examined by recording their OD600 values every 2 h over a period of 14 h. The OMPs and LPS of P. multocida were extracted as previously described [36]. The OMP concentration was detected using a BCA Protein Assay Kit (Thermo Scientific, Rockford, IL, USA). The protein samples were subsequently diluted in sample buffer [50 mM Tris, 20 % glycerol, 4 % sodium dodecyl sulfate (SDS), 0.005 % bromophenol blue, and 5 % β-mercaptoethanol] and boiled for 5 min at 95 °C. The samples were then subjected to 12.5 % SDS-polyacrylamide gel electrophoresis (PAGE) followed by Coomassie Brilliant Blue R-250 staining (Sigma-Aldrich). Additionally, 10 μl of the LPS supernatant was diluted 1:10 in loading buffer, and the mixture was then treated with 1 μl of proteinase K (20 mg/ml, Sigma-Aldrich) at room temperature for 1 h and analyzed by 12.5 % SDS-PAGE followed by silver staining [30].
Serum bactericidal assay
Duck serum was collected from healthy ducks and heat-inactivated via incubation for 30 min at 56 °C. The serum bactericidal activity against the P. multocida strains were then measured as previously described [39]. Briefly, bacteria were cultured overnight in BHI media to an OD600 of 0.8 to 0.9 at 37 °C and 180 rpm. The bacteria were re-suspended in phosphate-buffered saline (PBS) and diluted to a final concentration of 104 CFU/ml. Aliquots (100 μl) of the bacterial suspensions were added to 900 μl of duck serum or heat-inactivated duck serum and incubated for 3 h at 37 °C with shaking. After incubation, the samples were placed on ice to inhibit further bacteriolysis. Serial dilutions of the samples in PBS were cultured on BHI agar plates and incubated at 37 °C overnight. The growth rate was calculated as the CFU per ml at 3 h divided by the CFU per ml at 0 h. All tests were performed in triplicate.
Determination of the 50 % lethal dose (LD50) in ducks
All animal experiments in this study were conducted in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of China. All animal procedures were approved by the Animal Care and Use Committee of Sichuan Agricultural University (No. XF2014-18).
To determine the LD50 of P. multocida strains, duck infection studies were conducted as described previously [36]. Overnight cultures of P. multocida strains in BHI medium were diluted 1:100 in fresh medium and further cultured at 37 °C with shaking to an OD600 of 0.8–0.9. The numbers of viable bacteria were then counted, and the bacteria were diluted in 100 μl of PBS to obtain cultures of 102 to 1010 CFU/ml. Various doses of P. multocida 0818 or S416 (Δcrp) were then intranasally inoculated into groups of 2-week-old Sheldrake ducks. The clinical symptoms and health of the ducks were monitored over a period of 2 weeks after infection. The LD50 was calculated following the method described by Reed and Muench, and the experiment was repeated twice.
RNA extraction and sequencing
For preparation of bacterial RNA samples, P. multocida 0818 and S416 (Δcrp) were grown in BHI medium in triplicate with shaking. Once the OD600 of the cultures reached 0.8, the bacteria were harvested, and the total RNA from each sample was extracted and purified using the TRIzol reagent (Invitrogen, CA, USA). Contaminating DNA was removed from the total RNA samples with DNase I (NEB Inc., USA) at 37 °C for 10 min, and ribosomal RNA was eliminated using a MICROBExpress kit (Thermo Fisher Scientific Inc., CA, USA). The RNA quality and concentration were determined using a Nanodrop spectrophotometer (Thermo Fisher Scientific Inc., USA). A cDNA library was then constructed using a TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA, USA), and the Illumina Hiseq2500/MiSeq platform (Illumina) was used for RNA deep sequencing, which was conducted at Majorbio Bio-pharm Biotechnology Co., Ltd. (Shanghai, China).
Sequence analysis was performed as previously described [40]. In brief, clean reads were obtained from the sequenced raw data using FASTQC and NGS QC TOOLKIT and de novo assembled by Trinity software. The TGICL package was then applied to generate valid unigenes, and the Bowtie 2 and eXpress software programs were used for the mapping of the clean reads from each sample to unigenes based on the reference genome sequence of P. multocida Pm70. The number of mapped reads relative to each gene was measured using the RPKM method. The RSEM and edgeR software packages were subsequently applied for the screening of unigene transcripts with differential expression between P. multocida 0818 and S416.
Immunization and challenge
One-week-old Sheldrake ducks were intranasally immunized with 1 × 103 CFU of S416 or PBS twice at an interval of 10 days. The S416-immunized group included 16 ducks, and the PBS control group included 14 ducks. Serum and bile were collected from six randomly selected ducks from both groups on Day -3 and Day 20, respectively, and stored at -80 °C until analysis. For the challenge assay, the ducks in the S416-immunized and PBS groups were intranasally inoculated with a lethal dose of P. multocida strain 0818, approximately 1 × 107 CFU. The health status of the animals was monitored and recorded every day for two weeks post-challenge, and the deceased ducks were routinely subjected to bacterial isolation.
Enzyme-linked immunosorbent assay (ELISA)
The serum IgY and bile IgA levels were detected via indirect ELISA as previously described [36]. A 96-well ELISA microplate was coated with 1 × 1010 CFU heating-inactivated P. multocida 0818 or 0.25 μg/ml purified OMPs and incubated at 4 °C overnight. After three washes with PBST, the plate was then blocked with 2 % BSA in PBS. The serum samples were diluted 1:100 in PBS containing 1 % BSA, and the bile samples were diluted 1:40. After the plate was washed again, 100 μl of these dilutions was added to each well. After 1 h of incubation at 37 °C, the plate was incubated with 100 μl of 1:5000-diluted alkaline phosphatase (AP)-labeled mouse anti-duck IgY or IgA (AbD Serotec, Puchheim, Germany) at 37 °C for 1 h. AP solution (Sigma-Aldrich) was added for coloration, and the reaction was terminated by the addition of 100 μl of 0.2 M NaOH. The optical density (OD) value at 415 nm was measured using a microplate reader (Bio-Rad Laboratories, Richmond, CA, USA).
Statistical analyses
The GraphPad Prism 5 software package (Graph Software, San Diego, CA, USA) was used for the statistical analyses. The data are expressed as the means ± standard deviations (SD) and were evaluated using Student’s t test with significance levels set to 0.05 and 0.01. The animal experiments were performed at least twice, and the in vitro experiments were conducted independently three times in triplicate.
Results
Cloning and characterization of the crp gene of P. multocida
The coding region of the suspected crp gene (PM1157) in P. multocida strain 0818 was cloned via PCR, then sequenced and deposited in GenBank (accession number, KU507499). The PM1157 sequence was 630 bp in length and shared 73 % nucleotide identity with both S. Typhimurium crp (Gene ID, 1254989) and E. coli crp (Gene ID, 947867) over 604 nucleotides. Additionally, the 630 bp sequence of the P. multocida crp gene was predicted to encode a 209-amino acid polypeptide, which showed 86 and 87 % identity to S. Typhimurium Crp (Protein ID, NP_462369.1) and E. coli Crp (Protein ID, NP_417816.1) over 205 amino acids, respectively. The deletion of crp resulted in defects in maltose fermentation in S. Typhimurium [38]. To clarify the nature of the cloned sequence, we evaluated whether the PM1157 gene could restore maltose fermentation in S. Typhimurium crp mutant. Maltose fermentation was detected in S184 (S. Typhimurium Δasd) harboring pQK663 (control plasmid), S411 (S. Typhimurium Δasd Δcrp) harboring pQK663 and S411 harboring pQK163 (pQK663-PM1157). As shown in Fig. 1, S184 (pQK663) and S411 (pQK163) produced red clones, whereas S411 (pQK663) produced white clones on MacConkey maltose agar. Thus, the suspected crp gene (PM1157) complemented the Salmonella Δcrp mutant, allowing the utilization of maltose.
Detection of maltose fermentation in S. Typhimurium. Three S. Typhimurium strains, S184 (Δasd) harboring the control plasmid pQK663, S411 (ΔasdΔcrp) harboring pQK663 and S411 (ΔasdΔcrp) harboring the complementary plasmid pQK163 (PM1157), were cultured on MacConkey maltose agar to observe the colors of the clones
Construction of the non-polar Δcrp mutant in P. multocida 0818
To determine the role of crp (PM1157) in P. multocida, the Δcrp mutant S416 was constructed via suicide plasmid-mediated homologous recombination and characterized through PCR using three pairs of primers, 1&2, 3&4, and 5&6 (Fig. 2A). The DNA segment containing the PM1157 upstream sequence and a partial kanR cassette (3&4) and the DNA segment containing the PM1157 downstream sequence and a partial kanR cassette (5&6) were present in the S416 strain but not in the parent strain (P. multocida 0818), whereas the complete PM1157 sequence (1&2) was only present in the parent strain (Fig. 2B). The positive control 16S RNA could be amplified from both strains. Crp expression was also measured using a polyclonal anti-Crp antibody. Crp was expressed in the parent strain and the complementary strain S416 (pQK176), but not in S416 (Fig. 2C), demonstrating that the crp gene was successfully deleted in S416. Moreover, the crp mutation was stable for more than 20 passages (data not shown).
Construction of the Δcrp mutant in the P. multocida 0818 strain. a Schematic of the strategy used for deletion of the target crp gene (PM1157). The P. multocida crp gene was replaced with a kanR cassette via homologous recombination. Three pairs of primers, 1& 2, 3& 4, and 5& 6, were designed to select and characterize the mutant clones. b Characterization of the constructed Δcrp mutant via PCR. The parent strain and Δcrp mutant were identified using the primers 1&2, 3&4 and 5&6. M refers to the DNA marker; 16sRNA indicates amplification of the positive gene in both strains. c Detection of Crp expression in P. multocida stains. The parent strain, S416 (Δcrp) and S416 (pQK176) were grown in BHI media and collected at an OD600 of 1.0, and the expression of Crp was then measured in these strains with an anti-Crp antibody via western blotting. Crp refers to purified protein and served as a positive control
Phenotype characterization of the P. multocida crp mutant
To detect the influence of crp deletion, the growth curve, OMP and LPS profiles, and serum complement sensitivity were evaluated in the parent strain (P. multocida 0818), S416 (P. multocida Δcrp) and S416 (pQK176). The parent strain showed a typical growth curve, with a short lag phase (0–2 h), followed by a log phase during which major bacterial growth occurred (2–10 h) and then a stationary phase (10–14 h; Fig. 3A). In contrast, S416 grew more slowly after 2 h, particularly between 4 and 8 h. During this phase, the OD values of S416 were significantly lower than those of the parent strain (Fig. 3A). The complementary strain, S416, which harbors pQK176, partially restored the defective growth (Fig. 3A), but the OD values recorded at 8 h and 12 h remained lower than those of the parent strain.
Phenotype detection of the P. multocida Δcrp mutant. a Analysis of the growth curve. The parent strain, S416 (Δcrp) and S416 (pQK176) were grown in BHI broth or BHI broth supplemented with kanamycin, and the OD600 values were measured every 2 h over a period of 14 h. The data are expressed as the means ± SD, and the asterisks indicate significant differences compared with the parent strain. b and c Profiles of the OMPs and LPS of P. multocida. OMPs or LPSs were isolated from the parent strain or the Δcrp mutant and then analyzed by SDS-PAGE. Coomassie blue staining and silver staining were then performed to visualize the OMPs (b) and LPSs (c), respectively. M refers to the protein marker
The S416 strain displayed OMP and LPS profiles similar to those of the parent strain (Figs. 3B and C). Specifically, these profiles primarily consisted of proteins larger than 25 kDa and short-length LPS, respectively. Moreover, both the parent strain and S416 (pQK176) grew rapidly in either untreated or heat-treated duck serum, whereas S416 (Δcrp) was rapidly killed in untreated serum but grew in heat-treated serum (Table 3). The growth rates of S416 significantly differed between untreated and heat-treated serum (Table 3), demonstrating that the deletion of crp increased sensitivity to complement-mediated killing.
Determination of the virulence of wild-type P. multocida and the Δcrp mutant
To determine the effects of the crp mutation on bacterial virulence, the LD50 values of the parent strain and S416 (P. multocida Δcrp) were assessed in a duck animal model. The results showed that the LD50 of S416 was 7.4 × 106 CFU, which was 85-fold higher than that of the parent strain, whose LD50 was 8.66 × 104 (Table 4).
Identification of genes regulated by crp in P. multocida
Transcriptome sequencing was performed to screen for genes regulated by crp in P. multocida. Compared with the parent strain, 186 genes in addition to crp itself were differentially expressed in the S416 (Δcrp) strain during the exponential growth stage. Specifically, 92 of these genes were up-regulated, and 94 genes were down-regulated (see Additional file 1). Genes exhibiting fold-differences in transcription greater than 3.5 between the parent strain and the Δcrp mutant are listed in Table 5. A KEGG enrichment analysis showed that the regulated genes were significantly involved in six pathways, including two-component systems, arginine and proline metabolism, pyruvate metabolism, nitrogen metabolism, and oxidative phosphorylation (see Additional file 2).
Evaluation of immune responses and the protection rate conferred by the Δcrp mutant
The antibody responses induced by S416 (Δcrp) were detected by ELISA post-immunization. As shown in Fig. 4, no specific serum IgY or bile IgA against bacteria antigen were detected in both the S416-immunized group and the PBS group three days prior to immunization. In contrast to the PBS group, S416-immunized group induced significantly higher levels of serum IgY to whole bacteria antigen and OMPs 20 days post-immunization (Figs. 4A and B). In addition, the bile IgA levels against whole bacteria antigen and OMPs were significantly increased in the S416-immunized group compared with the PBS group 20 days post-immunization (Figs. 4C and D). Moreover, 60 % of the ducks in the immunized group survived and steadily gained weight after challenge, whereas all control ducks were dead within one week (Table 6). Thus, the Δcrp mutant induced 60 % protection against challenge with a dosage of 100-fold of the LD50 of wild-type P. multocida 0818 in ducks.
Antibody responses induced by the Δcrp mutant in ducks. Ducks were intranasally immunized twice with the Δcrp mutant at an interval of 10 days. The serum IgY responses specific to whole bacterial antigen (a) and OMPs (b) and the bile IgA responses specific to whole bacterial antigen (c) and OMPs (d) were then assessed 3 days before immunization and 20 days post-immunization via indirect ELISA. The data are expressed as the means ± SD and were analyzed at significance levels of 0.05 (*) and 0.01 (**)
Discussion
Global regulators play a vital role in the adaptation of bacteria to the environmental changes that are encountered during infection. Despite the broad spectrum of diseases caused by P. multocida and their worldwide economic impact, only a few regulators have been characterized in P. multocida, including Fis (nucleoid-associated proteins) [41], Fur (ferric uptake regulation) [42], PhoP [36] and FnrP [43]. These regulators are all associated with bacterial virulence. Here, we first identified the homologue of the crp gene in P. multocida and showed that PM1157 is the functional crp gene. Two pieces of evidence support this conclusion: 1) PM1157 exhibited a high degree of amino acid identity (86 %) with corresponding proteins from other bacteria, including E. coli and Salmonella enterica; and 2) the PM1157 gene of P. multocida was able to restore maltose fermentation in the Salmonella crp mutant. Thus, gene PM1157 of P. multocida is a crp gene that is interchangeable with Salmonella crp.
The resistance of S416 (P. multocida Δcrp) to duck serum complement was significantly reduced compared with that of the parent stain. Previous reports have demonstrated that the long O-antigen chain of Salmonella serovars or the presence of certain outer membrane proteins of some bacteria, such as Salmonella Rck and Haemophilus influenzae P5, contributes to complement resistance [44–46]. Capsular polysaccharide also prevents the complement-mediated clearance of Salmonella enterica serotype Typhi [47] and P. multocida [48]. Here, we showed that the LPS profile of S416 was similar to that of the parent strain, mainly containing short oligosaccharides. Several OMP genes, rather than capsule synthesis genes, were differentially expressed in the absence of the crp gene (see Additional file 1). Therefore, we speculated that some of these OMPs were responsible for the decrease in complement resistance, which should be confirmed in a later study. In addition, the virulence of the S416 strain decreased 85-fold after intranasal inoculation, and this reduction was much less pronounced in the P. multocida Δcrp mutant than in the Salmonella Δcrp mutant, for which virulence was reduced by five orders of magnitude [49]. Similar results have been observed for a crp mutation in Edwardsiella ictaluri [50], indicating that crp does not necessarily impact virulence, or that its impact on virulence is species-specific, or that other genes in P. multocida may compensate for a loss of crp to minimize adverse effects.
Crp–cAMP can directly control a minimum of 378 promoters and perhaps > 500 genes in E. coli [22]. In this study, crp was shown to influence the transcription of 186 genes, including 92 up-regulated genes and 94 down-regulated genes in P. multocida (see Additional file 1). This finding suggested that crp acts as both a positive and negative regulator. The majority of differentially expressed genes participate in metabolism, including carbon metabolism, arginine and proline metabolism, pyruvate metabolism, nitrogen metabolism and ABC transporters. Therefore, Crp plays an important role in the metabolism of P. multocida. The deletion of the crp gene also resulted in the down-regulation of two global regulators (fnr and iscR) and up-regulation of seven regulators (pgtC, qseB, arcA, ttrC, ttrB, rraA and isrR). Among these regulators, fnr, qseB and arcA have been shown to positively regulate the virulence of pathogenic bacteria. Deletion of fnr, qseB or arcA significantly reduces the virulence of S. Typhimurium [51], Aeromonas hydrophila [52] and Vibrio cholerae [53], respectively. Thus, the down-regulation of fnr observed in the P. multocida Δcrp mutant may partially account for the decreased virulence of this strain, and the up-regulation of qseB and arcA may have attenuated this decrease.
Vaccines are the most economical and effective means to control infectious disease. Compared with vaccines based on a subunit or dead bacteria, attenuated live vaccines are advantageous because they can induce long-term immunity and confer good protection [4]. A significant number of attenuated live P. multocida vaccines have been successfully developed by targeting capsule genes [54, 55], toxin genes [56] and aroA [57], which elicit protective responses in mice, livestock or poultry. Here, we evaluated the vaccine potential of the P. multocida Δcrp mutant. Because circumstantial evidence has implicated the respiratory tract as the main route of P. multocida infection, we selected the intranasal route to immunize ducks. The P. multocida Δcrp mutant induced potent serum IgY and bile IgA responses in ducks, indicating the high immunogenicity of the Δcrp mutant. Additionally, this mutant provided 60 % protection against challenge with the P. multocida virulent strain at a dosage of 100-fold of the LD50 (Table 6). This level of protection was lower than that conferred by the Salmonella crp mutant [13]. The immunization dose of an attenuated P. multocida strain has been demonstrated to be related to the level of protection against challenge [58]; animals receiving a higher vaccine dose (109 or 108 CFU of the attenuated strain) are less affected clinically, bacteriologically, and pathologically through wild-type challenge compared with the administration of a lower dose of 107 CFU [58]. Because the P. multocida Δcrp mutant was not fully attenuated, only low doses of this mutant (103 CFU) were used to immunize the ducks. We speculate that the low immunization dose might have been responsible for the limited protection observed. However, other studies have also shown that fully attenuated bacterial strains such as the Edwardsiella tarda aroA mutant fail to provide effective protection against virulent challenge even at a high dose of immunization, probably because of reduced immunogenicity or loss of protective antigens [59–61]. Therefore, it is vital to retain or improve bacterial immunogenicity while achieving attenuation through the deletion of virulence regulators. Furthermore, bacteria with mutations in two or more regulator genes are better attenuated than those with single-gene mutations in some cases and provide high vaccine potency [62]. Thus, mutations in other regulator genes need to be selected and introduced into the crp mutant to achieve full attenuation for vaccine development in a later study.
Conclusions
The PM1157 gene is the crp homologue of P. multocida. The deletion of the crp gene has an inhibitory effect on bacterial growth and bacterial resistance to serum complement, without affecting the electrophoretic bands of LPS and OMPs in vitro. Furthermore, the P. multocida crp mutant was attenuated and provided 60 % protection in ducks. The present study provides a basis that will allow the mechanisms of crp-regulated genes to be explored to ultimately develop a platform for an attenuated vaccine against P. multocida.
Abbreviations
AP, alkaline phosphatase; BHI, brain heart infusion; DAP, diaminopimelic acid; E. coli, Escherichia coli; ELISA, enzyme-linked immunosorbent assay; kanR, kanamycin resistance; LB, Luria-Bertani; LD50, 50 % lethal dose; LPS, lipopolysaccharide; OMPs, outer membrane proteins; P. multocida, Pasteurella multocida; PAGE, polyacrylamide gel electrophoresis; PBS, phosphate-buffered saline; S. Typhimurium, Salmonella enterica serovar Typhimurium; SDS, sodium dodecyl sulfate
References
Harper M, Boyce JD, Adler B. Pasteurella multocida pathogenesis: 125 years after Pasteur. FEMS Microbiol Lett. 2006;265(1):1–10.
Heddleston KL, Gallagher JE, Rebers PA. Fowl cholera: gel diffusion precipitin test for serotyping Pasteruella multocida from avian species. Avian Dis. 1972;16(4):925–36.
Christensen JP, Bisgaard M. Fowl cholera. Rev Sci Tech. 2000;19(2):626–37.
Ahmad TA, Rammah SS, Sheweita SA, Haroun M, El-Sayed LH. Development of immunization trials against Pasteurella multocida. Vaccine. 2014;32(8):909–17.
Dalebroux ZD, Miller SI. Salmonellae PhoPQ regulation of the outer membrane to resist innate immunity. Curr Opin Microbiol. 2014;17:106–13.
Munson GP. Virulence regulons of enterotoxigenic Escherichia coli. Immunol Res. 2013;57(1-3):229–36.
Moreira CG, Weinshenker D, Sperandio V. QseC mediates Salmonella enterica serovar typhimurium virulence in vitro and in vivo. Infect Immun. 2010;78(3):914–26.
Li J, Ma M, Sarker MR, McClane BA. CodY is a global regulator of virulence-associated properties for Clostridium perfringens type D strain CN3718. MBio. 2013;4(5):e00770–13.
Porcheron G, Dozois CM. Interplay between iron homeostasis and virulence: Fur and RyhB as major regulators of bacterial pathogenicity. Vet Microbiol. 2015;179(1-2):2–14.
Jansen G, Crummenerl LL, Gilbert F, Mohr T, Pfefferkorn R, Thanert R, et al. Evolutionary transition from pathogenicity to commensalism: Global regulator mutations mediate fitness gains through virulence attenuation. Mol Biol Evol. 2015;32(11):2883–96.
Okay S, Kurt KA. Comparative genome analysis of five Pasteurella multocida strains to decipher the diversification in pathogenicity and host specialization. Gene. 2015;567(1):58–72.
Desin TS, Koster W, Potter AA. Salmonella vaccines in poultry: past, present and future. Expert Rev Vaccines. 2013;12(1):87–96.
Hassan JO, Curtiss 3rd R. Development and evaluation of an experimental vaccination program using a live avirulent Salmonella typhimurium strain to protect immunized chickens against challenge with homologous and heterologous Salmonella serotypes. Infect Immun. 1994;62(12):5519–27.
Emmer M, deCrombrugghe B, Pastan I, Perlman R. Cyclic AMP receptor protein of E. coli: its role in the synthesis of inducible enzymes. Proc Natl Acad Sci U S A. 1970;66(2):480–7.
McKay DB, Steitz TA. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature. 1981;290(5809):744–9.
Lawson CL, Swigon D, Murakami KS, Darst SA, Berman HM, Ebright RH. Catabolite activator protein: DNA binding and transcription activation. Curr Opin Struct Biol. 2004;14(1):10–20.
Gosset G, Zhang Z, Nayyar S, Cuevas WA, Saier Jr MH. Transcriptome analysis of Crp-dependent catabolite control of gene expression in Escherichia coli. J Bacteriol. 2004;186(11):3516–24.
Notley-McRobb L, Death A, Ferenci T. The relationship between external glucose concentration and cAMP levels inside Escherichia coli: implications for models of phosphotransferase-mediated regulation of adenylate cyclase. Microbiology. 1997;143(Pt 6):1909–18.
Berg OG, von Hippel PH. Selection of DNA binding sites by regulatory proteins. II. The binding specificity of cyclic AMP receptor protein to recognition sites. J Mol Biol. 1988;200(4):709–23.
Baichoo N, Heyduk T. Mapping cyclic nucleotide-induced conformational changes in cyclicAMP receptor protein by a protein footprinting technique using different chemical proteases. Protein Sci. 1999;8(3):518–28.
Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP). J Mol Biol. 1999;293(2):199–213.
Shimada T, Fujita N, Yamamoto K, Ishihama A. Novel roles of cAMP receptor protein (CRP) in regulation of transport and metabolism of carbon sources. PLoS One. 2011;6(6):e20081.
Kelly SM, Bosecker BA, Curtiss 3rd R. Characterization and protective properties of attenuated mutants of Salmonella choleraesuis. Infect Immun. 1992;60(11):4881–90.
Rickman L, Scott C, Hunt DM, Hutchinson T, Menendez MC, Whalan R, et al. A member of the cAMP receptor protein family of transcription regulators in Mycobacterium tuberculosis is required for virulence in mice and controls transcription of the rpfA gene coding for a resuscitation promoting factor. Mol Microbiol. 2005;56(5):1274–86.
Skorupski K, Taylor RK. Cyclic AMP and its receptor protein negatively regulate the coordinate expression of cholera toxin and toxin-coregulated pilus in Vibrio cholerae. Proc Natl Acad Sci U S A. 1997;94(1):265–70.
Petersen S, Young GM. Essential role for cyclic AMP and its receptor protein in Yersinia enterocolitica virulence. Infect Immun. 2002;70(7):3665–72.
Heroven AK, Dersch P. Coregulation of host-adapted metabolism and virulence by pathogenic yersiniae. Front Cell Infect Microbiol. 2014;4:146.
Sheoran AS, Timoney JF, Tinge SA, Sundaram P, Curtiss 3rd R. Intranasal immunogenicity of a Delta cya Delta crp-pabA mutant of Salmonella enterica serotype Typhimurium for the horse. Vaccine. 2001;19(27):3787–95.
Chu CY, Wang SY, Chen ZW, Chien MS, Huang JP, Chen JJ, et al. Heterologous protection in pigs induced by a plasmid-cured and crp gene-deleted Salmonella choleraesuis live vaccine. Vaccine. 2007;25(41):7031–40.
Fomsgaard A, Freudenberg MA, Galanos C. Modification of the silver staining technique to detect lipopolysaccharide in polyacrylamide gels. J Clin Microbiol. 1990;28(12):2627–31.
Nandi B, Nandy RK, Mukhopadhyay S, Nair GB, Shimada T, Ghose AC. Rapid method for species-specific identification of Vibrio cholerae using primers targeted to the gene of outer membrane protein OmpW. J Clin Microbiol. 2000;38(11):4145–51.
May BJ, Zhang Q, Li LL, Paustian ML, Whittam TS, Kapur V. Complete genomic sequence of Pasteurella multocida, Pm70. Proc Natl Acad Sci U S A. 2001;98(6):3460–5.
Kang HY, Srinivasan J, Curtiss 3rd R. Immune responses to recombinant pneumococcal PspA antigen delivered by live attenuated Salmonella enterica serovar typhimurium vaccine. Infect Immun. 2002;70(4):1739–49.
Bosse JT, Durham AL, Rycroft AN, Kroll JS, Langford PR. New plasmid tools for genetic analysis of Actinobacillus pleuropneumoniae and other pasteurellaceae. Appl Environ Microbiol. 2009;75(19):6124–31.
Kong Q, Yang J, Liu Q, Alamuri P, Roland KL, Curtiss 3rd R. Effect of deletion of genes involved in lipopolysaccharide core and O-antigen synthesis on virulence and immunogenicity of Salmonella enterica serovar typhimurium. Infect Immun. 2011;79(10):4227–39.
Xiao K, Liu Q, Liu X, Hu Y, Zhao X, Kong Q. Identification of the avian pasteurella multocida phoP gene and evaluation of the effects of phoP deletion on virulence and immunogenicity. Int J Mol Sci. 2016;17(1):12.
Roland K, Curtiss 3rd R, Sizemore D. Construction and evaluation of a delta cya delta crp Salmonella typhimurium strain expressing avian pathogenic Escherichia coli O78 LPS as a vaccine to prevent airsacculitis in chickens. Avian Dis. 1999;43(3):429–41.
Curtiss 3rd R, Wanda SY, Gunn BM, Zhang X, Tinge SA, Ananthnarayan V, et al. Salmonella enterica serovar typhimurium strains with regulated delayed attenuation in vivo. Infect Immun. 2009;77(3):1071–82.
Chung JY, Wilkie I, Boyce JD, Townsend KM, Frost AJ, Ghoddusi M, et al. Role of capsule in the pathogenesis of fowl cholera caused by Pasteurella multocida serogroup A. Infect Immun. 2001;69(4):2487–92.
Wang A, Liu F, Chen S, Wang M, Jia R, Zhu D, et al. Transcriptome analysis and identification of differentially expressed transcripts of immune-related genes in spleen of gosling and adult goose. Int J Mol Sci. 2015;16(9):22904–26.
Steen JA, Steen JA, Harrison P, Seemann T, Wilkie I, Harper M, et al. Fis is essential for capsule production in Pasteurella multocida and regulates expression of other important virulence factors. PLoS Pathog. 2010;6(2):e1000750.
Bosch M, Tarrago R, Garrido ME, Campoy S, de Henestrosa AR F, de Rozas AM P, et al. Expression of the Pasteurella multocida ompH gene is negatively regulated by the Fur protein. FEMS Microbiol Lett. 2001;203(1):35–40.
Uhlich GA, McNamara PJ, Iandolo JJ, Mosier DA. Cloning and characterization of the gene encoding Pasteurella haemolytica FnrP, a regulator of the Escherichia coli silent hemolysin sheA. J Bacteriol. 1999;181(12):3845–8.
Bravo D, Silva C, Carter JA, Hoare A, Alvarez SA, Blondel CJ, et al. Growth-phase regulation of lipopolysaccharide O-antigen chain length influences serum resistance in serovars of Salmonella. J Med Microbiol. 2008;57(Pt 8):938–46.
Boyd MA, Tennant SM, Saague VA, Simon R, Muhsen K, Ramachandran G, et al. Serum bactericidal assays to evaluate typhoidal and nontyphoidal Salmonella vaccines. Clin Vaccine Immunol. 2014;21(5):712–21.
Rosadini CV, Ram S, Akerley BJ. Outer membrane protein P5 is required for resistance of nontypeable Haemophilus influenzae to both the classical and alternative complement pathways. Infect Immun. 2014;82(2):640–9.
Wilson RP, Winter SE, Spees AM, Winter MG, Nishimori JH, Sanchez JF, et al. The Vi capsular polysaccharide prevents complement receptor 3-mediated clearance of Salmonella enterica serotype Typhi. Infect Immun. 2011;79(2):830–7.
Boyce JD, Adler B. The capsule is a virulence determinant in the pathogenesis of Pasteurella multocida M1404 (B:2). Infect Immun. 2000;68(6):3463–8.
Zhang X, Kelly SM, Bollen W, Curtiss 3rd R. Protection and immune responses induced by attenuated Salmonella typhimurium UK-1 strains. Microb Pathog. 1999;26(3):121–30.
Santander J, Mitra A, Curtiss 3rd R. Phenotype, virulence and immunogenicity of Edwardsiella ictaluri cyclic adenosine 3’,5’-monophosphate receptor protein (Crp) mutants in catfish host. Fish Shellfish Immunol. 2011;31(6):1142–53.
Fink RC, Evans MR, Porwollik S, Vazquez-Torres A, Jones-Carson J, Troxell B, et al. FNR is a global regulator of virulence and anaerobic metabolism in Salmonella enterica serovar Typhimurium (ATCC 14028 s). J Bacteriol. 2007;189(6):2262–73.
Khajanchi BK, Kozlova EV, Sha J, Popov VL, Chopra AK. The two-component QseBC signalling system regulates in vitro and in vivo virulence of Aeromonas hydrophila. Microbiology. 2012;158(Pt 1):259–71.
Sengupta N, Paul K, Chowdhury R. The global regulator ArcA modulates expression of virulence factors in Vibrio cholerae. Infect Immun. 2003;71(10):5583–9.
Chung JY, Wilkie I, Boyce JD, Adler B. Vaccination against fowl cholera with acapsular Pasteurella multocida A:1. Vaccine. 2005;23(21):2751–5.
Boyce JD, Adler B. Acapsular Pasteurella multocida B:2 can stimulate protective immunity against pasteurellosis. Infect Immun. 2001;69(3):1943–6.
Kim T, Son C, Lee J, Kim K. Vaccine potential of an attenuated Pasteurella multocida that expresses only the N-terminal truncated fragment of P. multocida toxin in pigs. Can J Vet Res. 2012;76(1):69–71.
Homchampa P, Strugnell RA, Adler B. Cross protective immunity conferred by a marker-free aroA mutant of Pasteurella multocida. Vaccine. 1997;15(2):203–8.
Dagleish MP, Hodgson JC, Ataei S, Finucane A, Finlayson J, Sales J, et al. Safety and protective efficacy of intramuscular vaccination with a live aroA derivative of Pasteurella multocida B:2 against experimental hemorrhagic septicemia in calves. Infect Immun. 2007;75(12):5837–44.
Mo ZL, Li J, Li GY, Xiao P. Phenotypic characterization, virulence, and immunogenicity of Edwardsiella tarda LSE40 aroA mutant. Appl Microbiol Biotechnol. 2013;97(14):6325–35.
O’Callaghan D, Maskell D, Liew FY, Easmon CS, Dougan G. Characterization of aromatic- and purine-dependent Salmonella typhimurium: attention, persistence, and ability to induce protective immunity in BALB/c mice. Infect Immun. 1988;56(2):419–23.
Trebichavsky I, Splichalova A, Rychlik I, Hojna H, Muneta Y, Mori Y, et al. Attenuated aroA Salmonella enterica serovar Typhimurium does not induce inflammatory response and early protection of gnotobiotic pigs against parental virulent LT2 strain. Vaccine. 2006;24(20):4285–9.
Han X, Bai H, Tu J, Yang L, Xu D, Wang S, et al. Deletion of luxS further attenuates the virulence of the avian pathogenic Escherichia coli aroA mutant. Microb Pathog. 2015;88:39–47.
Acknowledgments
We thank Dr. Paul R. Langford for providing the pMC-Express plasmid, Dr. Dekang Zhu for providing wild-type P. multocida 0818, and Dr. Mingshu Wang and Dr. Kunfeng Sun for providing animal housing.
Funding
This research was supported by the Special Foundation for Young Scientists of Sichuan Province (No. 2014JQ0006) and the Foundation of the Educational Commission of Sichuan Province (No. 15ZA0018).
Availability of data and materials
The coding region of the suspected crp gene (PM1157) in P. multocida strain 0818 was cloned via PCR, then sequenced and deposited in GenBank (accession number, KU507499). All other data supporting the findings are contained within the manuscript or will be shared upon request.
Authors’ contributions
XZ carried out most of the experimental work, performed the statistical analysis, and wrote the manuscript. QL, KX, YH, XL participated in mutant construction, phenotype characterization, and animal experiments. YL participated in the design of the study and helped to revise the manuscript. QK and XZ secured funding and analyzed and interpreted the data. QL, QK conceived of the study, designed the experiments and revised the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
All animal procedures were approved by the Animal Care and Use Committee of Sichuan Agricultural University (No. XF2014-18).
Author information
Authors and Affiliations
Corresponding authors
Additional files
Additional file 1:
Total differentially expressed genes between the parent strain and the Δcrp mutant. (PDF 142 kb)
Additional file 2:
KEGG enrichment analysis of the differentially expressed genes in the Δcrp mutant. KOBAS software was used to analyze the crp-regulated genes in KEGG pathways. Each column in A and B indicates one pathway, and the abscissa represents the name and classification of the pathway. The column color refers to the significance, and the depth of the color directly correlates with the degree of significance. *, p < 0.05; **, p < 0.01; ***, p < 0.001. (TIF 367 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhao, X., Liu, Q., Xiao, K. et al. Identification of the crp gene in avian Pasteurella multocida and evaluation of the effects of crp deletion on its phenotype, virulence and immunogenicity. BMC Microbiol 16, 125 (2016). https://doi.org/10.1186/s12866-016-0739-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-016-0739-y