Abstract
Background
The spread of carbapenem resistant Enterobacteriaceae (CRE) is an emerging clinical problem, of great relevance in Europe and worldwide. The aim of this study was the molecular epidemiology of CRE isolates in Valle d’Aosta region, Italy, and the mechanism of carbapenem resistance.
Results
Sixty consecutive CRE samples were isolated from 52 hospital inpatients and/or outpatients from November 2013 to August 2014. Genotyping of microbial isolates was done by pulsed-field gel electrophoresis (PFGE) and multi-locus sequence typing (MLST), carbapenemases were identified by PCR and sequencing. Carbapenem resistance gene transfer was performed by filter mating, plasmids from parental and transconjugant strains were assigned to incompatibility groups by PCR-based replicon typing. Molecular characterization of CRE isolates assigned 25 Klebsiella pneumoniae isolates to PFGE types A1-A5 and sequencing type (ST) 101, 17 K. pneumoniae isolates to PFGE type A and ST1789 (a single locus variant of ST101), 7 K. pneumoniae isolates to PFGE types B or C and ST512, 2 K. pneumoniae isolates to PFGE type D and ST405, and 5 Escherichia coli isolates to PFGE type a and ST131. All K. pneumoniae ST101 and ST1789 isolates were extended-spectrum beta-lactamase (ESBL) producers and carried bla CTX-M-1 group gene; 4 K. pneumoniae ST101 isolates were resistant to colistin. Molecular analysis of beta-lactamase genes identified bla KPC-2 and bla CTX-M-group 1 into conjugative plasmid/s assigned to IncFII incompatibility group in ST101 and ST1789 K. pneumoniae isolates, bla KPC-3 into conjugative plasmid/s assigned to IncF incompatibility group in ST512 and ST405 K. pneumoniae isolates, bla VIM-1 into conjugative plasmid/s assigned to IncN incompatibility group in ST131 E. coli isolates.
Conclusions
The spread of CRE in Valle d’Aosta region was caused by the selection of KPC-2 producing K. pneumoniae ST101 and ST1789 epidemic clones belonging to clonal complex 101, KPC-3 producing K. pneumoniae epidemic clones assigned to ST512 and ST405, and VIM-1 producing E.coli ST131 epidemic clone. Carbapenem resistance, along with bla KPC-2, bla KPC-3 and bla VIM-1 carbapenemase genes, was transferred by conjugative plasmids assigned to IncFII, IncF, and IncN incompatibility groups, respectively, in filter mating experiments. The emergence of colistin resistance was observed in KPC-2 producing K. pneumoniae ST101 isolates.
Similar content being viewed by others
Background
The spread of resistance to carbapenems among Enterobacteriaceae has become a major public health problem. In fact, epidemics or endemic conditions of patients infected or colonized by Enterobacteriaceae non susceptible to carbapenems has been reported in the hospital and in the community setting [1–3]. Carbapenem resistant Enterobacteriaceae (CRE) are resistant to most β-lactams, including the carbapenems, and frequently carry additional antimicrobial resistance genes to other non-β-lactam antibiotics, which render them resistant to most antibiotics [1–3].
Two main mechanisms are responsible for carbapenem resistance in Enterobacteriaceae: reduced outer membrane permeability by porin loss in combination with the production of an extended spectrum beta-lactamase or of AmpC-type beta-lactamase; and the production of beta-lactamases capable of hydrolysing carbapenems (carbapenemases) [1–3]. Because acquired carbapenemases are encoded by transferable genes located in mobile elements such as plasmids and transposons, which may disseminate among different strains and species, carbapenemase-mediated resistance represents the major treat in CRE [1–6]. The occurrence of either class B metallo-beta-lactamases (IMP, VIM, NDM) or classes A (KPC) and D (OXA-48) serine carbapenemases have been reported in Europe and world-wide [1–6]. In Italy, Enterobacteriaceae producing VIM-type enzymes and more recently KPC-type enzymes have been described [7–10]. The increase of carbapenem resistance in Enterobacteriaceae has been sustained also by the selection of multidrug-resistant high risk clonal lineages, which successfully propagated in the hospital and the community settings [6, 11]. Also, the coexistence of carbapenem resistance and extended-spectrum beta-lactamase (ESBL) production has been frequently described [2, 12–14]. Moreover, the movement of colonized patients, mainly elderly, between hospital wards and long-term care settings may contribute to the spread of CRE [1–3, 8].
To control the spread of CRE in health care facilities and the transmission of CRE among patients, guidelines have been established from European Centre for Disease Prevention and Control for the screening and identification of CRE isolates among colonized patients [15], which have been adopted in Italy, both at regional and national level [16].
The aim of the current study was to analyze the molecular epidemiology of 60 consecutive CRE in the hospital wards and long-term care facilities of the USL Valle d’Aosta, and the mechanism of carbapenem resistance.
Methods
Setting and study period
Valle d’Aosta is the smallest region of Italy (3262 sq Kms), entirely located in the north western Alps region; there is one health agency and only one public Hospital, with 480 beds: there is one microbiology laboratory that processes samples from both Hospital and the territory. Moreover, a network of about 45 long-term care residencies for elders is distributed over the territory, accounting for another 1000 beds. The present study analyzed 60 consecutive isolates of CRE from 52 hospital inpatients and/or outpatients who were admitted to the USL Valle d’Aosta between November 2013 and August 2014.
Surveillance of CRE Enterobacteriaceae
Surveillance of CRE was performed according to European Centre for Disease Prevention and Control [15] and the Ministero della Salute, Italy [16]. Surveillance and infection control measures adopted for CRE positive patients included: rectal swab screening of both hospital inpatients and outpatients; isolation and cohorting of CRE positive hospital inpatients; recall and screening of CRE positive outpatients.
Bacterial strains and microbiological methods
All the Enterobacteriaceae isolates were identified using the Vitek 2 automatic system and the ID-GNB card for identification of Gram-negative bacilli according to the manufacturer’s instructions (bio-Merieux, Marcy l’Etoile, France). Two Raoultella spp., and one Enterobacter spp. were further identified as R. planticola, R. ornithinolytica and E. aerogenes by amplification and sequencing of the rpoB gene as previously described [17].
Antimicrobial susceptibility testing
Carbapenem resistance of Enterobacteriaceae was screened using the meropenem disk alone as previously described [18]. Susceptibility tests were performed using the Vitek 2 system and the AST-GN card (bioMe’rieux). Breakpoints values were those recommended by the EUCAST 2014 [19]. Escherichia coli ATCC 25922 was used as quality control strain. ESBL activity was evaluated using the double-disk synergy test as previously described [20]. Colistin susceptibility was evaluated by broth microdilution in Mueller–Hinton broth II (MHBII) according to Clinical and Laboratory Standards Institute guidelines [21].
Molecular analysis of antimicrobial resistance genes
All the isolates were subjected to polymerase chain reaction (PCR) to screen for the presence of bla KPC, bla IMP, bla VIM, bla NDM and bla OXA-48, as described previously [22]. Acinetobacter baumannii AC 54/97 [23], A. baumannii 161/07 [24] and Klebsiella pneumoniae D001 [18] were used as positive quality control strains for bla IMP-2, bla NDM-1 and bla OXA-48 carbapenemase genes, respectively. The presence of the correct size PCR product was confirmed by agarose gel electrophoresis. The full-length alleles of bla KPC and bla VIM were amplified using primers KPC-F2: GTATCGCCGTCTAGTTCTGC and KPC-Rm: CTTGTCATCCTTGTTAGGCG, primers 5’VIM1: ATGTTAAAAGTTATTAGTAGTT and 3’VIM1: CTACTCGGCGACTGAGCGATT, respectively. Direct sequencing of PCR products was performed using the ABI Prism BigDye Terminator v3.1 Ready Reaction cycle sequencing kit and the 3730 DNA analyzer (Applied Biosystems, Foster City, CA). Nucleotide sequences obtained were compared with the deduced amino acid sequence available in GenBank using tblastx [25]. The presence of bla CTX-M extended-spectrum beta-lactamase genes was investigated by multiplex PCR as described previously [26]. Escherichia coli SGH 20-ADM was used as positive quality control strain for bla CTX-M-15 gene [27]. The presence of the armA methylase gene was investigated by PCR using specific primers as described previously [28].
PFGE typing and dendrogram analysis
XbaI DNA macrorestriction of Enterobacteriaceae isolates was performed as previously reported [29]. PFGE conditions were as follows: pulse times ranged from 5 to 40 s over 24 h at6 · 0 V/cm and at 14C. The PFGE profiles obtained were converted to TIFF files and subjected to cluster analysis using the GelCompare II v. 4.6 software package (AppliedMaths, Sint-Martens-Latem, Belgium). Clustering was based on the unweighted pair-group method with arithmetic averages (UPGMA). The Dice correlation coefficient was used to analyze the similarities of the banding patterns with a tolerance of 1.5 %. The interpretation of chromosomal DNA restriction patterns was based on the criteria of Tenover et al. for closely related isolates [30]. Briefly, strains showing more than three DNA fragment variations and a similarity of <85 % at dendrogram analysis were considered to represent different PFGE types, while one to three-fragment differences and a similarity of >85 % upon dendrogram analysis were considered to represent PFGE pattern subtypes.
MLST analysis
MLST analysis of K. pneumoniae isolates was performed using the Institut Pasteur’s MLST scheme [31]. The allele sequences and sequence types (STs) were identified at http://bigsdb.web.pasteur.fr/klebsiella/klebsiella.html. MLST analysis of E. coli isolates was performed as previously described [32, 33], with the following minor modifications for annealing temperatures of the primers, respectively 54 °C for adk, fumC and icd genes, 58 °C for recA and mdh genes, 60 °C for purA gene, and 64 °C for gyrB gene. Allele sequence and MLST profile definitions were identified at http://mlst.warwick.ac.uk/mlst/dbs/Ecoli.
Conjugative transfer of carbapenem resistance and plasmid typing
Conjugation experiments were performed by filter mating experiments as previously described [34] using sodium-azide resistant E. coli J53 [35] as recipient strain on BHI agar plates in the presence of 16 mg/l imipenem and 100 mg/l sodium-azide. The frequency of transfer was calculated as the number of transconjugants divided by the number of surviving recipients. Plasmids from parental and transconjugant strains were assigned to incompatibility groups by PCR-based replicon typing (PBRT-kit) (Diatheva s.r.l., Fano, Italy) performed on total DNA using previously described primers and conditions [36, 37].
Ethical approval
The study was approved by the ethics committee of the Aosta Regional Hospital (protocol number 836/2015). All microbiological samples were taken as part of standard care procedures. No written informed consent was necessary for this type of study.
Results
CRE isolates included in the study
The molecular epidemiology of carbapenem-resistant Enterobacteriaceae was studied in the Valle d’Aosta region from November 2013 to August 2014. During this period, 60 consecutive CRE were isolated from 52 different patients, both from clinical samples and from surveillance screening. Of these patients, 6 were outpatients, 4 were admitted to nursing homes, while the remaining 42 were admitted to hospital’s internal wards (in particular, 19 in geriatrics, 4 in intensive care, 4 in neurology, 10 in medical departments and 5 in surgical wards). The isolated materials were 32 urine culture, 20 rectal swabs, 3 respiratory secretions, 2 blood culture, 1 liquid drainage, 1 peritoneal-fluid, and 1 wound swab. Fifty-one isolates were identified as K. pneumoniae, 6 were E. coli, 1 R. planticola, 1 R. ornithinolytica, and 1 E. aerogenes (Figs. 1 and 2 and Additional file 1: Table S1).
Genotypic analysis of carbapenem resistant K. pneumoniae isolates. Dendrogram analysis of digitized XbaI PFGE profiles of carbapenem resistant K. pneumoniae isolates is shown. Percentage of similarity at dendrogram analysis and sizes in kilobases (kb) of lambda DNA molecular mass markers are indicated above dendrogram and PFGE profiles, respectively. Isolate number, taxonomic identification, source, date of isolation, patient name, ward, PFGE type, MLST sequence type and carbapenemase are shown also. UC, urine culture; RS, rectal swab; WS, wound swab; DL, drainage liquid; EMO, blood culture; PF, peritoneal fluid; BA, bronchial aspirate; PFGE, pulsed-field gel electrophoresis; MLST, multilocus sequence typing; ST, sequence type
Genotypic analysis of carbapenem resistant E. coli isolates. Dendrogram analysis of digitized XbaI PFGE profiles of carbapenem resistant E. coli isolates is shown. Percentage of similarity at dendrogram analysis and sizes in kilobases (kb) of lambda DNA molecular mass markers are indicated above dendrogram and PFGE profiles, respectively. Isolate number, taxonomic identification, source, date of isolation, patient name, ward, PFGE type, MLST sequence type and carbapenemase are shown also. UC, urine culture; RS, rectal swab; WS, wound swab; DL, drainage liquid; EMO, blood culture; PF, peritoneal fluid; BA, bronchial aspirate; PFGE, pulsed-field gel electrophoresis; MLST, multilocus sequence typing; ST, sequence type
Antimicrobial susceptibility patterns of Enterobacteriaceae isolates
Both K. pneumoniae and E. coli isolates showed a multidrug-resistant antibiotype. In particular, they were resistant to imipenem, meropenem, ertapenem, beta-lactam/beta-lactamase inhibitor combinations (clavulanic acid/amoxicillin, piperacillin/tazobactam), third and fourth generation cephems, fluoroquinolones (ciprofloxacin), and trimethoprim-sulfamethoxazole; 38 K. pneumoniae isolates were resistant but 13 K. pneumoniae isolates were susceptible to aminoglycosides (amikacin and gentamicin); 46 K. pneumoniae isolates were resistant but 5 K. pneumoniae isolates were susceptible to fosfomycin; all E. coli isolates were susceptible to aminoglycosides and fosfomycin. Also, 42 K. pneumoniae isolates were identified as ESBL producers (Table 1 and Additional file 1: Table S1). Interestingly, all E. coli isolates and the majority of K. pneumoniae isolates were susceptible to colistin, but 4 K. pneumoniae isolates were resistant to colistin, minimal inhibitory concentrations (MICs) 16–125 mg/l (Table 1 and Additional file 1: Table S1). Raoultella spp and E. aerogenes isolates were resistant to carbapenems, beta-lactam/beta-lactamase inhibitor combinations, third and fourth generation cephems, fluoroquinolones and trimethoprim-sulfamethoxazole but susceptible to aminoglycosides, fosfomycin and colistin (Additional file 1: Table S1).
Molecular analysis of antimicrobial resistance genes of CRE isolates
PCR and sequence analysis of carbapenemase genes identified bla VIM-1 in 5 E.coli isolates, bla KPC-2 in 42 K. pneumoniae and 1 R. planticola isolates, and bla KPC-3 in 9 K. pneumonia isolates. No carbapenemase genes were identified in 1 R. ornithinolytica, 1 E. coli, and 1 E. aerogenes isolate (Figs. 1 and 2 and Additional file 1: Table S1). Also, bla CTX-M-1 group gene was amplified from all 42 ESBL-producing K. pneumoniae isolates; armA gene was amplified from all 38 K. pneumoniae isolates resistant to aminoglycosides but not from 13 K. pneumoniae isolates susceptible to aminoglycosides (Additional file 1: Table S1).
Molecular epidemiology of CRE isolates in the USL Valle d’Aosta
To investigate whether the isolation of CRE during the study period was caused by the spread of epidemic clones, all 60 CRE isolates were genotyped. Molecular typing by PFGE identified four major PFGE types in 51 K. pneumoniae isolates, which we categorized from A to D. Of the 42 K. pneumoniae isolates assigned to major PFGE type A, 17 isolates showed an identical macrorestriction pattern (PFGE type A), while 25 strains differed in the migration of one or two DNA fragments and were classified into subtypes A1-A5 (Fig. 1). PFGE types E and F were assigned to R. ornithinolytica and R. planticola isolates, respectively (data not shown). Also, PFGE analysis identified identical macrorestriction pattern (PFGE type “a”) in 5 E. coli isolates and PFGE types “b” and “c”, which differed in the migration of more than 3 bands, in 1 E. coli and 1 E. aerogenes isolate, respectively (Fig. 2). Genotype analysis using MLST identified ST101 for K. pneumoniae isolates assigned to PFGE subtypes A1-A5 and a novel ST1789 for K. pneumoniae isolates assigned to PFGE type A. Because ST101 and ST1789 were single locus variants, they were clustered in the same clonal complex (CC) 101 by eBURST analysis. On the contrary, MLST identified ST512 for K. pneumoniae isolates assigned to PGFE types B and C and ST405 for K. pneumoniae isolates assigned to PGFE type D. ST512 and ST405 differed by at least two loci from all other profiles found in the population under analysis and were regarded as singletons (Fig. 1). Based on all the above data, four distinct epidemic genotypes were identified in K. pneumoniae isolates, which we named ST1789/A, ST101/A1-A5, ST512/B-C, and ST405/D, respectively. MLST analysis identified ST131 for all E. coli isolates assigned to PFGE type “a” and subtype “a1”, which we named ST131/a epidemic genotype (Additional file 1: Table S1). All K. pneumoniae isolates assigned to ST101/A1-A5 and ST1789/A epidemic genotypes were ESBL producers and carried bla CTX-M-1 group gene. Also, 23 of 25 K. pneumoniae isolates assigned to ST101/A1-A5 epidemic genotype, 8 of 17 K. pneumoniae isolates assigned to ST1789/A epidemic genotype, and all K. pneumoniae isolates assigned to ST512/B-C epidemic genotype carried armA gene and were resistant to aminoglycosides (Tables S1). Four K. pneumoniae isolates assigned to ST101/A1-A5 epidemic genotype were resistant to colistin (Additional file 1: Table S1).
To further study the mechanism responsible for carbapenem resistance, we asked whether carbapenem resistance might have been transferred through conjugation. Resistance to imipenem and meropenem (MIC ≥16 mg/l) and ESBL activity along with bla KPC-2 and bla CTX-M-group 1 was transferred from K. pneumoniae isolates assigned to genotypes ST1789/A and ST101/A1 to E. coli J53 strain at frequencies ranging from 8.3 × 10−4 to 1.4 × 10−6 cfu/recipient cells. Also, resistance to imipenem and meropenem (MIC ≥16 mg/l) along with bla KPC-3 or bla VIM-1 was transferred from K. pneumoniae isolates assigned to genotypes ST512/B, ST512/C, ST405/D and E. coli isolate assigned to genotype ST131/a, respectively, to E. coli J53 at frequency ranging from 2.5 × 10−2 to 2.5 × 10−7 cfu/recipient cells. All transconjugants were resistant to sodium-azide and showed a PFGE profile identical with that of the recipient strain (Table 2). Also, we asked whether conjugative plasmids were responsible for the horizontal transfer of carbapenem resistance. PCR-based replicon typing identified FII replicon and IncFII incompatibility group plasmid/s in ESBL positive and carbapenem-resistant K. pneumoniae isolates and transconjugants producing bla KPC-2 and bla CTX-M-group 1 genes, while FIIk replicon and IncF incompatibility group plasmid/s in K. pneumoniae isolates and transconjugants producing bla KPC-3 gene. Moreover, the E. coli donor strain expressing bla VIM-1 gene and its transconjugant were shown to carry plasmid/s with N replicon and IncN incompatibility group (Table 2).
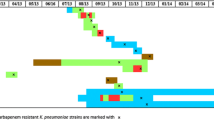
Molecular epidemiology of CRE isolates showed that the increase of CRE was caused by the spread of KPC-2 producing K. pneumoniae ST101/A1-A5 and ST1789/A epidemic genotypes, which belonged to the same CC101. In particular, 17 KPC-2 producing K. pneumoniae assigned to ST1789/A were isolated from 17 patients; 25 KPC-2 producing K. pneumoniae assigned to ST101/A1-A5 genotype were isolated from 21 patients, 3 patients showing repetitive isolation of K. pneumoniae ST101/A1-A3, one of which having isolation of K. pneumoniae ST101/A1-A3 in neurology ward and one month later as outpatient. Also, 2 patients showed isolation of ST1789/A and ST101/A2-A3. Seven KPC-3 producing K. pneumoniae ST512/B-C, 2 KPC-3 producing K. pneumoniae ST405/D, and 5 VIM-1 producing E. coli ST131/a were isolated from 7, 2, and 5 patients, respectively. Two different patients showed the isolation of ST512/B1 and ST405/D, and ST101/A1 and ST131/a (Fig. 1). As shown in Fig. 3, KPC-2 producing K. pneumoniae ST1789/A were isolated from 6, 5, 4 and 1 patients in nursing homes, geriatrics, medical wards, neurology, respectively, and 1 outpatient; KPC-2 producing K. pneumoniae ST101/A1-A5 were isolated from 12, 3, 2, 2, and 1 patients in geriatrics, neurology, intensive care unit, medical and surgical wards, respectively and in 5 outpatients (Fig. 3). KPC-3 producing K. pneumoniae ST512/B-C, ST405/D, and VIM-1 producing E. coli ST131/a epidemic genotypes were isolated from 2 patients in intensive care unit and 5 and 4 patients in medical and surgical wards, respectively; KPC-3 producing K. pneumoniae ST405/D and VIM-1 producing E. coli ST131/a genotypes were isolated from 3 patients in geriatrics and 2 outpatients, respectively (Fig. 3).
Discussion
In the present report, we studied the molecular epidemiology and the genetic basis of carbapenem resistance in CRE isolates in the USL Valle d’Aosta between November 2013 and August 2014. In accordance with previous data [1–7, 9, 10], the spread of CRE in Valle d’Aosta region was caused by the selection of few epidemic distinct genotypes of KPC-2 producing K. pneumoniae, KPC-3 producing K. pneumoniae, or VIM- producing E. coli. The majority of the isolates (42 of 60) were assigned to K. pneumoniae ST101/A1-A5 and ST1789/A genotypes, which belonged to the same CC101 and carried bla KPC-2 into IncFII conjugative plasmid/s. KPC-2 producing K. pneumoniae ST101/A1-A5 were isolated from patients in geriatrics, neurology, other medical wards, and from outpatients; KPC-2 producing K. pneumoniae ST1789/A were isolated from patients in geriatrics, medical wards, nursing homes, and one outpatient. The isolation of identical genotypes of CRE in long-term care facilities and adjacent hospital wards is in agreement with a previous study showing the isolation of identical carbapenemase among Enterobacteriaceae in an Italian tertiary-care hospital and adjacent long-term care facilities [8]. KPC-2 producing K. pneumoniae ST101 has already been reported as sporadic isolation in Brazil [38] and sporadic or epidemic isolation in Italy [9, 39–41], while KPC-2 producing K. pneumoniae ST1789 and nosocomial outbreak caused by KPC-2 producing K. pneumoniae ST101 and ST1789 belonging to CC101 were described for the first time herein. Also, sporadic isolation of KPC-3 producing K. pneumoniae ST101 has been reported in Southern Italy [42], thus suggesting that bla KPC-2 might have been acquired through horizontal gene transfer. In further support of this hypothesis, we demonstrated herein that K. pneumoniae ST101 and ST1789 isolates carried bla KPC-2 into IncFII conjugative plasmid/s. We also observed the spread of KPC-3 producing K. pneumoniae ST512/B and ST512/C genotypes. Because K. pneumoniae ST512 is a single locus variant of ST258, KPC-3 producing K. pneumoniae ST512 isolates can be assigned to the multidrug-resistant high risk CC-258, which was responsible of several epidemics in Europe and Italy [6, 9, 11, 42]. Our data showed also the spread of VIM-1 producing E. coli ST131 among CRE isolates. This is consistent with previous studies showing the emergence of hypervirulent E. coli ST131 clonal lineage [6] and VIM-like producing CRE in Europe and Northern Italy [4, 6–8, 10].
Several studies demonstrate that epidemic clones of CRE are selected in the community and hospital setting because of their multiple antimicrobial resistances [1–11]. The presence of carbapenemases and the acquisition of carbapenemases through horizontal gene transfer have been frequently reported [1–7]. Accordingly, our data showed that carbapenem resistance, along with bla KPC-2, bla KPC-3 and bla VIM-1 carbapenemase genes carried by conjugative plasmids assigned to IncFII, IncF, and IncN incompatibility groups, respectively, was transferred by filter mating experiments. Interestingly, bla KPC-3 carried by conjugative plasmid/s assigned to IncF incompatibility group was found in both K. pneumoniae ST512 and ST405 epidemic genotypes isolated in the study. Our data are in agreement with previous studies showing bla VIM-1 carbapenemase into conjugative plasmids assigned to IncN incompatibility group [43], bla KPC-2 into conjugative plasmids assigned to IncFII incompatibility group [5], bla KPC-3 into conjugative plasmids assigned to IncF incompatibility group [37]. However, the presence of bla KPC-2 carbapenemase into conjugative plasmids assigned to IncF incompatibility group has been described also [41]. The above all data suggest that conjugation was responsible for carbapenem resistance acquisition in CRE included in the study. The analysis of antimicrobial resistance profiles of CRE epidemic genotypes showed that they were microbial-drug resistant. In particular, we identified CTX-M1 group ESBL in KPC-2 producing ST101 and ST1789 K. pneumoniae isolates and corresponding transconjugants. In agreement with our data, the coexistence of KPC-2 carbapenemase and CTX-M-15 group 1 ESBL has been demonstrated [12–14]. Also, all 7 K. pneumoniae isolates assigned to ST512, 23 out of 25 isolates assigned to ST101, and 8 out 17 isolates assigned to ST1789 epidemic genotypes were resistant to aminoglycosides and showed the presence of armA gene (Additional file 1: Table S1). This is consistent with previous studies showing the isolation of extensively drug-resistant ArmA-and KPC-2-producing ST101 K. pneumoniae isolates in Italy [40, 41]. An interesting finding of our study was the occurrence of colistin resistance in four KPC-2 producing K. pneumoniae isolates assigned to ST101/A2-A4 genotypes. Our data are in agreement with recent studies showing the emergence of colistin resistance in KPC-2 producing K. pneumoniae isolates from the Netherlands [44] and KPC-2 or KPC-3 producing K. pneumoniae isolates from Italy [42, 45, 46]. Because KPC-2 or KPC-3 producing colistin resistant K. pneumoniae isolates from Italy and the Netherlands belong to ST258 or ST512 single locus variant [42, 44–46], the data shown herein suggest that colistin resistance may develop in different genetic backgrounds.
Conclusions
The spread of CRE in Valle d’Aosta region was caused by the selection of KPC-2 producing K. pneumoniae ST1789 and ST101 isolates, which were single locus variants and could be assigned to CC101. KPC-2 producing K. pneumoniae CC101 were isolated from patients in geriatrics and other hospital medical wards and in outpatients in the USL Valle d’Aosta. KPC-3 producing K. pneumoniae ST512 and ST405, and VIM-1 producing E.coli ST131 were isolated also. Carbapenem resistance, along with bla KPC-2, bla KPC-3 and bla VIM-1 carbapenemase genes, was transferred by conjugative plasmids assigned to IncFII, IncF, and IncN incompatibility groups, respectively, in filter mating experiments. The emergence of colistin resistance was observed in KPC-2 producing K. pneumoniae ST101 isolates.
Abbreviations
- BA:
-
Bronchial aspirate
- CC:
-
Clonal complex
- CRE:
-
Carbapenem resistant Enterobacteriaceae
- DL:
-
Drainage liquid
- EMO:
-
Blood culture
- ESBL:
-
Extended-spectrum beta-lactamase
- MIC:
-
Minimal inhibitory concentration
- MLST:
-
Multi-locus sequence typing
- PF:
-
Peritoneal fluid
- PFGE:
-
Pulsed-field gel electrophoresis
- RS:
-
Rectal swab
- ST:
-
Sequencing type
- UC:
-
Urine culture
- WS:
-
Wound swab
References
Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med. 2012;18:263–72.
Canton R, Akova M, Carmeli Y, Giske CG, Glupczynski Y, Gniadkowski M, et al. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin Microbiol Infect. 2012;18:413–31.
Temkin E, Adler A, Lerner A, Carmeli Y. Carbapenem-resistant Enterobacteriaceae: biology, epidemiology, and management. Ann NY Acad Sci. 2014;1323:22–42.
Miriagou V, Tzelepi E, Gianneli D, Tzouvelekis LS. Escherichia coli with a self-transferable, multiresistant plasmid coding for metallo-beta-lactamaseVIM-1. Antimicrob Agents Chemother. 2003;47:395–7.
Mataseje LF, Boyd DA, Willey BM, Prayitno N, Kreiswirth N, Gelosia A, et al. Plasmid comparison and molecular analysis of Klebsiella pneumoniae harbouring blaKPC from New York City and Toronto. J Antimicrob Chemother. 2011;66:1273–7.
Mathers AJ, Peirano G, Pitout JD. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev. 2015;28:565–91.
Aschbacher R, Doumith M, Livermore DM, Larcher C, Woodford N. Linkage of acquired quinolone resistance (qnrS1) and metallo-β-lactamase (blaVIM-1) genes in multiple species of Enterobacteriaceae from Bolzano, Italy. J Antimicrob Chemother. 2008;61:515–23.
Aschbacher R, Pagani L, Doumith M, Pike R, Woodford N, Spoladore G, et al. Metallo-beta lactamases among Enterobacteriaceae from routine samples in an Italian tertiary care hospital and long-term care facilities during 2008. Clin Microbiol Infect. 2011;17:181–9.
Giani T, Pini B, Arena F, Conte V, Bracco S, Migliavacca R, et al. Epidemic diffusion of KPC carbapenemase-producing Klebsiella pneumoniae in Italy: results of the first countrywide survey, 15 May to 30 June 2011. Euro Surveill. 2013; 18:20489. Available at http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20489.
Barbarini D, Russello G, Brovarone F, Capatti C, Colla R, Perilli M, et al. Evaluation of carbapenem-resistant Enterobacteriaceae in an Italian setting: report from the trench. Infect Genet Evol. 2014;30:8–14.
Bialek-Davenet S, Criscuolo A, Ailloud F, Passet V, Jones L, Delannoy-Vieillard A-S, et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis. 2014;20:1812–20.
Leavitt A, Carmeli Y, Chmelnitsky I, Goren MG, Ofek I, Navon- Venezia S. Molecular epidemiology, sequence types, and plasmid analyses of KPC-producing Klebsiella pneumoniae strains in Israel. Antimicrob Agents Chemother. 2010;54:3002–6.
Pournaras S, Poulou A, Voulgari E, Vrioni G, Kristo I, Tsakris A. Detection of the new metallo-beta-lactamase VIM-19 along with KPC-2, CMY-2 and CTX-M-15 in Klebsiella pneumoniae. J Antimicrob Chemother. 2010;65:1604–7.
Souli M, Galani I, Antoniadou A, Papadomichelakis E, Poulakou G, Panagea T, et al. An outbreak of infection due to beta-Lactamase Klebsiella pneumoniae Carbapenemase 2-producing K. pneumoniae in a Greek University Hospital: molecular characterization, epidemiology, and outcomes. Clin Infect Dis. 2010;50:364–73.
European Centre for Disease Prevention and Control. Antimicrobial resistance surveillance in Europe 2011. Annual report of the European Antimicrobial Resistance Surveillance Network (ears-net). Stockholm: ECDC; 2012.
Ministero della Salute - 2013: Sorveglianza e controllo delle infezioni da batteri produttori di carbapenemasi (CPE) (Circolare n 4968 26/02/2013). Available at http://www.salute.gov.it/portale/documentazione/p6_2_6.jsp?lingua=italiano&area27&btnCerca=cerca&iPageNo=4. Accessed 6 Nov 2015.
Mollet C, Drancourt M, Raoult D. rpoB sequence analysis as a novel basis for bacterial identification. Mol Microbiol. 1997;26:1005–11.
Pournaras S, Zarkotou O, Poulou A, Kristo I, Vrioni G, Themeli-Digalaki K, et al. A combined disk test for direct differentiation of carbapenemase-producing Enterobacteriaceae in surveillance rectal swabs. J Clin Microbiol. 2013;51:2986–90.
The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters. Version 4.0, valid from 2014-01-01. Available at: www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Breakpoint_table_v_4.0.pdf. Accessed 6 Nov 2015.
Drieux L, Brossier F, Sougakoff W, Jarlier V. Phenotypic detection of extended-spectrum ß- lactamase production in Enterobacteriaceae: review and bench guide. Clin Microbiol Infect. 2008;14 Suppl 1:90–103.
Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; twenty-second informational supplement. Approved standard M100-S22. Wayne: Clinical and Laboratory Standards Institute; 2012.
Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70:119–23.
Riccio ML, Franceschini N, Boschi L, Caravelli B, Cornaglia G, Fontana R, et al. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of blaIMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob Agents Chemother. 2000;44:1229–35.
Bonnin RA, Poirel L, Naas T, Pirs M, Seme K, Schrenzel J, et al. Dissemination of New Delhi metallo-β-lactamase-1-producing Acinetobacter baumannii in Europe. Clin Microbiol Infect. 2012;18:E362–5.
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402.
Woodford N, Fagan EJ, Ellington MJ. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum (beta)-lactamases. J Antimicrob Chemother. 2006;57:154–5.
Moubareck C, Daoud Z, Hakime NI, Hamzé M, Mangeney N, Matta H, et al. Countrywide spread of community- and hospital-acquired extended-spectrum b-lactamase (CTX-M-15)-producing Enterobacteriaceae in Lebanon. J Clin Microbiol. 2005;43:3309–13.
Brigante G, Migliavacca R, Bramati S, Motta E, Nucleo E, Manenti M, et al. Emergence and spread of a multidrug- resistant Acinetobacter baumannii clone producing both the carbapenemase OXA-23 and the 16S rRNA methylase ArmA. J Med Microbiol. 2012;6:653–61.
Villari P, Iacuzio L, Torre I, Nani E. Molecular epidemiology as an effective tool in the surveillance of infections in the neonatal intensive care unit. J Infect. 1998;37:274–81.
Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9.
Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43:4178–82.
Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60:1136–51.
Lau SH, Reddy S, Cheesbrough J, Bolton FJ, Willshaw G, Cheasty T, et al. Major uropathogenic Escherichia coli strain isolated in the northwest of England identified by multilocus sequence typing. J Clin Microbiol. 2008;46:1076–80.
Zarrilli R, Tripodi MF, Di Popolo A, Fortunato R, Bagattini M, Crispino M, et al. Molecular epidemiology of high-level aminoglycoside-resistant enterococci isolated from patients in a università hospital in southern Italy. J Antimicrob Chemother. 2005;56:827–35.
Jacoby GA, Han P. Detection of extended-spectrum beta-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J Clin Microbiol. 1996;34:908–11.
Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219–28.
Villa L, García-Fernández A, Fortini D, Carattoli A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother. 2010;65:2518–29.
Seki LM, Pereira PS, de Souza MP, Conceição Mde S, Marques EA, Porto CO, et al. Molecular epidemiology of KPC-2- producing Klebsiella pneumoniae isolates in Brazil: the predominance of sequence type 437. Diagn Microbiol Infect Dis. 2011;70:274–7.
Mammina C, Bonura C, Aleo A, Fasciana T, Brunelli T, Pesavento G, et al. Sequence type 101 (ST101) as the predominant carbapenem-non-susceptible Klebsiella pneumoniae clone in an acute general hospital in Italy. Int J Antimicrob Agents. 2012;39:543–5.
Mezzatesta ML, Gona F, Caio C, Adembri C, Dell’Utri P, Santagati M, et al. Emergence of an extensively drug-resistant ArmA- and KPC-2-producing ST101 Klebsiella pneumoniae clone in Italy. J Antimicrob Chemother. 2013;68:1932–4.
Frasson I, Lavezzo E, Franchin E, Toppo S, Barzon L, Cavallaro A, et al. Antimicrobial treatment and containment measures for an extremely drug-resistant Klebsiella pneumoniae ST101 isolate carrying pKPN101-IT, a novel fully sequenced bla KPC-2 plasmid. J Clin Microbiol. 2012;50:3768–72.
Bonura C, Giuffrè M, Aleo A, Fasciana T, Di Bernardo F, Stampone T, et al. An Update of the Evolving Epidemic of bla KPC Carrying Klebsiella pneumoniae in Sicily, Italy, 2014: Emergence of Multiple Non-ST258 Clones. PLoS One. 2015;10:e0132936.
Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother. 2009;53:2227–38.
Weterings V, Zhou K, Rossen JW, van Stenis D, Thewessen E, Kluytmans J, et al. An outbreak of colistin-resistant Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae in the Netherlands (July to December 2013), with inter-institutional spread. Eur J Clin Microbiol Infect Dis. 2015;34:1647–55.
Monaco M, Giani T, Raffone M, Arena F, Garcia-Fernandez A, Pollini S, et al. Colistin resistance superimposed to endemic carbapenem-resistant Klebsiella pneumoniae: a rapidly evolving problem in Italy, November 2013 to April 2014. Euro Surveill. 2014; 19:20939. Available at http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20939.
Giani T, Arena F, Vaggelli G, Conte V, Chiarelli A, De Angelis LH, et al. Large nosocomial outbreak of colistin-resistant KPC carbapenemase-producing Klebsiella pneumoniae by clonal expansion of an mgrB deletion mutant. J Clin Microbiol. 2015;53:3341–4.
Acknowledgements
We thank the team of the curators of the Institut Pasteur MLST system (Paris, France) for importing K. pneumoniae novel alleles, profiles and isolates at http://bigsdb.web.pasteur.fr. We thank Laurent Poirel, HFR - Hôpital Cantonal, Fribourg, Switzerland, Spyros Pournaras, University of Athens, Greece, Gian Maria Rossolini, University of Siena, Italy, and Ziad Daoud, University of Balamand, Tripoli, Lebanon, for providing NDM-1, OXA-48, IMP-2 and CTX-M-15 positive control strains, respectively.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RN and RZ designed the study; MDF, LP and MB performed the experiments; CG and MT collected the epidemiological data; CGG and PM collected the microbiological data; MDF, LP and RZ analyzed the data; RZ wrote the manuscript. All authors read and approved the final manuscript.
Mariateresa Del Franco and Laura Paone contributed equally to this work.
Additional file
Additional file 1: Table S1.
Phenotypic and genotypic features of Enterobacteriaceae included in the study. (XLSX 19 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Del Franco, M., Paone, L., Novati, R. et al. Molecular epidemiology of carbapenem resistant Enterobacteriaceae in Valle d’Aosta region, Italy, shows the emergence of KPC-2 producing Klebsiella pneumoniae clonal complex 101 (ST101 and ST1789). BMC Microbiol 15, 260 (2015). https://doi.org/10.1186/s12866-015-0597-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-015-0597-z