Abstract
Background
Liver fibrosis is a wound-healing response to tissue injury and inflammation hallmarked by the extracellular matrix (ECM) protein deposition in the liver parenchyma and tissue remodelling. Different cell types of the liver are known to play distinct roles in liver injury response. Hepatocytes and liver endothelial cells receive molecular signals indicating tissue injury and activate hepatic stellate cells which produce ECM proteins upon their activation. Despite the growing knowledge on the molecular mechanism underlying hepatic fibrosis in general, the cell-type-specific gene regulatory network associated with the initial response to hepatotoxic injury is still poorly characterized.
Results
In this study, we used thioacetamide (TAA) to induce hepatic injury in adult zebrafish. We isolated three major liver cell types - hepatocytes, endothelial cells and hepatic stellate cells - and identified cell-type-specific chromatin accessibility and transcriptional changes in an early stage of liver injury. We found that TAA induced transcriptional shifts in all three cell types hallmarked by significant alterations in the expression of genes related to fatty acid and carbohydrate metabolism, as well as immune response-associated and vascular-specific genes. Interestingly, liver endothelial cells exhibit the most pronounced response to liver injury at the transcriptome and chromatin level, hallmarked by the loss of their angiogenic phenotype.
Conclusion
Our results uncovered cell-type-specific transcriptome and epigenome responses to early stage liver injury, which provide valuable insights into understanding the molecular mechanism implicated in the early response of the liver to pro-fibrotic signals.
Similar content being viewed by others
Background
Liver injury is a rising public health concern, especially in European and North American countries. Its increasing prevalence leads to an expanding body of work regarding the molecular mechanisms present in advanced liver disease, however our knowledge about the earliest stages of liver injury is still limited. Liver injury is manifested by the formation of fibrous tissue as a result of ECM deposition at the site of injury [1]. Progressive fibrous scar formation may distort normal liver structure by formation of septa and nodules of regenerating hepatocytes (HEPs) leading to impaired portal blood flow and formation of cirrhotic architecture [2]. Liver cirrhosis is the end-stage of hepatic fibrosis affecting about 0.1% of the European population [1]. The most serious outcome of cirrhosis is hepatocellular carcinoma (HCC), constituting 70-90% of cases of primary liver cancer [1]. The predominant causes of liver fibrosis are chronic excessive alcohol consumption, viral hepatitis B and C and non-alcoholic fatty liver disease (NAFLD), the latter becoming a major concern with the increasing incidence of obesity in Europe and the USA [1].
Liver parenchymal cells, HEPs, are the most abundant cell subpopulation in this organ in mammals, constituting ca. 85% of the total liver cell mass [3]. Under physiological conditions, HEPs are responsible for a wide range of functions, including carbohydrate, fatty acid and protein metabolism as well as immune response [3]. Upon liver damage, HEPs are a source of reactive oxygen species, pro-inflammatory signals as well as cytokines, taking part in the activation of repair pathways [3].
Hepatic stellate cells (HSCs) comprise 8% of the total liver cell population [4]. Under normal physiological conditions, these mesenchymal cells reside in the space of Disse, maintaining a quiescent state, storing vitamin A in cytoplasmic lipid droplets [5]. Upon liver damage, HSCs are activated and transdifferentiate into myofibroblast-like cells. Their activation is triggered by multiple autocrine and paracrine signals, such as transforming growth factor (TGFβ), SMAD3, protein platelet-derived growth factor receptor (PDGF), vascular endothelial growth factor (VEGF) and connective tissue growth factor (CTGF) [6]. In an active state, HSCs are the primary ECM-producing cell population, resulting in the creation of a temporary scar tissue at the damaged site. Active HSCs produce cytokines and growth factors, promoting liver regeneration. In chronic liver disease, however, the reoccurring HSC activation may result in permanent scar formation, resulting in sections of non-functional liver tissue [5].
Endothelial cells in the liver are found mainly lining the inner walls of the sinusoidal blood vessels (liver sinusoidal endothelial cells - LSECs). LSECs are highly specialized, forming a permeable barrier by virtue of their fenestrae, between hepatocyte membranes and blood vessel lumen. The presence of fenestrae, combined with the absence of a basement membrane, contribute to making the LSECs the most endocytosis-capable cell population in the human body [7]. LSECs regulate the tone of hepatic blood vessels and maintain the quiescent state of HSCs [7].
In response to chronic hepatotoxic injury, various molecular and cellular factors interact with HEPs and LSECs, leading to sequential activation of HSCs [8]. This in turn initiates the perpetuation phase, hallmarked by proliferative, contractile and inflammatory phenotype characterized by increased production of ECM proteins including collagens, fibronectin, decorin, elastin and proteoglycans [2, 9]. The understanding of molecular mechanisms of hepatic fibrosis has markedly increased due to the availability of liver fibrosis models such as cell culture systems, rodent model systems and biopsied human material [10]. However, our knowledge of cell-type-specific gene regulatory networks and epigenetic hallmarks associated with the initial response to hepatotoxic injury is still lacking, mainly due to the challenges of studying cell interactions and their behaviour in a living organism. Such knowledge is crucial for accurate diagnosis and development of new therapeutic approaches targeting liver fibrosis and related disorders.
The zebrafish (Danio rerio) has emerged as a useful model organism for studying the mechanism of liver disease in vivo, both in larvae and adult individuals [11,12,13]. Despite the distinct architecture between mammalian and zebrafish liver, they contain similar main cell types, including HEPs, endothelial cells (ECs) and HSCs, with conserved function and gene expression profiles [5, 14, 15]. To dissect the molecular mechanisms regulating the initiation of hepatic fibrosis and understand the interplay between genetic and epigenetic signals in this process, we utilized the model of thioacetamide-induced liver injury in adult zebrafish and characterized cell-type-specific changes at both transcriptome and epigenome level in three main liver cell types. Thioacetamide (TAA) is a potent hepatotoxin that has been widely used to induce acute and chronic liver injury in rodent models [16,17,18]. There is a wide variation in the administration routes and time of exposure between studies, but most commonly a regimen of intraperitoneal injections of 100-200 mg/kg of body mass 2-3 times per week for over 6 weeks has been used to induce liver fibrosis and cirrhosis [19]. TAA has also been utilized to induce liver injury in zebrafish larvae, establishing it as a model for steatohepatitis [13]. The larvae used in the cited study were exposed to 0.025% TAA for 10 days starting at 72 h post-fertilization (hpf), when the embryonic liver becomes functional. At 5 days post-fertilization the embryos exhibited molecular markers of apoptosis and steatohepatitis, which continued until the end of the treatment. TAA has also been used in juvenile zebrafish, where intraperitoneal injections of 300 mg/kg b.m. three times a week induced steatosis [20].
We employed three transgenic zebrafish lines to isolate the respective cell populations: HEPs (Tg(fabp10a:dsRed)), HSCs (Tg(hand2:EGFP)), and ECs (Tg(kdrl:ras-mCherry)). We implemented a machine learning technique known as self-organizing maps (SOMs) to generate whole genome expression profiles of both physiological state and early response to liver injury from the three studied cell types [21]. The integration of this data with genome-wide open chromatin maps (ATAC-seq) from corresponding samples allowed to uncover specific gene and chromatin signatures of the studied cell populations. Our analysis revealed that early response of the liver to pro-fibrotic signals is manifested in cell-type specific transcriptome and epigenome rearrangements and identified molecular hallmarks of this process. This work provides a step towards understanding the initial stages of liver injury and may serve as a resource for further investigation aimed at developing new diagnostic and treatment tools.
Results
Identification of liver cell-type-specific transcriptional portraits under normal physiological condition
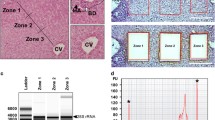
In order to characterize the molecular profiles representing the HEPs, HSCs, and ECs under physiological conditions, we utilized three transgenic lines Tg(fabp10a:dsRed), Tg(hand2:EGFP) and Tg(kdrl:Hsa.HRAS-mCherry) which express red (dsRed, mCherry) or green fluorescent proteins (GFP) in the corresponding cell types [14, 22, 23]. Whole livers were dissected from adult zebrafish from each of the transgenic lines used in this study (Fig. 1A). Fluorescent microscopy of liver from the corresponding transgenic lines confirmed the fluorescence observed in the corresponding cell types (Fig. 1B). We prepared cell suspensions and performed FACS according to previously established protocols (See Methods, Supp. Fig. 1). The number of RNA-seq reads corresponding to fluorescent reporters specific to each cell-type (Fig. 1B) was strongly enriched in fluorescent-positive samples, which confirmed the purity of FACS isolated samples (Fig. 1C). In order to ascertain the cell-type gene signatures, we performed differential expression comparisons between samples and identified the most enriched genes in each cell type (Fig. 2A, Supp. Table 2). The largest number of cell-specific genes were found in ECs (4553), then in HSCs (380) and in HEPs (126) (Supp. Table 2). These included known cell-specific markers for ECs (sox18 [24], sele [25], flt1 [26]) and HEPs (soat2 [27]) (Fig. 2B). On the other hand, genes related to fatty acid metabolism (fasn [28], fat3b, hmgcra [29], hmgcs1 [30], elovl4a [31]) and cholesterol biosynthesis (cyp51, sc5d, hmgcra, msmo1, nsdhl, hmgcs1, dhcr7) were upregulated in HSCs which are known to contain vitamin A lipid droplets [32] (Supplementary Table 2). Gene ontology (GO) analysis revealed the enrichment of genes related to angiogenesis in ECs, insulin-like growth factor receptor signalling genes and cellular phosphate ion homeostasis in HEPs and lipid transport and metabolism genes in HSCs (Fig. 2C). Taken together, the enrichment of known markers and the relevant GO terms in ECs, HEPs, and HSCs support the identity of the respective cell types.
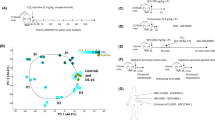
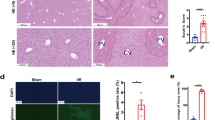
Transcriptional portraits of liver cells in response to TAA. a A scheme of the study. Adult transgenic zebrafish lines were treated with TAA (500 mg/kg) or control (saline) three times per week for 2 weeks. Livers were removed and fluorescent-positive cells were sorted by FACS. RNA-seq and ATAC-seq libraries were performed from sorted cells; b Transgenic zebrafish liver cryosection micrographs visualizing ECs (Tg(kdrl:Hsa.HRAS-mCherry)), HSCs (TgBAC(hand2:EGFP)) and HEPs (Tg(fabp10a:dsRed)) as indicated on the figure legends; c Number of transgene BLAST hits from fluorescent-negative and positive cells from transgenic zebrafish lines; d Microscopic images of histological H&E sections of control and TAA-treated animals indicating inflammation loci (arrowheads) and extracellular lipid droplets (asterisks); e Portraits of co-regulated over- or underexpressed metagenes as red and blue spots, respectively. The color gradient of the map visualizes over- and underexpression of the metagenes compared with the mean expression level in the pool of all samples studied; f Sample pairwise Pearson correlation heatmap on the clustered data; g Independent Component Analysis on clustered data
Liver cell signatures in quiescent and activated state. a Number of identified cell type specific genes at quiescent state in each cell type, logFC > 0, padj < 0.05; b Heatmaps of top 25 cell type specific genes at quiescent state in each cell type, logFC > 0, padj < 0.05; c GO over-representation analysis of identified cell type specific genes at quiescent state in each cell type; d Volcano plot of selected genes, involved in liver fibrosis and response to oxidative stress, under TAA treatment
TAA metabolism is reflected in the transcriptional shift in liver cells
We then sought to determine the transcriptional signatures of early hepatotoxic injury response in each of the three liver cell types. We induced liver injury using TAA at a concentration of 500 mg/kg of body mass. The short term TAA treatment induced mild histological changes with observed inflammation (Fig. 1D). We then collected whole livers from TAA-treated Tg(fabp10a:dsRed), TgBAC(hand2:EGFP) and Tg(kdrl:Hsa.HRAS-mCherry) fishes, isolated the corresponding cell types by FACS, and performed RNA-seq.
We evaluated cell-type-specific transcriptional response to TAA activation by looking at the expression of genes related to TAA metabolism and genes activated in response to liver injury and fibrogenesis (Fig. 2D, Supp. Table 3). The increased expression of genes related to cell redox homeostasis such as catalase (cat) [33], cytochromes (cyp2y3, cyp2p6) [34], superoxide dismutase 2 (sod2) [34], glutathione peroxidase 1a (gpx1a) [35] was observed in response to TAA, with the most striking response in ECs. Pro-fibrotic genes [8] including ECM proteins such as collagens (col1a1a, col1a2, col5a2a, col5a1, col6a3), decorin (dcn) as well as metallopeptidase inhibitor 2a (timp2a), integrin alpha V (itgav) and annexin 5b (anxa5b) were specifically upregulated in HSCs, in response to TAA (Fig. 2D).
TAA induces transcriptional reprogramming of hepatic endothelial cells
To provide a global view of the behaviour of correlated gene clusters in three hepatic cell types in response to TAA, we used self-organizing map based tool oposSOM R package [36]. The tool first constructed transcriptional portraits of all the samples, then a second unsupervised reduction step was performed, further reducing dimensionality to overexpression spots representing clusters (A-H, Supp. Table 4) of co-expressed metagenes which are highly expressed in, at minimum, one condition (Fig. 3A, B) [37]. To link overexpression with gene set overrepresentation in a sample- and spot-specific way, we visualized the metagene expression across samples on the heatmap (Fig. 3C) and performed the gene set overrepresentation analysis (Fig. 3D, E; Supp. Table 5). The gene expression portraits of both control and TAA-treated samples from each of the three cell types revealed that short-term TAA exposure induced strong changes in genome-wide expression landscapes between cell types in physiological state and upon TAA activation (Fig. 1E, F). Interestingly, the most striking changes induced by TAA treatment were observed in ECs (Fig. 1G).
Functional characterization of overexpression spots landscape. a Overexpression spots landscape. Logged expression values of each gene were transformed into differential expression values relative to the mean expression of the particular gene in the experimental series of samples considered. Overexpression spots are coloured in red; b Overexpression spots annotation to clusters from A to H; c Mean overexpression spots expression across samples; d Gene sets enrichment analysis on the clustered data. Overrepresentation p-values for each cluster are provided; e Meta-analysis of gene set enrichment performed by Metascape. Only significantly enriched terms are shown (padj < 0.05)
Analysis of the SOM clusters in ECs revealed an increase in expression of genes related to metabolic and redox processes as well as cellular transport (Fig. 3C, D - clusters B and F). We also observed downregulation of genes related to vasculature development as well as activation of immune response in ECs after treatment with TAA (Fig. 3C, D - clusters G and H; Supp. Fig. 6).
In HEPs, TAA treatment induced an increase in the expression of gene sets associated with regulation of metabolic processes, namely carboxylic acid and hydroxy compound metabolism, as well as intra- and intercellular transport when compared to their control counterparts (Fig. 3C, D - cluster B). In contrast, we observed a decreased expression of gene sets associated with the formation and function of endoplasmic reticulum as well as negative regulation of various growth binding factors (Fig. 3C, D - clusters E and G). We also observed a relative reduction of expression of genes associated with the G2/M cell cycle transition in TAA-treated HEPs (Fig. 3C, D - cluster D; Supp. Fig. 5).
Modest changes in gene expression were observed in HSCs. Analysis of clusters revealed that upregulated gene sets were associated with extracellular space and structure organization as well as protein hydrolysis (Fig. 3C, D - cluster A), which reflects the known role of HSCs in ECM formation during liver damage response [9]. Conversely, we observed downregulation of genes associated with G2/M cell cycle transition, endoplasmic reticulum, estrogen response and immune activation (Fig. 3C, D - clusters G and H).
Altogether, cell-type-specific transcriptome profile revealed transcriptional response to short term TAA exposure. All of the analyzed cell types were subject to TAA-induced transcriptional shifts, with the highest change observed in ECs. These were hallmarked by decrease of vascular-specific genes and the increase of fatty acid and carbohydrate metabolism genes as well as in immune response-associated genes.
TAA leads to genome-wide changes in chromatin regions enriched in binding sites for transcription factors regulating fatty acid metabolism and angiogenesis
Epigenetics has been acknowledged as an important player in liver fibrosis and regeneration [38,39,40], with a prospect of the development of epigenetic biomarkers and therapies. To investigate this aspect of liver damage, we ask whether epigenetic changes are involved in the earliest stages of liver fibrosis. To determine whether and to what extent epigenetic landscape in each liver cell type is altered during early stage liver injury, we characterized the changes in chromatin accessibility in HEPs, HSCs, and ECs upon TAA treatment.
We observed that in TAA-treated animals the most significant changes in chromatin state compared to control were observed in ECs, followed by HSCs and HEPs (Fig. 4A, B). ATAC-seq peaks distribution across the genome showed that the highest fraction of peaks (30-40%) was localized in the promoter (+/− 3 kb) regions (Fig. 4C, Supp. Table 7). Interestingly, the most significant changes in chromatin accessibility was observed in ECs, with the largest number of upregulated peaks found within the promoters of genes in clusters B (440 peaks) and F (74 peaks) and downregulated peaks in clusters G (120 peaks) and H (113 peaks) (Fig. 5A). The observed changes in chromatin accessibility correlates with changes observed in the transcriptional levels of genes within the corresponding clusters (increase in clusters B and F, and decrease in clusters G and H) (Fig. 4D). On the other hand, modest changes in chromatin accessibility were observed in the other two cell types. In HEPs, the highest change was observed in cluster B (30 up- and 18 downregulated). In HSC, 62 and 7 peaks were upregulated or downregulated in cluster B, respectively and 39 downregulated in cluster H.
Chromatin accessibility maps of liver cells. a Principal component analysis of ATAC-seq peaks across cell types and conditions; b Sample pairwise Pearson correlation heatmap of chromatin accessibility in ATAC-seq peaks across cell types and conditions; c ATAC-seq peak distribution across genomic categories; d Coverage heatmaps of ATAC-seq peaks localized in the promoters (− 3 to + 3 kb from TSS) of SOM clusters
TF motif enrichment in response to pro-fibrotic stimuli. a Metrics of differential promoter peaks (− 3 to + 3 kb from TSS) in SOM clusters; b Homer motif enrichment analysis in ECs differential peaks; c Homer motif enrichment analysis in HEPs differential peaks; d Homer motif enrichment analysis in HSCs differential peaks. Only enriched motifs with p-adjusted < 0.1 are shown
To identify potential regulators involved in TAA response in each cell type, we searched for transcription factor (TF) motifs enriched in differentially accessible promoter peaks from SOM cluster genes (Fig. 5B-D, Supp. Table 6). Significant enrichments (p-adjusted < 0.05, Supp. Table 6) were identified predominantly in five tested groups of regions: cluster B upregulated regions in ECs and HSCs, cluster G downregulated regions in ECs and cluster H downregulated regions in ECs and HSCs. In ECs, we observed significant enrichment in motifs of fatty acid metabolism nuclear receptors such as RXR [41], THRB [42], HNF4A [43] and PPARA [41] among peaks upregulated in cluster B. This is in accordance with the result of gene set overrepresentation analysis (Fig. 3D). A drop in chromatin accessibility was observed for ECs peaks located in the promoter of genes from cluster G. TFs motifs identified in this cluster belong to ETS family (ETV2, ERG, SPDEF, ETS1) and Sox family (Sox6, Sox17, Sox3) involved in cell differentiation, migration and proliferation [44,45,46]. In HSCs, we found enriched motifs of TFs involved in cellular glucose homeostasis such as FOXA3 [47], FOXK1 [48], FOXK2 [49] and cell differentiation such as RARA, TR4, FOXA1, FOXA3 [50]. In cluster H downregulated regions, both in EC and HSC, we also found enriched motifs of ETS family including ETV2, ERG, ELF5, ELF3, ETS1, EHF, SPIB, ELF4. Additionally, in HSCs we found enrichment of ATF4 and Chop motifs, which are known to be involved in response to endoplasmic reticulum stress [51, 52]. Notably, ETS TFs also regulate endothelial function and homeostasis [53]. Altogether, our results show increased chromatin accessibility in the promoter regions of gene clusters associated with fatty acid metabolism, especially in ECs, and decrease of accessibility in clusters related to endothelial homeostasis and inflammatory response.
ECs exhibit the highest gene regulatory response to TAA-induced liver injury
To further investigate cell type specific responses to TAA treatment we examined the character of promoter accessibility change in clusters most specific to each cell type. These included clusters B, G and H in ECs and cluster A in HSCs. In cluster B we observe the tendency in ECs towards increase in promoter accessibility upon treatment (Fig. 4D and Supp. Fig. 3B) combined with increase in expression (Fig. 3C). Among the genes that increase in accessibility, we focused on those that exemplify the largest gain in accessibility by selecting the top 25th percentile of change in accessibility and lower 25th percentile of read counts in the control sample (Fig. 6A). Among those were homologs of known human liver fibrosis markers such as Apolipoprotein A-IV [54] or Fibulin-5 [55] (Fig. 6C, D). In clusters G and H we observe a decrease in promoter accessibility (Fig. 4D, Supp. Fig. 3C, D) accompanied by reduced gene expression (Fig. 3C). To select genes with the most prominent loss of accessible regions in their promoter after treatment, we examined differentially accessible regions in the lower 25th percentile in terms of accessibility change and upper 25th percentile in read counts in the control sample (Fig. 6B, Supp. Fig. 4A). Among such genes in cluster G were EC marker kdrl and known vascular endothelial regulator etv2 [56] (Fig. 6E, Supp. Fig. 4C). In contrast, a limited number of changes were observed in promoter accessibility of HSCs in cluster A (Fig. 4D and Supp. Fig. 3A). Among the 5 genes within the top 25th percentile of accessibility changes and lower 25th percentile of read counts in control were col4a6 and elovl1a (Supp. Fig. 4B, D, E).
Cell type specific accessibility changes in response to TAA treatment in selected cell types and clusters. a Heatmap of selected genes in each cell type. Genes were selected based on accessibility patterns in cluster B; b Heatmap of selected genes in each cell type. Genes were selected based on accessibility patterns in cluster B; c Genomic browser snapshot at apoa4b.1 promoter localization with accessibility track expressed as reads per million. Highlighted peak was used as a selection criteria in a., its three most enriched motifs are shown next to the browser track; d Genomic browser snapshot at fbln5 promoter localization with accessibility track expressed as reads per million. Highlighted peak was used as a selection criteria in a., its three most enriched motifs are shown next to the browser track; e Genomic browser snapshot at kdrl promoter localization with accessibility track expressed as reads per million. Highlighted peak was used as a selection criteria in b., its three most enriched motifs are shown next to the browser track
Discussion
Liver fibrosis is a wound-healing response to tissue injury and inflammation hallmarked by the ECM protein deposition in the liver parenchyma and tissue remodelling [57]. The predominant causes of liver fibrosis are chronic excessive alcohol consumption, viral hepatitis B and C and non-alcoholic fatty liver disease (NAFLD), the latter becoming a major concern with the increasing incidence of obesity in Europe and the USA [1]. While these conditions have been widely studied [1], current knowledge of gene regulatory networks and epigenetic hallmarks associated with the early response to hepatotoxic injury is still lacking. It is crucial to study these primary changes in the cell types most affected by injury to improve the tools for diagnosis of early liver fibrosis and related disorders. In order to dissect the molecular mechanisms regulating the initiation of hepatic fibrosis and understand the interplay between genetic and epigenetic signals in this process, we utilized the model of TAA-induced liver injury in adult zebrafish and characterized cell-type-specific changes at both transcriptome and epigenome level in three main liver cell types: HEPs, HSCs and ECs.
The conservation of many metabolic pathways across vertebrate species renders the zebrafish a potent model organism in drug discovery studies. It has been extensively used to study liver development and injury [58, 59], and has been especially useful in establishing various toxicity models [60]. Many xenobiotics used to establish murine models of drug-induced liver injury have been found to be as effective in zebrafish, with an added advantage of the larvae being suitable for toxicological studies at 3 days post-fertilization, when mature liver parenchyma can be observed [60]. While the zebrafish liver architecture is distinct from its mammalian counterpart, the morphology, localization and gene expression profiles of HEPs, ECs and HSCs are similar [58, 60, 61].
The hepatotoxic properties of TAA in mice and rats induces oxidative stress resulting first in formation of intracellular lipid deposits in the liver parenchymal cells (hepatocyte ballooning), and later leading to HEPs damage and necrosis [62]. Bioactivation of TAA into its hepatotoxic counterpart, TASO2 [63], requires proteins from the cytochrome p450 complex, functional orthologs for many of which exist in zebrafish, including proteins with > 44.87% sequence similarity to CYP2E1, the protein thought to be directly responsible for TAA metabolism in humans [64]. Moreover, CYP2E1 function was reproduced in zebrafish tissue homogenates, albeit without identifying the specific enzyme responsible for the process [65].
In line with previous reports [5, 66], we observed that gene expression profiles of respective cell populations are similar to those exhibited by their mammalian counterparts. Specifically, our sorted cell populations were enriched for known cell specific markers and relevant GO terms. These results are in agreement with the established existence of conserved molecular pathways between species [58]. Moreover, our analysis of cell-type-specific transcriptional response to TAA treatment highlighted known molecular components of the TAA metabolism pathway such as elements of the cytochrome p450 superfamily (Supp. Table 3). The most striking transcriptional response to TAA was observed in the ECs, highlighting those cells as the most affected by the treatment. This is likely a consequence of high permeability of ECs and also reflects their driving role in hepatotoxic injury response [67]. ECs, particularly LSEC, due to their exceptional permeability and intimate contact with the blood stream [68], are at the frontline of the toxic stimuli sensing. During liver damage, endothelial dysfunction occurs at early phases, before fibrosis initiation [69,70,71], under many liver etiologies such as non-alcoholic fatty liver disease (NAFLD) and alcoholic liver damage. Some evidence shows that LSEC dysfunction occurs before other liver injury early markers including Kupffer cell activation, nitric oxide content reduction or TNFα, IL-6 and ICAM-1 up-regulation [67, 70, 72]. To accompany their high toxins susceptibility ECs play a regulatory role in the liver cellular response to an injuring factor [67]. The main target of this regulation are the hepatic stellate cells (HSC), but evidence was shown on ECs involvement in control of HEPs proliferation [73]. In chronic models of liver injury, ECs, specifically LSEC, can generate a strong immune response and became highly proinflammatory, while secreting a vast range of cytokines and chemokines including TNF-α, IL-6, IL-1, CCL2 [67]. In response to those stimuli as well as the damaging toxin, other cells co-participate in the liver cellular response regulation. Injured hepatocytes and inflammatory cells secrete inflammatory mediators, which further stimulate LSEC and the inflammatory response.
To assess TAA-induced transcriptional changes in more detail, we applied SOM to identify clusters of co-expressed genes in our transcriptome data. We found eight clusters that showed greatest variability between conditions. The largest of these, cluster B, showed highest upregulation in response to TAA treatment in ECs. Interestingly, this cluster consists of genes related to metabolic and redox processes, including 20 members of the cytochrome p450 superfamily. This suggests that cluster B represents the set of genes most directly responding to TAA treatment. The expression of CYP2E1 in LSECs was recently reported in the case of alcohol induced liver injury in mice [74]. Moreover, in agreement with the ability of ECs to regulate neighboring cells, eg. via angiocrine factors, we found many genes whose products are known to localize in the extracellular space in cluster B. This includes Apolipoprotein A-IV which has been recently identified as a potent liver fibrosis biomarker [54]. Conversely, clusters G and H showed strong downregulation upon TAA treatment. Of these, genes involved in extracellular structure organisation (cluster G) showed the strongest response in the ECs, while genes involved in immune response (cluster H) were commonly downregulated across all cell types. Contrary to previous reports [75, 76], we did not observe an upregulation of extracellular space-associated genes, especially matrix metalloproteinase genes (clusters A and C) in HEPs. This may be due to the differences in experimental design, as in contrast to the cited studies we investigated the earliest stages of liver injury. Other possible sources of divergent results may be the choice of hepatotoxin, as both cited studies employed CCl4. This result could also highlight the differences in model organisms of choice, as the cited studies have employed mice, rats and human cell lines.
The observed gene expression upregulation in response to treatment is accompanied by increased promoter accessibility. In agreement with RNA-seq data, we observe the largest chromatin rearrangements in ECs. This result suggests that chromatin remodeling is an important mechanism driving gene expression response to liver injury. Indeed, our motif enrichment analysis identified known motifs of transcriptional activators, such as the pioneer factors foxa1 and foxa3, to be enriched in the regions of increased accessibility. Curiously, the murine homolog of foxa3 has been implicated in promoting liver regeneration [77], while foxa1 is important for proper liver parenchyma development [78]. Changes in promoter accessibility in other cell types were less prominent, however the increase in chromatin accessibility was observed in HSCs’ col4a6 promoter region upon TAA treatment. This, taken together with the increased transcription of ECM genes in both ECs and HSCs can suggest that the initiation of ECM remodeling driven by both these cell types is triggered by hepatic injury.
Conclusions
We induced liver injury using TAA, an established potent hepatotoxin, in adult zebrafish. Using this system, we identified cell-type specific response to early hepatotoxic liver injury at the transcriptomic and regulatory level. We demonstrated that in zebrafish, the first major liver cell population exposed to hepatotoxin - ECs - is also the most affected at both transcriptomic and chromatin accessibility level at this stage of liver injury. Importantly, genes known to be key players in ECM remodelling as well as metabolic and redox processes were observed to be responsive to TAA-mediated liver injury, including some which undergo chromatin re-arrangement at their promoter regions. Besides revealing the global transcriptome and epigenome landscape of early liver injury, this work provides insight into the molecular processes involved in early stages of liver damage. It also promises the viability of employing approaches providing even more specific, in-depth information, such as single cell sequencing or long read sequencing. These could potentially allow researchers to identify subpopulations of cells within major cell types that are responsible for distinct signals and injury response patterns, or assess transcript modifications triggered by early liver injury.
Methods
TAA dose-response assessment
Treatment of adult zebrafish individuals with TAA at a concentration of 300 mg/kg b.m. which was previously reported for female zebrafish [20] did not result in morphological changes compared to saline-injected controls (Supp. Fig. 2), thus suggesting that a higher concentration of TAA is required to induce liver injury in adult fish. In order to establish the optimal TAA concentration for adult zebrafish, we first performed a range-finding experiment to identify the working dose for zebrafish embryos, which we would then use as a guideline for establishing the higher dose in adults. By performing the toxicity assay in embryos instead of adults we bypassed the need to sacrifice large numbers of animals. Embryos at 48 hpf (n = 18 for each concentration) were placed individually in 12-well plates. 5 concentrations were tested: 150 mg/l, 375 mg/l, 750 mg/l, 1500 mg/l and 3750 mg/l. The TAA solution was changed every 24 h for 72 h, at which point the embryo survival was estimated. A control group for each concentration was kept in E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) and changed every 24 h for the duration of the experiment. We found that treatment of embryos with 1500 mg/l of TAA for 72 h resulted in ~ 50% mortality, thereby approximating the embryonic LC50 for TAA at this concentration. To ensure an adequate amount of TAA delivered to the adult liver, we adopted the intraperitoneal injection strategy repeated 6 times over the span of 2 weeks, with a dose of 500 mg/kg of body mass per injection.
TAA administration and isolation of liver cell populations by fluorescence-activated cell sorting (FACS)
Zebrafish transgenic lines Tg(fabp10a:dsRed), Tg(hand2:EGFP) and Tg(kdrl:ras-mCherry) in AB wild-type background were maintained in the IIMCB zebrafish facility (License no. PL14656251) according to standard procedures. Adult females were anesthetized with MS-222 (Sigma-Aldrich, Germany) as previously described [79] and injected intraperitoneally with 500 mg/kg thioacetamide (TAA) or sterile water as a control 6 times over the course of 2 weeks. A single dose of TAA would not approach the estimated LC50 for embryos, but the overall exposure to the toxin would exceed the estimated LC50. Adult fish weighing less than 2 g prior to the injections were excluded due to welfare concerns. Prior to toxin administration, the injection spot was wiped down with 1% povidone iodine to further limit the risk of infection. Overall, 15 fishes were injected with TAA. An additional 6 were injected with saline as a control. Fishes injected with TAA survived to the end of the 2-week treatment with 20% mortality (n surviving = 12). All saline-injected fishes survived the procedure. Experimental protocol for the treatment of animals in this study follows the guidelines approved by First Warsaw Local Ethics Committee for Animal Experimentation (file 15/2015). Livers were dissected and digested in Hank’s solution (1× HBSS, 2 mg/mL BSA, 10 mM Hepes pH 8.0) containing 0.05% trypsin (Sigma-Aldrich, Germany) and 2% collagenase (Sigma-Aldrich, Germany). Cell suspension was centrifuged at 500 g for 10 min at 4 °C. Cell pellet was resuspended in FACSmax (Amsbio, UK) and passed through a sterile 0.22 μm cell strainer (VWR, USA). Fluorescent cells were sorted by using FACSAria II cytometer (BD Biosciences, USA).
RNA-seq
For RNA sequencing 100,000 fluorescent liver cells were sorted directly to TRIzol LS (Thermo Fisher Scientific, USA). After ethanol precipitation RNA was depleted of DNA by using DNase I treatment and purified on columns by using RNA Clean & Concentrator™-5 (Zymo Research, USA). RNA integrity was measured by RNA ScreenTape on the Agilent 2200 TapeStation system (Agilent Technologies, USA). RNA Integrity Number (RIN) was in the range from 8.5 to 10 for all the samples used for RNA-seq. Ribosomal RNA removal from 10 ng of total RNA was performed using RiboGone Kit (Clontech Laboratories, USA). cDNA synthesis for next-generation sequencing (NGS) was performed by SMARTer Universal Low Input RNA Kit (Clontech Laboratories, USA) as recommended by the manufacturer. DNA libraries were purified with Agencourt AMPure XP PCR purification beads (Beckman Coulter, USA) and DNA fragment distribution was assessed by using D1000 ScreenTape and Agilent 2200 TapeStation system (Agilent Technologies, USA). KAPA library quantification kit (Kapa Biosystems, USA) was used for qPCR-based quantification of the libraries obtained. Paired-end sequencing (2 × 75 bp reads) was performed with NextSeq 500 sequencing system (Illumina, USA).
ATAC-seq
For ATAC-seq 60,000 fluorescent liver cells were sorted to Hank’s solution (1× HBSS, 2 mg/mL BSA, 10 mM Hepes pH 8.0), centrifuged for 5 min at 500×g and prepared for chromatin tagmentation as previously described [80]. NEBNext High-Fidelity 2 × PCR Master Mix (New England Biolabs, USA) and custom HPLC-purified primers containing Illumina-compatible indexes were used to prepare DNA sequencing libraries as previously described [81]. DNA libraries were purified with Agencourt AMPure XP PCR purification beads (Beckman Coulter, USA) and DNA fragment distribution was assessed by using D1000 ScreenTape and Agilent 2200 TapeStation system (Agilent Technologies, USA). KAPA library quantification kit (Kapa Biosystems, USA) was used for qPCR-based quantification of the libraries obtained. Paired-end sequencing (2 × 75 bp reads) was performed with NextSeq500 sequencing system (Illumina, USA).
Bioinformatics analysis
Raw RNA-seq and ATAC-seq reads were quality checked using Fastqc (0.11.8). Adapters were removed using Cutadapt (1.18) [82]. RNA-seq reads matching ribosomal RNA were removed using rRNAdust [83] and remaining reads were aligned to the zebrafish reference genome (GRCz11) using STAR (2.6) [84]. ATAC-seq reads were aligned to the zebrafish reference genome (GRCz11) using Bowtie2 (2.3.4.3) [85]. Reads quality filtering was performed using SAMtools (1.9) [86]. Read and alignment quality reports were prepared in Multiqc (1.6). To identify nucleosome free regions (NFRs) ATAC-seq reads originating from fragments not longer than 128 bp were retained and shifted by + 4 / -5 bp depending on the alignment strand using alignmentSieve utility from deepTools suite (3.2.0) [87]. Those reads were further used for peak calling using Macs2 (2.1.0.2) [88] subcommands. Shortly for each of the three replicates per base enrichment p-value track was calculated using the Poisson test. Then p-values tracks from replicates were combined using Fisher method. After Benjamini - Hochberg multiple testing correction, peaks were called on obtained tracks with q-value cutoff of 1e-5. Further obtained BED files were manipulated using Bedtools (2.27.1) [89] to discard NFRs overlapping low complexity regions as defined in the Ensembl’s [90] reference genome (GRCz11). Enriched motifs in NFRs were identified using Homer (4.10) [91]. Downstream bioinformatics analysis were performed in R 3.4.4 using several Bioconductor [92] packages. Cell type specific genes at quiescent state, were identified using DESeq2 [93] by comparing gene expression in specific cell type with gene expression in the other two. High-dimensional portraying of gene expression profiles was performed using oposSOM [36]. Differential gene expression analysis and differential accessibility analysis was performed using DESeq2 [93]. ATAC-seq peaks were processed and visualized using ChIPseeker [94], clusterProfiler [95], rtracklayer [96] and Gviz [97].
Histology and fluorescent microscopy
Adult females were sacrificed by overdosing MS-222 (Sigma-Aldrich, Germany) as previously described [98]. Samples were fixed in Dietrich’s fixative [98], dehydrated in ethanol and embedded in JB-4 resin (Sigma-Aldrich, Germany) for 3 h at 4 °C. Liver histology was examined microscopically in sections (4 μm thick) after hematoxylin and eosin (Sigma-Aldrich, Germany) staining using a modified protocol with increased staining and wash times to account for the lower staining efficiency in JB-4 resin. To detect fluorescence of GFP, mCherry and RFP, livers were fixed in 4% formaldehyde, incubated overnight in 20% sucrose, frozen in OCT solution (Leica Biosystems, France) and viewed under fluorescence microscope after sectioning (section thickness = 15 μm).
Availability of data and materials
RNA-seq and ATAC-seq data have been submitted to the NCBI Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE145565.
Change history
04 August 2023
Karim Abu Nahia’s Family name was tagged incorrectly in the article metadata. The article has been updated to rectify the error.
Abbreviations
- ECM:
-
Extracellular matrix
- TAA:
-
Thioacetamide
- HEP:
-
Hepatocyte
- HCC:
-
Hepatocellular carcinoma
- NAFLD:
-
Non-alcoholic fatty liver disease
- HSC:
-
Hepatic stellate cell
- LSEC:
-
Liver sinusoidal endothelial cell
- EC:
-
Endothelial cell
- SOM:
-
Self-organising map
- FACS:
-
Fluorescence-activated cell sorting
- GO:
-
Gene ontology
- TF:
-
Transcription factor
References
Blachier M, Leleu H, Peck-Radosavljevic M, Valla D-C, Roudot-Thoraval F. The burden of liver disease in Europe: a review of available epidemiological data. J Hepatol. 2013;58:593–608.
Baranova A, Lal P, Birerdinc A, Younossi ZM. Non-invasive markers for hepatic fibrosis. BMC Gastroenterol. 2011;11:91.
Tu T, Calabro SR, Lee A, Maczurek AE, Budzinska MA, Warner FJ, et al. Hepatocytes in liver injury: victim, bystander, or accomplice in progressive fibrosis? J Gastroenterol Hepatol. 2015;30:1696–704.
Baratta JL, Ngo A, Lopez B, Kasabwalla N, Longmuir KJ, Robertson RT. Cellular organization of normal mouse liver: a histological, quantitative immunocytochemical, and fine structural analysis. Histochem Cell Biol. 2009;131:713–26.
Yin C, Evason KJ, Asahina K, Stainier DYR. Hepatic stellate cells in liver development, regeneration, and cancer. J Clin Invest. 2013;123:1902–10.
Gandhi CR. Hepatic stellate cell activation and pro-fibrogenic signals. J Hepatol. 2017;67:1104–5.
Poisson J, Lemoinne S, Boulanger C, Durand F, Moreau R, Valla D, et al. Liver sinusoidal endothelial cells: physiology and role in liver diseases. J Hepatol. 2017;66:212–27.
Lefebvre P, Lalloyer F, Baugé E, Pawlak M, Gheeraert C, Dehondt H, et al. Interspecies NASH disease activity whole-genome profiling identifies a fibrogenic role of PPARα-regulated dermatopontin. JCI Insight. 2017;2:e92264.
Tsuchida T, Friedman SL. Mechanisms of hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol. 2017;14:397–411.
Iredale JP. Models of liver fibrosis: exploring the dynamic nature of inflammation and repair in a solid organ. J Clin Invest. 2007;117:539–48.
Sapp V, Gaffney L, EauClaire SF, Matthews RP. Fructose leads to hepatic steatosis in zebrafish that is reversed by mechanistic target of rapamycin (mTOR) inhibition. Hepatol Baltim Md. 2014;60:1581–92.
Sadler KC, Amsterdam A, Soroka C, Boyer J, Hopkins N. A genetic screen in zebrafish identifies the mutants vps18, nf2 and foie gras as models of liver disease. Development. 2005;132:3561–72.
Amali AA, Rekha RD, Lin CJ-F, Wang W-L, Gong H-Y, Her G-M, et al. Thioacetamide induced liver damage in zebrafish embryo as a disease model for steatohepatitis. J Biomed Sci. 2006;13:225–32.
Yin C, Evason KJ, Maher JJ, Stainier DYR. The basic helix-loop-helix transcription factor, heart and neural crest derivatives expressed transcript 2, marks hepatic stellate cells in zebrafish: analysis of stellate cell entry into the developing liver. Hepatol Baltim Md. 2012;56:1958–70.
Langheinrich U. Zebrafish: a new model on the pharmaceutical catwalk. BioEssays News Rev Mol Cell Dev Biol. 2003;25:904–12.
Ramaiah SK, Apte U, Mehendale HM. Cytochrome P4502E1 induction increases thioacetamide liver injury in diet-restricted rats. Drug Metab Dispos Biol Fate Chem. 2001;29:1088–95.
Hajovsky L, Hu G, Koen Y, Sarma D, Cui W, Moore DS, et al. Metabolism and toxicity of thioacetamide and thioacetamide s-oxide in rat hepatocytes. Chem Res Toxicol. 2012;25:1955–63.
Akhtar T, Sheikh N. An overview of thioacetamide-induced hepatotoxicity. Toxin Rev. 2013;32:43–6.
Wallace M, Hamesch K, Lunova M, Kim Y, Weiskirchen R, Strnad P, et al. Standard operating procedures in experimental liver research: thioacetamide model in mice and rats. Lab Anim. 2015;49(1_suppl):21–9.
Rekha RD, Amali AA, Her GM, Yeh YH, Gong H-Y, Hu S-Y, et al. Thioacetamide accelerates steatohepatitis, cirrhosis and HCC by expressing HCV core protein in transgenic zebrafish Danio rerio. Toxicology. 2008;243:11–22.
Wirth H, von Bergen M, Binder H. Mining SOM expression portraits: feature selection and integrating concepts of molecular function. BioData Min. 2012;5:18.
Her GM, Chiang C-C, Chen W-Y, Wu J-L. In vivo studies of liver-type fatty acid binding protein (L-FABP) gene expression in liver of transgenic zebrafish (Danio rerio). FEBS Lett. 2003;538:125–33.
Chi NC, Shaw RM, De Val S, Kang G, Jan LY, Black BL, et al. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes Dev. 2008;22:734–9.
Yao Y, Yao J, Boström KI. SOX transcription factors in endothelial differentiation and endothelial-mesenchymal transitions. Front Cardiovasc Med. 2019;6. https://doi.org/10.3389/fcvm.2019.00030.
Goncharov NV, Nadeev AD, Jenkins RO, Avdonin PV. Markers and biomarkers of endothelium: when something is rotten in the state. Oxidative Med Cell Longev. 2017;2017:e9759735.
Shay S, Ahuva I, Shira N-Y, Caryn G, Debra G-W, Simcha Y, et al. A novel human-specific soluble vascular endothelial growth factor receptor 1. Circ Res. 2008;102:1566–74.
Marshall SM, Gromovsky AD, Kelley KL, Davis MA, Wilson MD, Lee RG, et al. Acute Sterol O-Acyltransferase 2 (SOAT2) knockdown rapidly mobilizes hepatic cholesterol for fecal excretion. PLoS One. 2014;9:e98953.
Jayakumar A, Tai MH, Huang WY, al-Feel W, Hsu M, Abu-Elheiga L, et al. Human fatty acid synthase: properties and molecular cloning. Proc Natl Acad Sci U S A. 1995;92:8695–9.
Yeh Y-S, Jheng H-F, Iwase M, Kim M, Mohri S, Kwon J, et al. The mevalonate pathway is indispensable for adipocyte survival. iScience. 2018;9:175–91.
Rokosz LL, Boulton DA, Butkiewicz EA, Sanyal G, Cueto MA, Lachance PA, et al. Human cytoplasmic 3-hydroxy-3-methylglutaryl coenzyme a synthase: expression, purification, and characterization of recombinant wild-type and Cys129 mutant enzymes. Arch Biochem Biophys. 1994;312:1–13.
Yao Y, Sun S, Wang J, Fei F, Dong Z, Ke A-W, et al. Canonical Wnt signaling remodels lipid metabolism in Zebrafish hepatocytes following Ras oncogenic insult. Cancer Res. 2018;78:5548–60.
Hautekeete M, Geerts A. Limited evidence for redistribution of vitamin a from the liver to oesophageal mucosa in chronic liver disease in humans. Leiden: Cells Hepatic Sinusoid; 1997. p. 54–7.
Albadri S, Naso F, Thauvin M, Gauron C, Parolin C, Duroure K, et al. Redox signaling via lipid peroxidation regulates retinal progenitor cell differentiation. Dev Cell. 2019;50:73–89.e6.
Park K-H, Kim S-H. Low dose of chronic ethanol exposure in adult zebrafish induces hepatic steatosis and injury. Biomed Pharmacother Biomedecine Pharmacother. 2019;117:109179.
Timme-Laragy AR, Goldstone JV, Imhoff BR, Stegeman JJ, Hahn ME, Hansen JM. Glutathione redox dynamics and expression of glutathione-related genes in the developing embryo. Free Radic Biol Med. 2013;65. https://doi.org/10.1016/j.freeradbiomed.2013.06.011.
Löffler-Wirth H, Kalcher M, Binder H. oposSOM: R-package for high-dimensional portraying of genome-wide expression landscapes on bioconductor. Bioinforma Oxf Engl. 2015;31:3225–7.
Wirth H, Löffler M, von Bergen M, Binder H. Expression cartography of human tissues using self organizing maps. BMC Bioinformatics. 2011;12:306.
Moran-Salvador E, Mann J. Epigenetics and liver fibrosis. Cell Mol Gastroenterol Hepatol. 2017;4:125–34.
Leung A, Parks BW, Du J, Trac C, Setten R, Chen Y, et al. Open chromatin profiling in mice livers reveals unique chromatin variations induced by high fat diet *. J Biol Chem. 2014;289:23557–67.
Wang AW, Wang YJ, Zahm AM, Morgan AR, Wangensteen KJ, Kaestner KH. The dynamic chromatin architecture of the regenerating liver. Cell Mol Gastroenterol Hepatol. 2020;9:121–43.
Kersten S. Peroxisome proliferator activated receptors and lipoprotein metabolism. PPAR Res. 2008;2008. https://doi.org/10.1155/2008/132960.
Pramfalk C, Pedrelli M, Parini P. Role of thyroid receptor β in lipid metabolism. Biochim Biophys Acta (BBA) - Mol Basis Dis. 1812;2011:929–37.
Reddy S, Yang W, Taylor DG, Shen X, Oxender D, Kust G, et al. Mitogen-activated protein kinase regulates transcription of the ApoCIII gene. Involvement of the orphan nuclear receptor HNF4. J Biol Chem. 1999;274:33050–6.
Sarkar A, Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12:15–30.
Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34.
Remy P, Baltzinger M. The Ets-transcription factor family in embryonic development: lessons from the amphibian and bird. Oncogene. 2000;19:6417–31.
Lin B, Morris DW, Chou JY. The role of HNF1alpha, HNF3gamma, and cyclic AMP in glucose-6-phosphatase gene activation. Biochemistry. 1997;36:14096–106.
He L, Gomes AP, Wang X, Yoon SO, Lee G, Nagiec MJ, et al. mTORC1 promotes metabolic reprogramming by the suppression of GSK3-dependent Foxk1 phosphorylation. Mol Cell. 2018;70:949–960.e4.
The UniProt Consortium. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–9.
Gaudet P, Livstone MS, Lewis SE, Thomas PD. Phylogenetic-based propagation of functional annotations within the gene ontology consortium. Brief Bioinform. 2011;12:449–62.
Chami M, Oulès B, Szabadkai G, Tacine R, Rizzuto R, Paterlini-Bréchot P. Role of SERCA1 truncated isoform in the proapoptotic calcium transfer from ER to mitochondria during ER stress. Mol Cell. 2008;32:641–51.
Su N, Kilberg MS. C/EBP homology protein (CHOP) interacts with activating transcription factor 4 (ATF4) and negatively regulates the stress-dependent induction of the asparagine synthetase gene. J Biol Chem. 2008;283:35106–17.
Shah AV, Birdsey GM, Randi AM. Regulation of endothelial homeostasis, vascular development and angiogenesis by the transcription factor ERG. Vasc Pharmacol. 2016;86:3–13.
Wang P-W, Hung Y-C, Wu T-H, Chen M-H, Yeh C-T, Pan T-L. Proteome-based identification of apolipoprotein A-IV as an early diagnostic biomarker in liver fibrosis. Oncotarget. 2017;8:88951–64.
Bracht T, Schweinsberg V, Trippler M, Kohl M, Ahrens M, Padden J, et al. Analysis of disease-associated protein expression using quantitative proteomics—Fibulin-5 is expressed in association with hepatic fibrosis. J Proteome Res. 2015;14:2278–86.
Oh S-Y, Kim JY, Park C. The ETS factor, ETV2: a master regulator for vascular endothelial cell development. Mol Cell. 2015;38:1029–36.
Bataller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115:209–18.
Wilkins BJ, Pack M. Zebrafish models of human liver development and disease. Compr Physiol. 2013;3:1213–30.
Kim S-H, Wu S-Y, Baek J-I, Choi SY, Su Y, Flynn CR, et al. A post-developmental genetic screen for Zebrafish models of inherited liver disease. PLoS One. 2015;10. https://doi.org/10.1371/journal.pone.0125980.
Goessling W, Sadler KC. Zebrafish: an important tool for liver disease research. Gastroenterology. 2015;149:1361–77.
Wrighton PJ, Oderberg IM, Goessling W. There is something fishy about liver cancer: Zebrafish models of hepatocellular carcinoma. Cell Mol Gastroenterol Hepatol. 2019;8:347–63.
Hou W, Syn W-K. Role of metabolism in hepatic stellate cell activation and fibrogenesis. Front Cell Dev Biol. 2018;6. https://doi.org/10.3389/fcell.2018.00150.
Mehendale HM, Chilakapati J. 9.29 - Thioacetamide. In: CA MQ, editor. Comprehensive toxicology. 2nd ed. Oxford: Elsevier; 2010. p. 627–38. https://doi.org/10.1016/B978-0-08-046884-6.01029-0.
Pritchard MT, Apte U. Chapter 2 - models to study liver regeneration. In: Apte U, editor. Liver regeneration. Boston: Academic Press; 2015. p. 15–40. https://doi.org/10.1016/B978-0-12-420128-6.00002-6.
Hartman JH, Kozal JS, Di Giulio RT, Meyer JN. Zebrafish have an ethanol-inducible hepatic 4-nitrophenol hydroxylase that is not CYP2E1-like. Environ Toxicol Pharmacol. 2017;54:142–5.
Chu J, Sadler KC. A new school in liver development: lessons from Zebrafish. Hepatol Baltim Md. 2009;50:1656–63.
Lafoz E, Ruart M, Anton A, Oncins A, Hernández-Gea V. The endothelium as a driver of liver fibrosis and regeneration. Cells. 2020;9. https://doi.org/10.3390/cells9040929.
Braet F, Spector I, De Zanger R, Wisse E. A novel structure involved in the formation of liver endothelial cell fenestrae revealed by using the actin inhibitor misakinolide. Proc Natl Acad Sci U S A. 1998;95:13635–40.
DeLeve LD, Wang X, Kanel GC, Atkinson RD, McCuskey RS. Prevention of hepatic fibrosis in a murine model of metabolic syndrome with nonalcoholic steatohepatitis. Am J Pathol. 2008;173:993–1001.
Horn T, Christoffersen P, Henriksen JH. Alcoholic liver injury: defenestration in noncirrhotic livers--a scanning electron microscopic study. Hepatol Baltim Md. 1987;7:77–82.
Pasarín M, La Mura V, Gracia-Sancho J, García-Calderó H, Rodríguez-Vilarrupla A, García-Pagán JC, et al. Sinusoidal endothelial dysfunction precedes inflammation and fibrosis in a model of NAFLD. PLoS One. 2012;7:e32785.
Tateya S, Rizzo NO, Handa P, Cheng AM, Morgan-Stevenson V, Daum G, et al. Endothelial NO/cGMP/VASP signaling attenuates Kupffer cell activation and hepatic insulin resistance induced by high-fat feeding. Diabetes. 2011;60:2792–801.
Greene AK, Wiener S, Puder M, Yoshida A, Shi B, Perez-Atayde AR, et al. Endothelial-directed hepatic regeneration after partial hepatectomy. Ann Surg. 2003;237:530–5.
Yang Y, Sangwung P, Kondo R, Jung Y, McConnell MJ, Jeong J, et al. Alcohol-induced Hsp90 acetylation is a novel driver of liver sinusoidal endothelial dysfunction and alcohol-related liver disease. J Hepatol. 2021;75:377–86.
Calabro SR, Maczurek AE, Morgan AJ, Tu T, Wen VW, Yee C, et al. Hepatocyte produced matrix metalloproteinases are regulated by CD147 in liver fibrogenesis. PLoS One. 2014;9:e90571.
del Carmen Garcíade León M, Montfort I, Tello Montes E, López Vancell R, Olivos García A, González Canto A, et al. Hepatocyte production of modulators of extracellular liver matrix in normal and cirrhotic rat liver. Exp Mol Pathol. 2006;80:97–108.
Wangensteen KJ, Zhang S, Greenbaum LE, Kaestner KH. A genetic screen reveals Foxa3 and TNFR1 as key regulators of liver repopulation. Genes Dev. 2015;29:904–9.
Le Lay J, Kaestner KH. The fox genes in the liver: from organogenesis to functional integration. Physiol Rev. 2010;90:1–22.
Matthews M, Varga ZM. Anesthesia and euthanasia in zebrafish. ILAR J. 2012;53:192–204.
Pawlak M, Kedzierska KZ, Migdal M, Nahia KA, Ramilowski JA, Bugajski L, et al. Dynamics of cardiomyocyte transcriptome and chromatin landscape demarcates key events of heart development. Genome Res. 2019;29:506–19.
Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21.29.1–9.
Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–2.
Hasegawa A, Daub C, Carninci P, Hayashizaki Y, Lassmann T. MOIRAI: a compact workflow system for CAGE analysis. BMC Bioinformatics. 2014;15:144.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9:357–9.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinforma Oxf Engl. 2009;25:2078–9.
Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–91.
Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137.
Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2.
Howe KL, Achuthan P, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, et al. Ensembl 2021. Nucleic Acids Res. 2021;49:D884–91.
Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89.
Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, et al. Orchestrating high-throughput genomic analysis with bioconductor. Nat Methods. 2015;12:115–21.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Yu G, Wang L-G, He Q-Y. ChIPseeker: an R/bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–3.
Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS J Integr Biol. 2012;16:284–7.
Lawrence M, Gentleman R, Carey V. rtracklayer: an R package for interfacing with genome browsers. Bioinforma Oxf Engl. 2009;25:1841–2.
Hahne F, Ivanek R. Visualizing genomic data using Gviz and bioconductor. Methods Mol Biol Clifton NJ. 2016;1418:335–51.
Ellis JL, Yin C. Histological analyses of acute alcoholic liver injury in Zebrafish. JoVE J Vis Exp. 2017;(123):55630.
Acknowledgments
We acknowledge the IIMCB Zebrafish Core Facility for service and fish maintenance. We thank E. Ober for the kind gift of Tg(fabp10a:dsRed).
Funding
This work has been supported by National Science Centre, Poland, SONATA grant number 2014/15/D/NZ5/03421. MP was supported by the Ministry of Science and Higher Education, Poland, and National Science Centre, Poland, OPUS grant number 2018/29/B/NZ2/01010 and Foundation For Polish Science TEAM grant number POIR.04.04.00-00-5C84/17. FG was supported by Polish National Science Centre grants 2018/31/N/NZ5/03214 and 2020/36/T/NZ5/00610. ET and MM are recipients of the Postgraduate School of Molecular Medicine doctoral fellowship for the program “Next generation sequencing technologies in biomedicine and personalized medicine”. The project no. POIR.04.04.00-00-1AF0/16-00/ carried out within the First TEAM programme of the Foundation for Polish Science co-financed by the European Union under the European Regional Development Fund supports CW and KAN.
Author information
Authors and Affiliations
Contributions
MP and ET performed in vivo experiments and collected biological material. KZK performed preliminary experiments and optimized the protocols. LB performed and KP supervised FACS analysis. ET performed histological staining and took microscopic images. KAN prepared NGS libraries and performed sequencing. MP and MM performed bioinformatics and statistical analysis. MP and MM contributed to the design of the study and interpreted data. FG analyzed and interpreted the data. MP, MM and ET prepared the figures. MP and CLW conceived the study. MP, ET, MM and CLW wrote the manuscript. MP and CLW are senior corresponding authors. All authors have read and approved the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All experimental protocols were approved by First Warsaw Local Ethics Committee for Animal Experimentation (file 15/2015). All methods were carried out in accordance with relevant guidelines and regulations and reported in accordance with ARRIVE guidelines for the reporting of animal experiments.
Consent for publication
Not applicable.
Competing interests
Authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Migdał, M., Tralle, E., Abu Nahia, K. et al. Multi-omics analyses of early liver injury reveals cell-type-specific transcriptional and epigenomic shift. BMC Genomics 22, 904 (2021). https://doi.org/10.1186/s12864-021-08173-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-021-08173-1