Abstract
Background
The emerged human infection with avian influenza A (H7N9) virus in China since 2013 has aroused global concerns. There is great demand for simple and rapid diagnostic method for early detection of H7N9 to provide timely treatment and disease control. The aim of the current study was to develop a rapid, accurate and feasible reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay for detection of H7N9 virus.
Results
The detection limits of the H7- and N9-specific RT-LAMP assay were both approximately 0.2 PFU per reaction. No cross-reactivity was observed with other subtype of influenza viruses or common respiratory viral pathogens. The assay worked well with clinical specimens from patients and chickens, and exhibited high specificity and sensitivity.
Conclusions
The H7/N9 specific RT-LAMP assay was sensitive and accurate, which could be a useful alternative in clinical diagnostics of influenza A (H7N9) virus, especially in the hospitals and laboratories without sophisticated diagnostic systems.
Similar content being viewed by others
Background
Since February 2013,a novel avian influenza A (H7N9) virus has emerged in China, resulting in human infections [1]. As of 7 Sept 2014, the Chinese CDC reported 452 laboratory-confirmed human cases [2]. According to the etiology and gene traceable results, H7N9 avian flu virus is a new recombinant virus [3]-[5]. Human infection with avian influenza A (H7N9) virus usually results in an influenza-like illness (ILI) with symptoms such as fever, cough with little to no sputum production, accompanied by headache, muscular soreness, and general malaise [6]. Most patients presented with rapidly progressing severe lower respiratory tract infections. Considering that the novel avian H7N9 is characteristic of mammalian adapted, scientific community widespreadly concerns that the emerging reassorted virus could cause a new influenza pandemic [7],[8].
Several newly published researches indicated that the H7N9 virus exhibited human-type receptor-binding ability and could replicate in mammals [9]-[11]. H7N9 virus could invade epithelial cells in human lower respiratory tract and pneumonocytes [11]. These biological characterizations of H7N9 virus increase the pandemic risk especially that the virus acquires the ability of transmitting readily among humans, and the lack of pre-existing immunity to virus of this subtype among humans [12]. Therefore, the risk of a pandemic caused by avian H7N9 virus requires rapid detection methods.
Currently, the available detection methods for H7N9 virus include virus isolation and real-time RT-PCR assay [6]. The serological detection and real-time PCR were recommended to detect avian influenza A (H7N9) in 2013 by World Health Organizatin (WHO) [13]. Considering the requirement for specific equipment and trained operators of real-time RT-PCR assay, there is great demand for simple, rapid and sensitive diagnosis method for early detection of H7N9 to provide timely treatment and disease control. The loop-mediated isothermal amplification (LAMP) method, first described in 2000, has been proved to be a rapid, accurate, and cost-effective diagnostic method for infectious diseases [14]-[16]. Previously, the LAMP methods have been applied for detection of different subtypes of influenza viruses, including avian H3, H5, H7, H9, the 2009 H1N1 influenza virus, seasons type A or B influenza virus, and H3 swine influenza virus [17]-[23]. All these assays showed high specificity, efficiency, and sensitivity that were similar to or even higher than real-time PCR assays. These studies exhibited the potential of LAMP as a routine diagnostic method for influenza infections.
In this study, a H7N9-specific RT-LAMP assay targeting the HA or NA gene of avian influenza A (H7N9) virus was developed and evaluated with clinical throat swab and avian samples. The results demonstrated that the RT-LAMP assay was sensitive and accurate, which could be a useful alternative in clinical diagnostics of influenza A (H7N9) virus, especially in the hospitals and laboratories without sophisticated diagnostic systems.
Results
The primer sets of H7 or N9 specific RT-LAMP comprise two outer primers (F3 and B3), two inner primers (FIP and BIP), and one loop primer LF that recognize seven distinct regions on the target sequence of HA or NA gene. Considering that the genomic stability of the emerging reassorted H7N9 was still unclear, the target sequences of the primers were optimized to avoid covering the hot spot in HA or NA gene, such as receptor binding domain or membrane fusion loop in HA gene. After evaluation with different dilutions of H7N9 viral RNA, the primer set (Table 1) was selected.
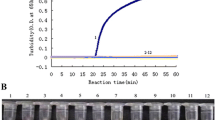
The detection limits of the H7 and N9 specific RT-LAMP assay were both 0.2 PFU per reaction (Figure 1A and C), which was 10-fold-higher sensitive than that of real-time RT-PCR assay recommended by WHO (Table 2). Furthermore, the cross-reactivity tests with seven respiratory viruses also revealed the high specificity of the H7/N9 RT-LAMP assay.
Sensitivity and specificity of the H7/N9 -specific RT-LAMP assay. Amplification curves of the H7/N9-special RT-LAMP were performed with 10-fold serial dilutions of viral RNA (from 200 to 0.002 PFU per reaction). Specificities of H7 and N9 RT-LAMP were tested by using direct visual detection of RT-LAMP with RNA extracted from viral culture of H1N1, H3N2, H5N1, PIV3, H9N2, RSV, HAdV-3 and H7N9. (A) Detection limit of H7-specific RT-LAMP was 0.2 PFU per reaction. (B) The color changes under natural or UV light were only observed in the tube of H7N9 viral RNA by using H7-specific RT-LAMP assay. (C) Detection Limit of N9-specific RT-LAMP was 0.2 PFU per reaction. (D) The color changes were observed for N9-specific RT-LAMP reaction with H7N9 viral RNA. No color change was seen in RT-LAMP with other subtype of influenza viral RNA and other three respiratory viruses under natural or UV light.
Discussion
The RT-LAMP assay could finish within 45 minutes. By using fluorescent detection reagent, the positive amplification could be observed by color change. As shown in the Figure 1B and D, the color of positive reaction (H7N9) change from orange to green under natural light, while the positive reaction also produced bright green fluorescence under UV light. For negative sample such as H1N1, H3N2, H5N1, PIV3, H9N2, RSV and HAdV-3 virus, the color of reaction mixture did not change either under normal light or under UV light.
The H7/N9-specific RT-LAMP assay was further evaluated using clinical samples and chicken samples compared with rRT-PCR assay. The RT-LAMP results showed that 17 samples were tested to be positive and the other 10 samples were negative for H7N9, respectively, which could be confirmed by rRT-PCR assay. No false positive and false negative were observed. By using these 27 samples, 100% diagnostic sensitivities were achieved with H7 or N9 specific RT-LAMP assay compared with rRT-PCR assays, which showed that RT-LAMP results were consistent with that of rRT-PCR. The LAMP method aimed to detect 2013 avian influenza H7N9 showed reliable results in the detection of avian and human clinical samples.
Previously, two RT-LAMP systems had been developed for detection of avian influenza subtype H7 virus [17],[23]. Currently, some other RT-LAMP assays for detection of H7N9 virus were also developed [24]-[28]. These assays showed excellent sensitivities and specificities. Our H7/N9-specific RT-LAMP assays, as well as those published RT-LAMP methods, were very useful for the early detection of suspected H7N9 infection, and could contribute to the prevention and control of H7N9 epidemics and potential pandemic in China and other asian countries.
Conclusions
The current study described a rapid, accurate and feasible H7/N9 specific RT-LAMP assay with perfect sensitivities and high specificities. It was simple and easy to perform. This assay have the potential to provide useful tools for detection of novel H7N9 virus especially in resource-limited setting, which will play important roles in the prevention and control of spread of novel H7N9 infection.
Methods
Samples
A total of 5 clinical throat-swab samples were collected from suspected patients with H7N9 infection. All samples were collected between 2 days and 7 days after the onset of illness. The clinical samples used in this study were appropriately anonymized. All individuals participating in the study gave written informed consent. Local ethical approval was obtained and guidelines were followed for the use of clinical material and accession to diagnostic results. A total of 22 mixed chicken stool samples were collected randomly from live poultry market in Nanjing, Jiangsu and got permission by the chicken farmer and administrator of poultry market. The stool samples were added by phosphate-buffered saline (PBS; 0.02 mol/L; pH 7.2) to make a 10% stool suspension and then centrifuged at 13,000 g for 5 min. The suspension was subsequently processed for RNA extraction.
The collection of samples for the present study was approved by the Ethical Committee of the State Key Laboratory of Pathogen and Biosecurity (Beijing, China), and the experimental procedures were carried out in strict accordance with guidelines on animal ethics and welfare.
Viruses
The H7N9 virus strain A/Anhui/1/2013, kindly provided by Chinese national influenza center (CNIC), was used as reference virus in the current study. Seven respiratory viruses, including human influenza virus H1N1 virus strain A/California/07/2009, H3N2 virus strain A/Beijing/073103/2009, H5N1 virus strain A/Vietnam/1194/2004, avian H9N2 virus strain A/chicken/Nanjing/1/2013, and other important respiratory viruses including type 3 parainfluenza virus (PIV3) strain BJ/01/2005, respiratory syncytial virus (RSV) strain BJ/01/2004 and human adenovirus type 3 (HAdV-3) strain GB, were used to evaluate the specificity of the assay.
Primers design of RT-LAMP
The nucleotide sequences of complete genome of H7N9 strains were retrieved from the GenBank and GISAID (Global Initiative on Sharing All Influenza Data), and aligned by using the ClustalX multiple sequence alignment program. The RT-LAMP assay primers were designed on the basis of the H7N9 HA or NA alignments by using the LAMP primer designing software PrimerExplorer (http://primerxplorer.jp/e/). The feasibility of all sets of primers was then subsequently validated by BLAST program (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and ClustalW software [29]. The primers were synthesized by Life Technologies (Shanghai) Ltd.
RNA extraction
Total RNA was extracted from 200 μl of the clinical samples or viral culture supernatant by using the RNeasy® Mini Kit (Qiagen, Hilden, Germany) according to the instructions of the manufacturer. The RNA was eluted in a final volume of 50 μl of RNase-free water and stored at -80°C until used.
RT-LAMP
The RT-LAMP reaction was carried out by using the Loopamp RNA amplification kit (Eiken Chemical, Tokyo, Japan). The reaction system contained 5 μl of total RNA, 40 pmol each of the primers FIP and BIP, 5 pmol each of the outer primers F3 and B3, 20 pmol of LF, 12.5 μl of 2 × Reaction Mix, 1 μl of Enzyme Mix and 1 μl of Fluorescent regent. The reaction mixture was incubated at 63°C for 45 min in a Loopamp real-time turbidimeter LA-320 (Eiken Chemical, Tokyo, Japan) and followed by heating at 80°C for 5 min to terminate the reaction. The optical density data of each reaction was real-time recorded every 6 s. The threshold of the turbidity for positive sample was defined at 0.1. The time of positivity (Tp) was determined when the turbidity value increased above the threshold.
Real-time RT-PCR
As compared with the RT-LAMP detection, real-time RT-PCR (rRT-PCR) was carried out with the set of specific primers and probe for the detections of influenza A H7N9 virus recommended by WHO. The rRT-PCR was performed with the One-Step PrimeScript™ RT-PCR Kit (Takara, Dalian, China) in the LightCycler 2.0 system (Roche, Mannheim, Germany) in a 20 μl mixture containing 2 μl of total RNA, 10 μl of 2 × One Step RT-PCR Buffer III, 0.4 μl of Ex Taq, 0.4 μl of PrimeScript™ RT Enzyme Mix II, 0.8 μM of forward primer, 0.8 μM of reverse primer and 0.8 μM of probe. The reaction was performed for 5 min at 42°C, followed by 20 s at 95°C, with a subsequent 40 cycles of amplification (95°C for 5 s, 55°C for 20 s). Fluorescence was recorded at 55°C.
Sensitivity and specificity of the assays
The sensitivities of the RT-LAMP assay and rRT-PCR assay were analyzed using 10-fold serially dilutions of viral RNA. The H7N9 virus strain A/Anhui/1/2013 was used to evaluate the sensitivity of RT-LAMP assay developed in the current study. The concentrations of viral RNA were serial diluted from 200 PFU to 0.002 PFU per reaction, respectively. The specificity of the assay was evaluated by cross-reactivity tests with seven respiratory viruses, including human influenza H1N1 virus, H3N2 virus, H5N1 virus, avian H9N2 virus, PIV3, RSV and HAdV-3.
Evaluation with clinical samples
The H7/N9-specific RT-LAMP assay was evaluated using clinical samples from suspected patients with H7N9 infection and chicken stool samples collected from local live poultry markets. A total of 27 samples were tested for the presence of H7N9 virus by using the RT-LAMP assay and the real-time RT-PCR assay. Cp value of rRT-PCR less than 35 was judged as positive. Positive standard was identified as emergence of the positive curve or the color change within 45 minutes in the RT-LAMP assay.
Authors’ contributions
JL, TJ and CFQ: designed the study, did laboratory testing, analyzed the test results. TJ, QYZ, EDQ and CFQ co-wrote and edited the manuscript. XFL, YZ, JL, YQD, QGN, SYZ, YH participated in the laboratory testing. All authors read and approved the final manuscript.
Abbreviations
- RT-LAMP:
-
Reverse-transcription loop-mediated isothermal amplification
- PFU:
-
Plaque Forming Unit
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- PIV:
-
Parainfluenza virus
- RSV:
-
Respiratory syncytial virus
- HAdV:
-
Human adenovirus
- Ct:
-
Cycle threshold value
- Tp:
-
time of positivity
References
Emergence of avian influenza A(H7N9) virus causing severe human illness - China, February-April 2013. MMWR Morb Mortal Wkly Rep. 2013, 62 (18): 366-371.
China CDC: Weekly China influenza surveillance report. In.; 2014., [http://www.cnic.org.cn/uploadfile/2014/0915/20140915024005922.pdf]
Liu D, Shi W, Shi Y, Wang D, Xiao H, Li W, Bi Y, Wu Y, Li X, Yan J, Liu W, Zhao G, Yang W, Wang Y, Ma J, Shu Y, Lei F, Gao GF: Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet. 2013, 381 (9881): 1926-1932. 10.1016/S0140-6736(13)60938-1.
Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q: Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013, 368 (20): 1888-1897. 10.1056/NEJMoa1304459.
Kageyama T, Fujisaki S, Takashita E, Xu H, Yamada S, Uchida Y, Neumann G, Saito T, Kawaoka Y, Tashiro M: Genetic analysis of novel avian A(H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill. 2013, 18 (15): 20453-
China CDC: Diagnostic and treatment protocol for human infections with avian influenza A (H7N9). In.; 2013., [http://www.chinacdc.cn/en/research_5311/Guidelines/201304/t20130425_80443.html]
Uyeki TM, Cox NJ: Global concerns regarding novel influenza A (H7N9) virus infections. N Engl J Med. 2013, 368 (20): 1862-1864. 10.1056/NEJMp1304661.
The Lancet: From SARS to H7N9: will history repeat itself?Lancet 2013, 381(9875):1333.,
Chen E, Chen Y, Fu L, Chen Z, Gong Z, Mao H, Wang D, Ni MY, Wu P, Yu Z: Human infection with avian influenza A(H7N9) virus re-emerges in China in winter 2013. Euro Surveill 2013, 18(43)
Fouchier RA, Kawaoka Y, Cardona C, Compans RW, Garcia-Sastre A, Govorkova EA, Guan Y, Herfst S, Orenstein WA, Peiris JS, Perez DR, Richt JA, Russell C, Schultz-Cherry SL, Smith DJ, Steel J, Tompkins SM, Topham DJ, Treanor JJ, Tripp RA, Webby RJ, Webster RG: Gain-of-function experiments on H7N9. Science. 2013, 341 (6146): 612-613. 10.1126/science.1243325.
Zhou J, Wang D, Gao R, Zhao B, Song J, Qi X, Zhang Y, Shi Y, Yang L, Zhu W, Bai T, Qin K, Lan Y, Zou S, Guo J, Dong J, Dong L, Wei H, Li X, Lu J, Liu L, Zhao X, Huang W, Wen L, Bo H, Xin L, Chen Y, Xu C, Pei Y, Yang Y: Biological features of novel avian influenza A (H7N9) virus. Nature. 2013, 499 (7459): 500-503. 10.1038/nature12379.
Watanabe T, Kiso M, Fukuyama S, Nakajima N, Imai M, Yamada S, Murakami S, Yamayoshi S, Iwatsuki-Horimoto K, Sakoda Y, Takashita E, McBride R, Noda T, Hatta M, Imai H, Zhao D, Kishida N, Shirakura M, de Vries RP, Shichinohe S, Okamatsu M, Tamura T, Tomita Y, Fujimoto N, Goto K, Katsura H, Kawakami E, Ishikawa I, Watanabe S, Ito M: Characterization of H7N9 influenza A viruses isolated from humans. Nature. 2013, 501 (7468): 551-555. 10.1038/nature12392.
WHO: Real-time RT-PCR protocol for the detection of avian influenza A(H7N9) virus. In.: WHO; 2013., [http://www.who.int/influenza/gisrs_laboratory/cnic_realtime_rt_pcr_protocol_a_h7n9.pdf]
Imai M, Ninomiya A, Minekawa H, Notomi T, Ishizaki T, Tashiro M, Odagiri T: Development of H5-RT-LAMP (loop-mediated isothermal amplification) system for rapid diagnosis of H5 avian influenza virus infection. Vaccine. 2006, 24 (44-46): 6679-6682. 10.1016/j.vaccine.2006.05.046.
Jayawardena S, Cheung CY, Barr I, Chan KH, Chen H, Guan Y, Peiris JS, Poon LL: Loop-mediated isothermal amplification for influenza A (H5N1) virus. Emerg Infect Dis. 2007, 13 (6): 899-901. 10.3201/eid1306.061572.
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T: Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28 (12): E63-10.1093/nar/28.12.e63.
Bao H, Wang X, Zhao Y, Sun X, Li Y, Xiong Y, Chen H: Development of a reverse transcription loop-mediated isothermal amplification method for the rapid detection of avian influenza virus subtype H7. J Virol Methods. 2012, 179 (1): 33-37. 10.1016/j.jviromet.2011.08.023.
Chen HT, Zhang J, Sun DH, Ma LN, Liu XT, Cai XP, Liu YS: Development of reverse transcription loop-mediated isothermal amplification for rapid detection of H9 avian influenza virus. J Virol Methods. 2008, 151 (2): 200-203. 10.1016/j.jviromet.2008.05.009.
Gu H, Qi X, Li X, Jiang H, Wang Y, Liu F, Lu S, Yang Y, Liu F: Rapid and specific detection of H3 swine influenza virus using reverse transcription loop-mediated isothermal amplification method. J Appl Microbiol. 2010, 108 (4): 1145-1154. 10.1111/j.1365-2672.2009.04520.x.
Kubo T, Agoh M, Le Mai Q, Fukushima K, Nishimura H, Yamaguchi A, Hirano M, Yoshikawa A, Hasebe F, Kohno S, Morita K: Development of a reverse transcription-loop-mediated isothermal amplification assay for detection of pandemic (H1N1) 2009 virus as a novel molecular method for diagnosis of pandemic influenza in resource-limited settings. J Clin Microbiol. 2010, 48 (3): 728-735. 10.1128/JCM.01481-09.
Ma XJ, Shu YL, Nie K, Qin M, Wang DY, Gao RB, Wang M, Wen LY, Han F, Zhou SM, Zhao X, Cheng YH, Li DX, Dong XP: Visual detection of pandemic influenza A H1N1 Virus 2009 by reverse-transcription loop-mediated isothermal amplification with hydroxynaphthol blue dye. J Virol Methods. 2010, 167 (2): 214-217. 10.1016/j.jviromet.2010.03.027.
Peng Y, Xie Z, Liu J, Pang Y, Deng X, Xie Z, Xie L, Fan Q, Feng J, Khan MI: Visual detection of H3 subtype avian influenza viruses by reverse transcription loop-mediated isothermal amplification assay. Virol J. 2011, 8: 337-10.1186/1743-422X-8-337.
Postel A, Letzel T, Frischmann S, Grund C, Beer M, Harder T: Evaluation of two commercial loop-mediated isothermal amplification assays for detection of avian influenza H5 and H7 hemagglutinin genes. J Vet Diagn Invest. 2010, 22 (1): 61-66. 10.1177/104063871002200110.
Bao H, Zhao Y, Wang Y, Xu X, Shi J, Zeng X, Wang X, Chen H: Development of a reverse transcription loop-mediated isothermal amplification method for the rapid detection of subtype H7N9 avian influenza virus. Biomed Res Int. 2014, 2014: 525064-
Ge Y, Wu B, Qi X, Zhao K, Guo X, Zhu Y, Qi Y, Shi Z, Zhou M, Wang H, Cui L: Rapid and sensitive detection of novel avian-origin influenza A (H7N9) virus by reverse transcription loop-mediated isothermal amplification combined with a lateral-flow device. PLoS One. 2013, 8 (8): e69941-10.1371/journal.pone.0069941.
Nakauchi M, Takayama I, Takahashi H, Tashiro M, Kageyama T: Development of a reverse transcription loop-mediated isothermal amplification assay for the rapid diagnosis of avian influenza A (H7N9) virus infection. J Virol Methods. 2014, 204: 101-104. 10.1016/j.jviromet.2014.03.028.
Nie K, Zhao X, Ding X, Li XD, Zou SM, Guo JF, Wang DY, Gao RB, Li XY, Huang WJ, Shu YL, Ma XJ: Visual detection of human infection with influenza A (H7N9) virus by subtype-specific reverse transcription loop-mediated isothermal amplification with hydroxynaphthol blue dye. Clin Microbiol Infect. 2013, 19 (8): E372-E375. 10.1111/1469-0691.12263.
Zhang J, Feng Y, Hu D, Lv H, Zhu J, Cao M, Zheng F, Zhu J, Gong X, Hao L, Srinivas S, Ren H, Qi Z, Li B, Wang C: Rapid and sensitive detection of H7N9 avian influenza virus by use of reverse transcription-loop-mediated isothermal amplification. J Clin Microbiol. 2013, 51 (11): 3760-3764. 10.1128/JCM.01907-13.
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG: Clustal W and Clustal X version 2.0. Bioinformatics. 2007, 23 (21): 2947-2948. 10.1093/bioinformatics/btm404.
Acknowledgments
We thank Prof. Yue-Long Shu (CNIC) for kindly providing the H7N9 viruses. All the sequences are available from GISAID or GenBank, and we thank the contributors for these sequences. This study was supported in part by the National 973 Project of China (No. 2012CB518904), National Major Research Special Foundation of China (no. 2013ZX10004805 and no. 2014ZX10004002), and National Natural Science Foundation of China (no. 81000722 and no. 81273324).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Liu, J., Nian, QG., Li, J. et al. Development of reverse-transcription loop-mediated isothermal amplification assay for rapid detection of novel avian influenza A (H7N9) virus. BMC Microbiol 14, 271 (2014). https://doi.org/10.1186/s12866-014-0271-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-014-0271-x