Abstract
Background
Aspergillus flavus, one of the causative agents of human fungal keratitis, can be phagocytosed by human corneal epithelial (HCE) cells and the conidia containing phagosomes mature into phagolysosomes. But the immunological responses of human corneal epithelial cells interacting with A. flavus are not clear. In this study, we report the expression of immune response related genes of HCE cells exposed to A. flavus spores using targeted transcriptomics.
Methods
Human corneal epithelial cell line and primary cultures were grown in a six-well plate and used for coculture experiments. Internalization of the conidia was confirmed by immunofluorescence microscopy of the colocalized endosomal markers CD71 and LAMP1. Total RNA was isolated, and the quantity and quality of the isolated RNA were assessed using Qubit and Bioanalyzer. NanoString nCounter platform was used for the analysis of mRNA abundance using the Human Immunology panel. R-package and nSolver software were used for data analysis. KEGG and FunRich 3.1.3 tools were used to analyze the differentially expressed genes.
Results
Different morphotypes of conidia were observed after 6 h of coculture with human corneal epithelial cells and found to be internalized by epithelial cells. NanoString profiling showed more than 20 differentially expressed genes in immortalized human corneal epithelial cell line and more than ten differentially expressed genes in primary corneal epithelial cells. Distinct set of genes were altered in their expression in cell line and primary corneal epithelial cells. KEGG pathway analysis revealed that genes associated with TNF signaling, NF-KB signaling, and Th17 signaling were up-regulated, and genes associated with chemokine signaling and B cell receptor signaling were down regulated. FunRich pathway analysis showed that pathways such as CDC42 signaling, PI3K signaling, and Arf6 trafficking events were activated by the clinical isolates CI1123 and CI1698 in both type of cells.
Conclusions
Combining the transcript analysis data from cell lines and primary cultures, we showed the up regulation of immune defense genes in A. flavus infected cells. At the same time, chemokine signaling and B cell signaling pathways are downregulated. The variability in the expression levels in the immortalized cell line and the primary cultures is likely due to the variable epigenetic reprogramming in the immortalized cells and primary cultures in the absence of any changes in the genome. It highlights the importance of using both cell types in host-pathogen interaction studies.
Similar content being viewed by others
Introduction
Aspergillus flavus is the second most common cause of invasive and non-invasive aspergillosis [1, 2]. A. flavus produces nonmotile conidia, and under extreme conditions, sclerotia were produced [3, 4]. The conidia are larger in size (3 to 6 μm) than A. fumigatus (2 to 3.5 μm), that helps in their deposition in the upper respiratory tract, which could cause fungal sinusitis [5, 6]. In tropical countries A. flavus is the predominant pathogen causing fungal keratitis of the cornea [5] along with Fusarium solani. Fungal keratitis is the leading cause of ocular morbidity and blindness, especially in tropical countries [7], and it has been estimated that the annual incidence is 113 per 100,000 population in Madurai, South India [8, 9]. Infection occurs predominantly when there is a loss of epithelial integrity owing to trauma or physical injury to the cornea [10]. Treatment options to combat fungal infections are limited, and the only approved drug for topical ophthalmic application for fungal keratitis is natamycin [11]. In vitro studies show voriconazole is better than natamycin as an antifungal antibiotic [12], however, in a clinical trial, natamycin is shown to be superior for treating fungal keratitis [13].
Interaction of A. fumigatus with macrophages and lung epithelial cells has been studied extensively, and these cells can internalize the fungal spores by phagocytosis. Macrophages internalize about 80% of the A. fumigatus conidia, and almost all the conidia are positive for lysosome associated membrane glycoprotein 1 (LAMP1) in the phagosomes at 60 min of infection [14]. Unlike macrophages, roughly 30% of A. fumigatus spores are internalized by lung epithelial cell line A549 [15], and roughly half the internalized conidia colocalize with endosomal markers LAMP1 and CD63 after 24 h of infection. Interestingly the internalized conidia survive and germinate inside the host phagosomes by 24 h [16]. The data indicate that the engulfed conidia matured by acquiring late endosomal marker LAMP1 or CD63 in macrophages and epithelial cells. Corneal epithelial cells upregulate the expression of receptors such as dectin-1 [17], TLR-2 and TLR4 [18] upon stimulation with inactivated A. fumigatus. Proinflammatory cytokines such as CXCL1, TNF-α, and IL-6 and activation of P38 MAPK are induced through lectin-like oxidized low-density lipoprotein receptor 1 (LOX-1) in corneal epithelial cells mixed with killed A. fumigatus [19]. These studies confirm the innate function of human corneal epithelial (HCE) cells towards fungal pathogen through expression and secretion of cytokines, and chemokines.
The transcriptional response of lung epithelial cells against A. fumigatus has revealed increased expression of genes involved in innate immune response, chemotaxis, and inflammatory response [20,21,22,23,24]. But the knowledge on the transcriptional response of corneal epithelial cells against fungal infection is not known. We have shown recently that the corneal epithelial cells engulf the A. flavus conidia via actin-dependent process [25]. Phagosomes with conidia mature by acquiring early and late endosomal markers, and we also showed the acidification of the endosomes carrying the conidia. In this report, the response of corneal epithelial cells after A. flavus infection was studied at the transcriptional level using the immunology panel of the NanoString platform using a coculture experimental setup. The nCounter analysis revealed the differential expression of genes involved in TNF signaling, Th17 differentiation, NF-kB signaling, and B cell receptor signaling. In addition, we observed differences in the gene expression profile between cell line and primary cultures in coculture experiments. Together, the data clearly show that the corneal epithelial cells, an earliest cell type of cornea which encounter the invading pathogen, are the first line of defense against fungal infection and can regulate the host’s innate immune response.
Methodology
Cell culture
Simian virus (SV) 40 immortalized Human Corneal Epithelial cell line (RCB2280) was obtained from Riken Cell Bank, Japan, in 2012. The cells were cultured in DMEM/F12, supplemented with 5% FBS, 5 µg/ml insulin, 5 ng/ml human epidermal growth factor, and 0.5% Pencillin-Streptomycin at 37 °C under 95% humidity and 5% CO2 as described by Araki Sasaki et al. [26]. Primary Human Corneal Epithelial cells were generated from limbal explants from cadaver eye globes. Human donor’s eyes were obtained from Rotary Aravind International Eye Bank, Madurai, with donor age less than 70 years with no history of ocular trauma and infection. The written informed consent was obtained for the donor eyes from their family. The tissues were handled in accordance with the Tenets of the declaration of Helsinki. The Institutional Review Board of the Aravind Medical Research Foundation, Madurai, approved this study (ID NO. IRB2011001BAS). The cornea was excised from the eye and the limbal explants from the peripheral cornea were placed in the six-well plates and cultured in DMEM/F12 along with hydrocortisone, sodium selenite, insulin, hEGF, gentamicin, amphotericin, DMSO, and transferrin until it reaches confluency [27].
Preparation of A. flavus conidia
Clinical isolates of A. flavus were obtained from keratitis patients at Aravind Eye Hospital, Madurai. A. flavus (CI1698) is an isolate from ulcer healed patients by anti-fungal therapy (healed case), and A. flavus (CI1123) is an isolate that was refractory to antifungal therapy (surgery case). All the fungal cultures were grown in Czapek Dox agar (M075, HiMedia, L.B.S.Marg, Mumbai) plates for seven days at 30 ºC and spores were collected using PBS with 0.05% tween-20. The mycelium from the spore suspension was removed by using a 0.45µ filter (Supor® 450 membrane disc filters, 0.45 μm-47mm, Pall, USA) and spore suspensions were counted using the Neubauer Hemocytometer chamber (Z359629, Merck, Germany). The freshly harvested spores were used for infecting corneal epithelial cells and the remaining spores were aliquoted as 108/ml in 20% glycerol and stored at -80 ºC freezer [28].
Immunofluorescence analysis
RCB2280 cells were cultured in 22 × 22mm coverslips and mixed with conidia (CI1698) at a multiplicity of ten and incubated for 6 h [25]. Following infection, cells were washed with 1X PBS, fixed with optimized fixation and permeabilization methods for CD71 (4% PFA and 0.1% triton X-100) and LAMP1 (MeOH-Acetone). After blocking, cells were incubated with mouse monoclonal antibodies of CD71 (transferrin receptor) (sc-65882, Santa Cruz Biotechnology, Texas, USA) or LAMP1 (sc-20011, Santa Cruz Biotechnology, Texas, USA) (1:50) for 1 h at room temperature. Finally, the cells were stained with goat anti-mouse IgG conjugated with dylight 550 (1:50) for 1 h at room temperature. Coverslips were mounted on glass slides using vectashield mounting medium containing DAPI and sealed with nail polish. The images were acquired using Leica TCS SP8 confocal laser scanning microscopy and analyzed using LAS AF Lite software.
RNA preparation
1 × 105 RCB2280 cells were seeded in a six-well plate and were grown up to confluence. Limbal explants from cadaver eye globes were plated in six-well plates and incubated for two weeks to obtain confluent growth. A. flavus spores (CI1698 & CI1123) were mixed with the cultured cells at a multiplicity of ten and incubated for six hours in a serum-free medium [21]. At the end of the incubation period, the cells were washed with RNase free 1X PBS and lysed with 350 µl RLT buffer. The lysates were passed through a 20-gauge needle for 5-10 times and stored at -80 ºC freezer. Isolation of RNA from control and infected cells was performed according to the manufacturer’s (74104, Qiagen, Germany) instructions and the DNA from the isolated RNA was removed using TURBO DNA free kit (Invitrogen™ AM1907, California, US) as per the instructions.
NanoString analysis
RNA integrity was determined using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) and Qubit high sensitivity assay (Q32855, Thermo Fischer Scientific, Massachusetts, US). mRNA transcript abundance was analyzed from 100 ng of extracted RNA using the NanoString nCounter Human Immunology panel (XT-CSO-HIM2-12 & NAV # 115000062) from two separate experiments (RCB2280 cells and primary cells). The Human Immunology panel consisted of 595 genes (580 well-annotated immune response genes and 15 housekeeping genes). Briefly, 70 µl of hybridization buffer was added to Reporter CodeSet to prepare the master mix. To set up the hybridization reactions, each sample tube contained 8 µl of master mix were mixed with 5 µl of the extracted RNA sample. Capture ProbeSet (2 µl) was added to each tube, and samples were hybridized at 65˚C for 18 h. The hybridized samples were separated using the NanoString nCounter SPRINT automated Profiler. Tripartite complexes (captured probes with targets) were aligned on the microscopic surface of the cartridge and was scanned using Maximum resolution (Max FOV) in the nCounter Digital Analyzer to generate RCC files (as per the sprint manual MAN-10017-08). Raw data were normalized using positive control genes (POS_A, POS_B, POS_C, POS_D, POS_E, POS_F) and the house keeping genes (ABCF1, ALAS1, EEF1G, G6PD, GAPDH, GUSB, HPRT1, OAZ1, POLR1B, POLR2A, PPIA, SDHA, TBP, TUBB, RPL19) using geNorm in the advanced nSolver analysis 4.0 software (MAN-C0019-08), and lowly expressed genes were filtered based on p-value. p-value less than 0.05 were considered as differentially expressed genes (Table S1 and S2) [23].
Enzyme-linked immunosorbent assay
Human IL-8/CXCL8 DuoSet ELISA kit was used to measure IL-8 concentration in the culture supernatant of RCB2280 cells infected with CI1123. RCB2280 cells were cultured in 12-well plate and infected with live CI1123 conidia for 12 h, 16 h, 20 and 24 h. The conditioned medium was collected, filtered using 0.22-micron filter and centrifuged at 12,000 rpm for 10 min. The supernatant was collected and stored at -80ºC freezer. The concentration of IL-8 was determined as described by the manufacturer. The incubation time with the substrate was optimized and found to be six minutes. Absorbance was measured at 450 nm with reference wavelength of 540 nm using Spectramax microplate reader.
Data analysis
MA plot for the differentially expressed genes was generated using R-package. Pathway analysis was done using KEGG and FunRich 3.1.3 software tools. Cytoscape 3.8.1 tool was used to create the functional network for the differentially expressed genes. Uniprot was used to analyze the functions of differentially expressed genes. Differentially expressed genes from NanoString data were determined using Student’s t-test (p<0.05).
Results
Interaction of A. flavus conidia with human corneal epithelial cells
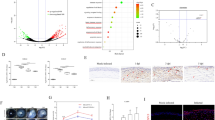
Morphotypes of conidia, swollen conidia, and germinated conidia were examined after coculturing the conidia with RCB2280 cells (B&C) or primary cultures (E&F) for 6 h. Clinical isolates 1698 and 1123 were used in these experiments. Almost all the bound conidia were germinated in the cocultures of RCB2280 cells, whereas a smaller number of conidia were germinated in primary HCE cells mixed with conidia (Fig. 1 see arrows). This result implies that the primary cultures can delay the germination of infected conidia.
Morphology of uninfected and A. flavus infected HCE cells. Microscopic images of primary HCE cells grown on 6-well culture plates were infected with (A) no infection (B) A. flavus CI1698 (C) A. flavus CI1123 and HCE cell line (RCB2280) infected with (D) no infection (E) A. flavus CI1698 (F) A. flavus CI1123 at 200X magnification. The arrow shows the A. flavus conidia
Colocalization of early and late endosomal markers with the A. flavus conidia. RCB2280 cells were infected with A. flavus (CI1698) swollen spores for 4 h and stained for endosomal markers. (A) A. flavus in early endosomes (CD71) (B) A. flavus in late endosomes (LAMP1). The endosomal markers (Red) were detected using dylight 550 conjugated goat anti-mouse secondary antibody. From left to right, panels show bright field, fluorescence image of the blue channel (DAPI), fluorescence image of red channel (endosomes), merged overlay of all fluorescent channels, White arrow shows the germinated conidia inside the early and late endosomes
Further, a fraction of the conidia and germinated conidia were found to be colocalized with the early endosomal marker (CD71) and late endosomal marker (LAMP1) in CI1698 cocultures (Fig. 2). The data indicate that corneal epithelial cells internalized the conidia and the phagosomes containing conidia were matured.
Differential gene expression of HCE cells upon infection with A. flavus
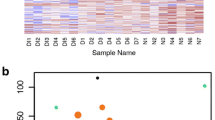
Venn diagram shows the overlap of differentially expressed genes of RCB2280 cells and primary cells in response to A. flavus infection (Fig. 3a). Expression of six genes, SLC2A1, TNFAIP3, ICAM1, RUNX1, ETS1, and IKZF3 were altered in RCB2280 cells infected with CI1698 and CI1123. On the other hand, transcripts of the LGALS3 gene encoding galectin-3 were the only gene whose expression was up-regulated in primary HCE cells exposed to CI1698 and CI1123. CI1123 induced the expression of KIR activating subgroup 3 genes, while CI1698 induced the expression of PLAUR and ETS1 genes in both the cells. ETS-1 gene was found to be up-regulated in RCB2280 cells cocultured with CI1698 and CI1123 and primary HCE cells infected with CI1698. Around 20 genes were found to be differentially regulated in RCB2280 cells, while the expression of about ten genes were altered in primary cells (Table S3). The number of up-regulated genes was more than the down-regulated genes except in primary HCE cells infected with CI1123 (Fig. 3b).
Differential regulation of immune response genes in HCE cells in response to A. flavus. (a) Venn diagram shows the comparison of differentially expressed genes between RCB2280 cells and primary HCE cells infected with CI1698 and CI1123. (b) Bar graph for the up and down-regulated genes in RCB2280 cells and primary HCE cells infected with A. flavus
Differential gene expression in HCE cells infected with CI1698
The MA plot shows the differential mRNA abundance in RCB2280 cells and primary HCE cells infected with CI1698 (Fig. 4a and b). Among the 580 immune response genes quantified, 25 genes were altered in their expression by CI1698 infection of RCB2280 cells, whereas the expression of only 12 genes was altered in primary HCE cells under similar experimental conditions. Of the 25 genes, the expression of 16 genes was upregulated, and nine genes were downregulated in RCB2280 cells with a significant p-value (p<0.05).
Figure 5 indicates the differentially expressed genes with corresponding log2 fold change. CSF2 was increased close to log2 fold change of 2, and LILRB2 was reduced with log2 fold change of -2 in RCB2280 cells. In primary HCE cells, eight genes were up-regulated, and four genes were down-regulated compared to uninfected control (Fig. 6). CD5 and CD6 were the ones showing the maximum variation in expression levels in primary cells after infection. The function of differentially expressed genes of RCB2280 cells and primary HCE cells infected with CI1698 is represented in Table 1.
Pathway analysis using KEGG showed the upregulation of genes involved in TNF signaling, cell adhesion pathway, NF-kB signaling, and Th17 differentiation in the coculture of RCB2280 and conidia of CI1698, whereas multiple genes involved in the B cell receptor signaling and JAK-STAT signaling were downregulated (Fig. 7a). In the coculture experiments using primary cultures and CI1698, hematopoietic cells, the lineage pathway is also upregulated, and chemokine signaling is downregulated along with other pathways (Fig. 7b). The experiment using primary culture or RCB2280, infected with CI1698, show similar pattern of alteration in gene expression profile.
KEGG pathway analysis of up and down-regulated genes of HCE cells infected with CI1698. (a) RCB2280 cells infected with CI1698 for 6 h. (b) Primary HCE cells infected with CI1698 for 6 h. Red bar - pathways associated with up-regulated genes; Green bar - pathways associated with down-regulated genes
Functionally grouped network of Gene Ontology enrichment analysis using FunRich 3.1.3 tool showed pathways enriched in the cocultures of RCB2280 and primary cultures. The number of genes in the enriched pathways is much higher in RCB2280 cell line cocultures (Fig. 8).
The functionally grouped network of Gene Ontology enrichment analysis. Differentially expressed mRNA transcripts of RCB2280 cells and primary HCE cells infected with CI1698 using FunRich 3.1.3 and Cytoscape 3.8.1 tool. Green circle - RCB2280 cells infected with CI1698; Blue circle - primary HCE cells infected with CI1698; Red rectangle - Biological pathways
Differential gene expression in HCE cells infected with CI1123
Figure 9 shows the MA plot for the differential mRNA transcripts for HCE cells in response to CI1123. Only 22 genes and 11 genes were differentially expressed in RCB2280 cells (Fig. 9a) and primary HCE cells (Fig. 9b), respectively. In RCB2280 cells, the expression of 14 genes was increased, and eight genes showed decreased expression compared to the control. In contrast, in primary HCE cells, the expression of only four genes was increased, and the expression of seven genes was decreased.
Figures 10 and 11 show the expression level of differentially regulated genes in RCB2280 cells and primary HCE cells. IL8 was the top up-regulated gene in RCB2280 cells, and LILRB2 was a top down-regulated gene in primary HCE cells. The functions of differentially expressed genes of RCB2280 cells and primary HCE cells infected with CI1123 is shown in Table 2. KEGG pathway analysis revealed that the up-regulated genes were involved in Th17 signaling and TNF signaling, and down-regulated genes were involved in NF-kB signaling in RCB2280 cells (Fig. 12a). In primary HCE cells, up-regulated genes were involved in cytokine-cytokine receptor interaction, and down-regulated genes were involved in B cell receptor signaling and chemokine signaling (Fig. 12b).
Functionally grouped network of Gene Ontology enrichment analysis of differentially expressed mRNA transcripts using FunRich 3.1.3, and Cytoscape 3.8.1 tool was represented in Fig. 13. The differentially expressed mRNA transcripts of RCB2280 cells were enriched in CDC42 signaling, Arf6 trafficking events, and IFN gamma pathway, and primary HCE cells were enriched in innate immune system and cytokine activity.
The functionally grouped network of Gene Ontology enrichment analysis. Differentially expressed mRNA transcripts of RCB2280 cells and primary HCE cells infected with CI1123 using FunRich 3.1.3 and Cytoscape 3.8.1 tool. Purple circle - RCB2280 cells infected with CI1698; Blue circle - primary HCE cells infected with CI1123; Red rectangle -Biological pathways
Response of HCE cells to A. flavus clinical isolates
CI1698 or CI1123 commonly altered eight genes in HCE cells (cell line and primary HCE cells), and the function of these genes is shown in Table 3. FunRich 3.1.3 pathway analysis showed that pathways such as CDC42 signaling, PI3K signaling, and Arf6 trafficking events were equally activated by CI1123 and CI1698 in RCB2280 cells and primary HCE cells. Cytokine-related pathways such as TNF-α/NF-kB, IL23, and TLR 4 signaling were overrepresented in CI1123 infected RCB2280 cells. Similarly, CI1123 infection activates TLR4 signaling and TNF signaling in primary HCE cells, Whereas CI1698 activated complement cascade and p38 MAPK pathway in RCB2280 cells and IL23 mediated signaling in primary HCE cells (Fig. 14a and b). KEGG pathway analysis shows that upregulated genes in response to CI1698 and CI1123 were associated with cytokine-cytokine receptor interaction, TNF signaling, NF-kB signaling pathway, JAK-STAT signaling pathway, Th17 differentiation, and down-regulated genes were associated with chemokine signaling, B-cell receptor signaling pathway, Th1, and Th2 cell differentiation, etc. (Fig. 14c and d).
Pathway analysis of HCE cells in response to A. flavus clinical isolates. (a) Biological pathways of RCB2280 cells infected with CI1698 and CI1123 for 6 h. Grey bar - RCB2280 cells infected with CI1123; Black bar - RCB2280 cells infected with CI1698. (b) Biological pathways of primary HCE cells infected with CI1698 and CI1123. Dark green bar - Primary HCE cells infected with CI1123; Light green bar - Primary HCE cells infected with CI1698. (c) Up-regulated immune genes associated pathways in HCE cells infected with A. flavus clinical isolates. (d) Down-regulated immune genes associated pathways in HCE cells infected with A. flavus clinical isolates
Validation of upregulation of IL-8 expression by ELISA
IL-8 expression was up-regulated in RCB2280 cells infected with CI1123 at the transcript level as shown above. To confirm this result, Human IL-8/CXCL8 DuoSet ELISA kit was used to determine IL-8 protein concentration in CI1123 infected RCB2280 cells. A time dependent increase in the IL-8 amount was observed. IL-8 was elevated from 12 h and reached maximum at 24 h (Fig. 15). IL-8 level was increased by ten-fold in infected RCB2280 cells compared to uninfected control. This data confirms the transcript data and indicates that the mRNA level and protein level are comparable in the infected cells.
Expression of IL-8 in RCB2280 cells infected with CI1123. RCB2280 cells were cultured in 12-well plate till confluency and infected with CI1123 for 12 h, 16 h, 20h and 24 h. Bar graph representing the concentration of IL-8 at different time point. Black bar -Control; Grey bar - RCB2280 cells infected with CI1123. The data were analyzed using two-way ANOVA with Bonferroni multiple comparison test (ns-no significance; **** p<0.0001)
Discussion
Our present study shows that the targeted, relevant transcriptome response of human corneal epithelial cells exposed to clinical isolates of A. flavus is not identical in the cell line and primary cultures. At six hours post-infection, different morphotypes of A. flavus such as conidia and germinated conidia were observed internal and external to the corneal epithelial cells; however, we found more germinated conidia in the coculture of RCB2280 cells with A. flavus compared to a similar experiment using primary cultures. The reduction in the germination in primary cells compared to cell line is not due to the multiplicity of infection since the MOI was kept constant in both cases. The defense pathways specifically activated in the primary cells are the likely cause of the inhibition of germination. It is also important to note that the conidia induce the activation of host response either by internalization or due to the binding of the conidia to the surface receptors. Additional experiments are needed to clarify this. Studies have shown that the internalized A. fumigatus spores survive inside lung epithelial cells due to phagosome maturation failure and the germlings escape by phagolysosome-plasma membrane fusion without rupturing the host cells and penetrating the neighboring cells [29]. A recent report has shown that airway epithelial cells (A549) and bronchial epithelial cells (16HBE) internalize A. fumigatus conidia and digest the engulfed conidia inside the phagolysosome, and the killing is more in bronchial epithelial cells [30]. However, it is difficult to extrapolate these results to our data since the pathogen as well as the host are different in these studies.
Even though the efficiency of phagocytosis of the added conidia are similar [25], the number of differentially expressed genes in primary HCE cells was lower than RCB2280 cells under similar coculture experimental conditions. This data indicates that the primary cultures respond differently to fungal infection. It has been shown previously that the host cell’s immune response is more robust towards the germinated conidia compared to ungerminated conidia, which are covered by immunologically inert hydrophobins [31]. Taken together, our data imply that the primary culture prevented the immune activation by fungal conidia by preventing the germination, leading to the masking of activating signals on the conidial surface. The actual mechanism involved in the robust immune response against fungal infection by primary cells compared to cell line is not known and additional experiments are needed to explain this especially when the number of genes altered in infected primary cultures are less.
NanoString approach was used to analyze the immune response in air-liquid interface (ALI) cultures of primary HBECs upon exposure to A. fumigatus for 6 h, which showed 41 differentially expressed genes out of 730 immune genes [23]. We found 37 differentially expressed genes in corneal epithelial cells exposed to A. flavus. Morton et al., analyzed the expression of 117 genes on immune arrays and showed 16 differentially expressed genes in iDCs exposed to A. fumigatus [32]. Together, these results show that the host-pathogen interaction is complex and depends on several conditions, including the culture conditions, host cell type, and the fungal species used.
RNA seq approach has been used to study corneal epithelial cells [33, 34], but, to our knowledge, there are no studies on transcriptome analysis of corneal epithelial cells infected with fungi. Therefore, we compared the differentially expressed immune gene data with the gene expression profile of A. fumigatus infected bronchial epithelial cells [23]. This study also used NanoString analysis of mRNA but using ALI cultures for bronchial epithelial cells compared to single monolayer and detected 41 differentially expressed genes. Although these genes were found in our dataset, their expression level was not altered in the corneal epithelial cells upon A. flavus infection. This is not surprising since even in the same host cell (bronchial epithelium), A. fumigatus and A. niger alter the expression of different genes [35]. Immune-related genes IL8, CSF2, and NFKBIA, were increased in their expression level in A. flavus infected corneal epithelial cells, as has been shown in the case of A. fumigatus infected iDC [32]. IL8 is upregulated with the maximum fold change in RCB2280 cells infected with CI1123 in the NanoString data and ELISA. IL8 is a chemotactic factor that attracts immune cells, principally neutrophils, to the site of infection and is known to be secreted by several innate cells, including respiratory epithelial cells [36] and corneal epithelial cells upon A. fumigatus infection [37]. Several studies show IL8 is one of the inflammatory cytokines that remarkably elevated during fungal infection [20, 38, 39]. NFKBIA, which limits the immune response by inhibiting the nucleus translocation of NF-kB/REL complexes, was found to be upregulated in RCB2280 cells exposed to CI1123. In the case of A. fumigatus infection of A549 cells, NFKBIA is reported to be upregulated in A. fumigatus infected A549 cells at 8 h of infection as shown by Affymetrix gene microarray [40]; however, in a slightly different experimental system, A549 cells cultured in the transwell membrane and infected with A. fumigatus for 6 h, NFKBIA has been shown to be downregulated [41].
CSF2 (GM- GSF) is one of the highly up-regulated genes in RCB2280 cells infected with CI1698. CSF2 is crucial for the recruitment of monocytes and is involved in the neutrophil and monocyte antifungal activity against A. fumigatus in mice model of aspergillosis [42]. Primary airway epithelial cells cocultured with A. fumigatus for 6 h show an increase in the expression of CSF2 mRNA by microarray. However, the expression of CSF2 mRNA has not been altered in A. fumigatus cocultured with transformed bronchial epithelial cells (16HBE14o) (22). Our study found altered expression of CSF2 in only one clinical isolate infecting the corneal epithelial cell line. These results confirm that the variation between the outcome of fungal infection depends on the clinical isolate used as well as the host cell type.
Six genes were overlapped between RCB2280 cells infected with CI1698 and CI1123 including SLC2A1, TNFAIP3, ICAM1, RUNX1, ETS1, and IKZF3. SLC2A1 encodes a glucose-transporter in the mammalian blood-brain barrier. It has been shown to up-regulate in SV40 immortalized cells cultured in air-liquid interface compared to native corneal epithelial tissue [43] and reported to regulate the glycolytic metabolism in bacterial infection [44]. TNFAIP3 is involved in inflammatory response and is shown to upregulate in A549 exposed to A. fumigatus [20, 40]. ICAM1, a glycoprotein is expressed in human corneal epithelial cells from limbal explants and involved in cell motility. It is reported that the inflammatory cytokines modulate the ICAM1 expression in primary corneal epithelial cells and promote epithelial injury by binding to inflammatory cell receptors [45]. Runt-related transcription factor play a role in development of hematopoietic stem cells and blood cells. It has been shown to elevate in tuberculosis patients and plays a role in phagosome maturation [46]. Ikaros, a zinc finger transcription factor, IL-10 expression in CD4+ cells. It regulates the development and differentiation of CD4+ T helper cells and also regulates cytokines, cytokine receptors and cytokine signaling pathways [47, 48].
ETS1 gene was elevated upon infection by A. flavus clinical isolates. ETS1 is a transcription factor that regulates the expression of cytokine and chemokine genes [49]. Increased expression of IL6, IL8, IL23A, and IL19 is likely to be the result of altered ETS1 gene expression in infected epithelial cells. LGALS3, a gene that encodes galectin-3, a beta-galactoside binding protein, has been shown to reduce the fungal burden in case of C. neoformans [50] and A. fumigatus [51] in mice model. It is the only gene commonly altered in primary HCE cells in response to CI1698 and CI1123. Both the clinical isolates respond differently in immortalized cell line and primary HCE cells. This is in agreement with our previous report where it has been shown that the virulence of different clinical isolates of A. flavus is different in G. mellonella model [28].
PLAUR encoding the Urokinase receptor (uPAR) is involved in multiple functions [52] and produced by several cell types. Their role in wound healing, activation of neutrophils, and innate immune response imply that they are involved in the fungal infection as well. Viral infection of type II alveolar epithelial cell line A549 induces PLAU and PLAUR at early time points after infection [53]. PLAUR has been shown as one of the hub genes that strongly interact with other differentially expressed genes and reported as one of the key genes to be involved in Pneumonia caused by gram-positive bacteria [54]. The data implies that PLAUR is likely to be involved in fungal infections as well. PLAUR was found to be up-regulated in the epithelial cells infected with clinical isolate CI1698. Our results show that the PLAUR pathway of wound repair is common to lung epithelium and corneal epithelium.
Leukocyte immunoglobin-like receptors (LILRs) consists of 11 receptors with extracellular Ig-like domains, categorized into inhibitory (LILRB1-B5) and activating (LILRA1-A6 except A3) and reported to be expressed mostly in neutrophils [55]. LILRB2 suppress immune cell activity and mainly reported in viral, bacterial and parasite infection [56]. Recently it has been shown that LILRB2 is up-regulated in corneal tissues of bacterial keratitis. In this study, it was down-regulated in RCB2280 cells infected with CI1698 and primary cells infected with CI1123. CD6 and CD5 genes were highly upregulated genes in primary HCE cells stimulated with CI1698. CD5 and CD6 are the scavenger receptor, family members, and they recognize bacterial, fungal, viral, and parasite components [57]. CD5 has a protective role against fungal infection by recognizing and binding to fungal species via β-glucan [58], while CD6 recognizes and binds to bacterial LPS [59]. It will be interesting to examine the signaling event activated by this interaction in epithelial cells and their role in the antifungal response.
The comparison of immune gene expression between uninfected immortalized and un infected primary corneal epithelial cells showed many immune genes are abundantly expressed in primary corneal epithelial cells compared to immortalized cell line (data not shown). The response to A. flavus differs between the cells as there is a difference in the immune gene content in both the cells. Immortalized cells are diverged from primary cultures due to their genome instability [60]. So, the results were validated using primary cultures which are closer to the native condition.
There are other genes whose expression level is altered in fungal infection of corneal epithelium; however, their role in antifungal defense is not clear. Overall, the cumulative data from multiple sets of infection conditions presented in this report show that the genes involved in NF-kB signaling, TNF signaling, Th17 cell differentiation, B cell receptor signaling, complement, and coagulation cascade, cytokine-cytokine receptor interaction, and chemokine signaling were altered upon fungal infection of corneal epithelial cells. The results reported in this paper confirm that the epithelial cells are involved in antifungal defense actively by modulating defense pathways by cytokine production and inflammation. Further, our study also shows that the fungal strain and the host cell types show distinct mRNA expression profile changes during fungal infection.
Conclusions
This is the first study examining the immune response of primary corneal epithelial cells and epithelial cell line exposed to A. flavus conidia from ocular clinical isolates. Our targeted transcriptome data using the NanoString approach showed that the gene expression profiles of primary cultures and cell lines of corneal epithelium are not identical. We detected that the genes associated with NF-kB signaling, TNF signaling, and Th17 signaling were upregulated, and the genes related to B cell receptor signaling and chemokine signaling were downregulated in one of the cells. These results show that the corneal epithelial cells could elaborate and influence immune and inflammatory responses during fungal infection.
Availability of data and materials
The datasets generated and analyzed are included in the article and additional files. NanoString RNA transcript data were deposited in the Gene Expression Omnibus (GEO) of NCBI under accession number GSE175824 with the following link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE175824.
References
Amaike S and Keller NP. Aspergillus flavus. Annu Rev Phytopathol. 2011; 49:107–33. https://doi.org/10.1146/annurev-phyto-072910-095221.
Krishnan S, Manavathu EK and Chandrasekar PH. Aspergillus flavus: an emerging non-fumigatus Aspergillus species of significance. Mycoses. 2009; 52:206–22. https://doi.org/10.1111/j.1439-0507.2008.01642.x.
Horn BW, Gell RM, Singh R, Sorensen RB and Carbone I. Sexual reproduction in Aspergillus flavus sclerotia: Acquisition of novel alleles from soil populations and uniparental mitochondrial inheritance. PLoS One. 2016; 11: e0146169. https://doi.org/10.1371/journal.pone.0146169.
Chang PK, Scharfenstein LL, Mack B and Ehrlich KC. Deletion of the Aspergillus flavus orthologue of A. nidulans fluG reduces conidiation and promotes production of sclerotia but does not abolish aflatoxin biosynthesis. Appl Environ Microbiol. 2012; 78:7557–63. https://doi.org/10.1128/AEM.01241-12.
Hedayati MT, Pasqualotto AC, Warn PA, Bowyer P, Denning DW. Aspergillus flavus: human pathogen, allergen and mycotoxin producer. Microbiology. 2007; 153: 1677–92. https://doi.org/10.1099/mic.0.2007/007641-0.
Pasqualotto AC. Differences in pathogenicity and clinical syndromes due to Aspergillus fumigatus and Aspergillus flavus. Med Mycol. 2009; 47. https://doi.org/10.1080/13693780802247702.
Lin CC, Lalitha P, Srinivasan M, Prajna NV, McLeod SD, Acharya NR, et al. Seasonal Trends of microbial keratitis in South India. Cornea. 2012; 31:1123–27. https://doi.org/10.1097/ICO.0b013e31825694d3.
Gonzales CA, Srinivasan M, Whitcher JP, Smolin G. Incidence of corneal ulceration in Madurai District, South India. Ophthalmic Epidemiol. 1996; 3:159–66. https://doi.org/10.3109/09286589609080122.
Srinivasan M, Gonzales CA, George C, Cevallos V, Mascarenhas JM, Asokan B, et al. Epidemiology and aetiological diagnosis of corneal ulceration in Madurai, South India. Br J Ophthalmol. 1997; 81:965–71. https://doi.org/10.1136/bjo.81.11.965.
Johnson AC, Heinzel FP, Diaconu E, Sun Y, Hise AG, Golenbock D et al. Activation of Toll-Like Receptor (TLR)2, TLR4, and TLR9 in the mammalian cornea induces MyD88-dependent corneal inflammation. Invest Ophthalmol Vis Sci. 2005; 46:589–95. https://doi.org/10.1167/iovs.04-1077
Sun CQ, Lalitha P, Prajna NV, Karpagam R, Geetha M, O’Brien KS et al. Association between in vitro susceptibility to natamycin and voriconazole and clinical outcomes in fungal keratitis. Ophthalmology. 2014; 121:1495-00.e1. https://doi.org/10.1016/j.ophtha.2014.03.004.
Lalitha P, Shapiro BL, Srinivasan M, Prajna NV, Acharya NR, Fothergill AW, et al. Antimicrobial susceptibility of Fusarium, Aspergillus, and other filamentous fungi isolated from keratitis. Arch Ophthalmol. 2007; 125:789–93. doi: https://doi.org/10.1001/archopht.125.6.789.
Prajna NV, Krishnan T, Mascarenhas J, Rajaraman R, Prajna L, Srinivasan M, et al. The mycotic ulcer treatment trial: a randomized trial comparing natamycin vs voriconazole. JAMA Ophthalmol. 2013; 131: 422–29. https://doi.org/10.1001/jamaophthalmol.2013.1497.
Ibrahim-Granet O, Philippe B, Boleti H, Boisvieux-Ulrich E, Grenet D, Stern M, et al. Phagocytosis and Intracellular Fate of Aspergillus fumigatus conidia in alveolar macrophages. Infect Immun. 2003; 71:891–03. https://doi.org/10.1128/iai.71.2.891-903.2003
Wasylnka JA, Moore MM. Uptake of Aspergillus fumigatus conidia by phagocytic and nonphagocytic cells in vitro: Quantitation using strains expressing Green Fluorescent Protein. Infect Immun. 2002; 70:3156–63. https://doi.org/10.1128/iai.70.6.3156-3163.2002.
Wasylnka JA, Moore MM. Aspergillus fumigatus conidia survive and germinate in acidic organelles of A549 epithelial cells. J Cell Sci. 2003; 116:1579–87. https://doi.org/10.1242/jcs.00329.
Li C, Zhao GQ, Che CY, Li N, Xu Q, Wang Q, et al. Expression of dectin-1 during fungus infection in human corneal epithelial cells. Int J Ophthalmol. 2014; 7:34–37. doi: https://doi.org/10.3980/j.issn.2222-3959.2014.01.06
Zhao J, Wu XY, Yu FSX. Activation of Toll-like receptors 2 and 4 in Aspergillus fumigatus keratitis. Innate Immun. 2009; 15: 155–168. doi: https://doi.org/10.1177/1753425908101521.
Li C, Zhao G, Che C, Lin J, Li N, Hu L, et al. The role of LOX-1 in innate immunity to Aspergillus fumigatus in corneal epithelial cells. Invest Ophthalmol Vis Sci. 2015; 56: 3593–3603. https://doi.org/10.1167/iovs.14-15989.
Chen F, Zhang C, Jia X, Wang S, Wang J, Chen Y, et al. Transcriptome profiles of human lung epithelial cells A549 interacting with Aspergillus fumigatus by RNA-Seq. PLoS One. 2015; 10: e0135720. https://doi.org/10.1371/journal.pone.0135720.
Gomez P, Hackett TL, Moore MM, Knight DA, Tebbutt SJ. Functional genomics of human bronchial epithelial cells directly interacting with conidia of Aspergillus fumigatus. BMC Genomics. 2010;11: 358. https://doi.org/10.1186/1471-2164-11-358.
Oosthuizen JL, Gomez P, Ruan J, Hackett TL, Moore MM, Knight DA et al. Dual organism transcriptomics of airway epithelial cells interacting with conidia of Aspergillus fumigatus. PLoS One. 2011; 6: e20527. https://doi.org/10.1371/journal.pone.0020527
Toor A, Culibrk L, Singhera GK, Moon KM, Prudova A, Foster LJ, et al. Transcriptomic and proteomic host response to Aspergillus fumigatus conidia in an air-liquid interface model of human bronchial epithelium. PLoS One. 2018; 13: e0209652. https://doi.org/10.1371/journal.pone.0209652.
Kale SD, Ayubi T, Chung D, Tubau-Juni N, Leber A, Dang HX, et al. Modulation of immune signaling and metabolism highlights host and fungal transcriptional responses in mouse models of invasive pulmonary aspergillosis. Sci Rep. 2017; 7:17096. https://doi.org/10.1038/s41598-017-17000-1.
Arunachalam D, Namperumalsamy VP, Prajna L, Kuppamuthu D. Human corneal epithelial cells internalize Aspergillus flavus spores by actin mediated endocytosis. Infect Immun. 2021; 89: e00794-20. https://doi.org/10.1128/IAI.00794-20.
Araki-Sasaki K, Ohashi Y, Sasabe T, Hayashi K, Watanabe H, Tano Y, et al. An SV40-immortalized human corneal epithelial cell line and its characterization. Invest Ophthalmol Vis Sci. 1995; 36:614–21.
Arpitha P, Prajna NV, Srinivasan M, Muthukkaruppan V. A subset of human limbal epithelial cells with greater nucleus-to-cytoplasm ratio expressing high levels of p63 possesses slow-cycling property. Cornea. 2008; 27:1164–70. https://doi.org/10.1097/ICO.0b013e3181814ce6.
Selvam RM, Nithya R, Devi PN, Shree RSB, Nila MV, Demonte NL, et al. Exoproteome of Aspergillus flavus corneal isolates and saprophytes: Identification of proteoforms of an oversecreted alkaline protease. J Proteomics. 2015; 115:23–35. https://doi.org/10.1016/j.jprot.2014.11.017.
Seidel C, Moreno-Velásquez SD, Ben-Ghazzi N, Gago S, Read ND, Bowyer P. Phagolysosomal survival enables non-lytic hyphal escape and ramification through lung epithelium during Aspergillus fumigatus infection. Front. Microbiol. 2020; 11:1955. doi: https://doi.org/10.3389/fmicb.2020.01955.
Ben-Ghazzi N, Moreno-Velásquez S, Seidel C, Thomson D, Denning DW, Read ND, et al. Characterization of Aspergillus fumigatus endocytic trafficking within airway epithelial cells using high-resolution automated quantitative confocal microscopy. J Fungi (Basel). 2021; 7:454. doi: https://doi.org/10.3390/jof7060454.
Aimanianda V, Bayry J, Bozza S, Kniemeyer O, Perruccio K, Elluru SR, et al. Surface hydrophobins prevents immune recognition of airborne fungal spores. Nature. 2009; 460:1117–21. https://doi.org/10.1038/nature08264.
Morton CO, Varga JJ, Hornbach A, Mezger M, Sennefelder H, KneitZ S, et al. The temporal dynamics of differential gene expression in Aspergillus fumigatus interacting with human immature dendritic cells in vitro. PLoS One. 2011; 6: e16016. https://doi.org/10.1371/journal.pone.0016016.
You J, Corley SM, Wen L, Hodge C, HÓ§llhumer R, Madigan MC, et al. RNA-Seq analysis and comparison of corneal epithelium in keratoconus and myopia patients. Sci Rep. 2018;8:389. https://doi.org/10.1038/s41598-017-18480-x.
Kaplan N, Wang J, Wray B, Patel P, Yang W, Peng H, et al. Single-cell RNA transcriptome helps define the limbal/corneal epithelial stem/early transit amplifying cells and how autophagy affects this population. Invest Ophthalmol Vis Sci. 2019; 60:3570–83. https://doi.org/10.1167/iovs.19-27656.
Escobar N, Valdes ID, Keizer EM, Ordonez SR, Ohm RA, Wosten HAB, et al. Expression profile analysis reveals that Aspergillus fumigatus but not Aspergillus niger makes type II epithelial lung cells less immunological alert. BMC Genomics. 2018; 19:534. https://doi.org/10.1186/s12864-018-4895-3.
Balloy V, Sallenave JM, Wu Y, Touqui L, Latge JP, Si-Tahar M, et al. Chignard M. Aspergillus fumigatus-induced interleukin-8 synthesis by respiratory epithelial cells is controlled by the phosphatidylinositol 3-kinase, p38 MAPK, and ERK1/2 pathways and not by the toll-like receptor-MyD88 pathway. J Biol Chem. 2008; 283:30513–21. https://doi.org/10.1074/jbc.M803149200.
Peng XD, Zhao GQ, Lin J, Jiang N, Xu Q, Zhu CC, et al. Fungus induces the release of IL-8 in human corneal epithelial cells, via Dectin-1-mediated protein kinase C pathways. Int J Ophthalmol. 2015; 8:441–47. https://doi.org/10.3980/j.issn.2222-3959.2015.03.02.
Rast TJ, Kullas AL, Southern PJ, Davis DA. Human epithelial cells discriminate between commensal and pathogenic interactions with Candida albicans. PLoS One. 2016;11: e0153165. https://doi.org/10.1371/journal.pone.0153165.
Bellanger AP, Millon L, Khoufache K, Rivollet D, Bieche I, Laurendeau I, et al. Aspergillus fumigatus germ tube growth and not conidia ingestion induces expression of inflammatory mediator genes in the human lung epithelial cell line A549. J Med Microbiol. 2009; 58:174–79. https://doi.org/10.1099/jmm.0.005488-0.
Sharon H, Amar D, Levdansky E, Mircus G, Shadkchan Y, Shamir R et al. PrtT-regulated proteins secreted by Aspergillus fumigatus activate MAPK signaling in exposed A549 lung cells leading to necrotic cell death. PLoS One. 2011; 6: e17509. https://doi.org/10.1371/journal.pone.0017509.
Morton CO, Fliesser M, Dittrich M, Mueller T, Bauer R, Kneitz S, et al. Gene expression profiles of human dendritic cells interacting with Aspergillus fumigatus in a bilayer model of the alveolar epithelium/endothelium interface. PLoS One. 2014; 9: e98279. https://doi.org/10.1371/journal.pone.0098279.
Kasahara S, Jhingran A, Dhingra S, Salem A, Cramer RA and Hohl TM. Role of granulocyte-macrophage colony-stimulating factor signaling in regulating neutrophil antifungal activity and the oxidative burst during respiratory fungal challenge. J Infect Dis. 2016: 213:1289–98. https://doi.org/10.1093/infdis/jiw054.
Greco D, Vellonen KS, Turner HC, Häkli M, Tervo T, Auvinen P, et al. Gene expression analysis in SV-40 immortalized human corneal epithelial cells cultured with an air-liquid interface. Mol Vis. 2010; 16:2109–20.
Hackett EE, Charles-Messance H, O’Leary SM, Gleeson LE, Muñoz-Wolf N, Case S, et al. Mycobacterium tuberculosis limits host glycolysis and IL-1β by restriction of PFK-M via microRNA-21. Cell Rep. 2020; 30:124–136. doi: https://doi.org/10.1016/j.celrep.2019.12.015.
Yannariello-brown J, Hallberg CK, Häberle H, Brysk MM, Jiang Z, Patel JA, et al. Cytokine modulation of human corneal epithelial cell ICAM-1 (CD54) expression. Exp Eye Res. 1998; 67:383–93. doi: https://doi.org/10.1006/exer.1998.0514.
Koh HJ, Kim YR, Kim JS, Yun JS, Kim S, Young S, et al. CD82 hypomethylation is essential for tuberculosis pathogenesis via regulation of RUNX1-Rab5/22. Exp Mol Med. 2018; 50:1–15. doi: https://doi.org/10.1038/s12276-018-0091-4.
Powell MD, Read KA, Sreekumar BK, Oestreich KJ. Ikaros zinc finger transcription factors: regulators of cytokine signaling pathways and CD4+ T helper cell differentiation. Front. Immunol. 2019; 10:1299. doi: https://doi.org/10.3389/fimmu.2019.01299.
Ridley ML, Fleskens V, Roberts CA, Lalnunhlimi S, Alnesf A, O’Byrne AM, et al. IKZF3/Aiolos is associated with but not sufficient for the expression of IL-10 by CD4+ T cells. J Immunol.2020; 204:2940–2048. doi: https://doi.org/10.4049/jimmunol.1901283.
Nagarajan P, Chin SS, Wang D, Liu S, Sinha S, Garrett-Sinha LA. Ets1 blocks terminal differentiation of keratinocytes and induces expression of matrix metalloproteases and innate immune mediators. J Cell Sci. 2010; 123:3566–75. https://doi.org/10.1242/jcs.062240.
Almeida F, Wolf JM, da Silva TA, DeLeon-Rodriguez CM, Rezende CP, Pessoni AM, et al. Galectin-1 impacts Cryptococcus neoformans infection through direct antifungal effects. Nat Commun. 2017; 8:1968. doi: https://doi.org/10.1038/s41467-017-02126-7.
Snarr BD, St-Pierre G, Ralph B, Lehoux M, Sato Y, Rancourt A, et al. Galectin-3 enhances neutrophil motility and extravasation into the airways during Aspergillus fumigatus infection. PLoS Pathog. 2020; 16: e1008741. https://doi.org/10.1371/journal.ppat.1008741
Mahmood N, Mihalcioiu C, Rabbani SA. Multifaceted role of the urokinase-type plasminogen activator (uPA) and its receptor (uPAR): diagnostic, prognostic, and therapeutic applications. Front Oncol. 2018; 8:24. https://doi.org/10.3389/fonc.2018.00024
Martinez I, Lombardía L, García-Barreno B, Domínguez O, Melero JA. Distinct gene subsets are induced at different time points after human respiratory syncytial virus infection of A549 cells. J Gen Virol. 2007; 88: 570–581. https://doi.org/10.1099/vir.0.82187-0.
Jia R, Yang J, Cui Y, Guo D, Li T. Gene expression analysis for pneumonia caused by gram-positive bacterial infection. Exp Ther Med. 2018; 15:3989–3996. https://doi.org/10.3892/etm.2018.5904.
Lewis Marffy AL, McCarthy AJ. Leukocyte Immunoglobulin-Like Receptors (LILRs) on Human Neutrophils: Modulators of Infection and Immunity. Front. Immunol. 2020; 11:857. doi: https://doi.org/10.3389/fimmu.2020.00857
Abdallah F, Coindre S, Gardet M, Meurisse F, Naji A, Suganuma N, et al. Leukocyte immunoglobulin-like receptors in regulating the immune response in infectious diseases: A window of opportunity to pathogen persistence and a sound target in therapeutics. Front. Immunol. 2021; 12:717998. doi: https://doi.org/10.3389/fimmu.2021.717998
Andrés MV, Casadó-Llombart S, Català C, Leyton-Pereira A, Lozano F, Aranda F. Soluble CD5 and CD6: Lymphocytic class I scavenger receptors as immunotherapeutic agents. Cells. 202; 9:2589. https://doi.org/10.3390/cells9122589.
Vera J, Fenutrίa R, Canadas O, Figueras M, Mota R, Sarrias MR, et al. The CD5 ectodomain interacts with conserved fungal cell wall components and protects from zymosan-induced septic shock-like syndrome. Proc Natl Acad Sci U S A. 2009; 106:1506–11. https://doi.org/10.1073/pnas.0805846106.
Sarrias MR, Farnόs M, Mota R, Sanchez-Barbero F, Ibanez A, Gimferrer I, et al. CD6 binds to pathogen-associated molecular patterns and protects from LPS-induced septic shock. Proc Natl Acad Sci U S A. 2007; 104: 11724–11729. https://doi.org/10.1073/pnas.0702815104.
Furuya K, Wu T, Orimoto A, Sugano E, Tomita H, Kiyono T, et al. The transcriptome of wild-type and immortalized corneal epithelial cells. Sci Data. 8:126. doi: https://doi.org/10.1038/s41597-021-00908-9.
Acknowledgements
We thank Dr. O. G Ramprasad and Dr. C. Gowri Priya for their help in the confocal microscopy experiments and Dr. J. Jeya Maheshwari for her critical comments and suggestions. We acknowledge Theracues Innovations Private Limited, Bangalore, for the NanoString analysis. Mr. D. Saravanan, Manager, Rotary Aravind International Eye Bank, Madurai, India, for sample collection.
Funding
This work was supported by the Department of Biotechnology, Government of India, New Delhi- No BT/01/CEIB/11/VI/09. Sun pharma grant for the financial support. Indian Council of Medical Research provided fellowship to Ms. A. Divya.
Author information
Authors and Affiliations
Contributions
Conception and design: DK. Design of experimental work: DA, DK. Cell culture and fungal co-culture: DA. Nano String analysis, data analysis: LM, AM, GR. ELISA- SMR, DA, DK. Interpretation of data and formal analysis: DA, DK. Writing: DK, DA. Critical revision: VPN, LP, GR, DK, DA. Final approval of manuscript: DK, DA, SMR, LM, AM, GR, VPN, LP, all authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Participants provided their written informed consent for the donor eyes. The study was approved by Institutional Review Board of the Aravind Medical Research Foundation, Madurai (ID NO. IRB2011001BAS). The study was performed in accordance with the Declaration of Helsinki. We confirm that all methods were performed in accordance with the relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
Authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
RNA transcript data for 595 genes profiled using NanoString’s Immunological Panel for RCB2280 cells infected with clinical isolates of A. flavus.
Additional file 2: Table S2.
RNA transcript data for 595 genes profiled using NanoString’s Immunological Panel for primary HCE cells infected with clinical isolates of A. flavus.
Additional file 3: Table S3.
Differentially expressed genes of RCB2280 cells and primary HCE cells infected with A. flavus CI1698 and CI1123.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Arunachalam, D., Ramanathan, S.M., Menon, A. et al. Expression of immune response genes in human corneal epithelial cells interacting with Aspergillus flavus conidia. BMC Genomics 23, 5 (2022). https://doi.org/10.1186/s12864-021-08218-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-021-08218-5