Abstract
Background
Intramuscular fat (IMF) is associated with meat quality and insulin resistance in animals. Research on genetic mechanism of IMF decomposition has positive meaning to pork quality and diseases such as obesity and type 2 diabetes treatment. In this study, an IMF trait segregation population was used to perform RNA sequencing and to analyze the joint or independent effects of genes and long intergenic non-coding RNAs (lincRNAs) on IMF.
Results
A total of 26 genes including six lincRNA genes show significantly different expression between high- and low-IMF pigs. Interesting, one lincRNA gene, named IMF related lincRNA (IRLnc) not only has a 292-bp conserved region in 100 vertebrates but also has conserved up and down stream genes (< 10 kb) in pig and humans. Real-time quantitative polymerase chain reaction (RT-qPCR) validation study indicated that nuclear receptor subfamily 4 group A member 3 (NR4A3) which located at the downstream of IRLnc has similar expression pattern with IRLnc. RNAi-mediated loss of function screens identified that IRLnc silencing could inhibit both of the RNA and protein expression of NR4A3. And the in-situ hybridization co-expression experiment indicates that IRLnc may directly binding to NR4A3. As the NR4A3 could regulate the catecholamine catabolism, which could affect insulin sensitivity, we inferred that IRLnc influence IMF decomposition by regulating the expression of NR4A3.
Conclusions
In conclusion, a novel functional noncoding variation named IRLnc has been found contribute to IMF by regulating the expression of NR4A3. These findings suggest novel mechanistic approach for treatment of insulin resistance in human beings and meat quality improvement in animal.
Similar content being viewed by others
Background
Intramuscular fat (IMF) refers to the amount of fat located in skeletal muscle fibers [1]. Excess accumulation of IMF has been reported to be associated with diseases, such as type 2 diabetes and insulin resistance in humans [2]. In animals, as an important determinant of meat quality, IMF content directly influences flavor and juiciness and indirectly influences tenderness and meat color [1]. Moreover, pork IMF contains more unsaturated fatty acids (∼10–15% of total fatty acids) than beef and lamb [3]. Long-chain polyunsaturated fatty acids (LC-PUFA) such as omega-3 PUFA, eicosapentaenoic (EPA, 20:5n-3), and docosahexaenoic (DHA, 22:6n-3) acids are well accepted having beneficial effects on human brain development and cardiovascular disease [4, 5]. Both extremely high and extremely low IMF content is undesirable in consumed meat [1]. Thus, IMF is an important factor for human health.
It is generally accepted that IMF is a complex trait that is influenced by multiple genes or quantitative trait loci (QTLs). To date, a total of 709 QTLs have been reported to be associated with pig IMF content (PigQTLdb, http://www.animalgenome.org/cgi-bin/QTLdb/SS/index, released at April 26, 2020) [6]. However, the locations of these QTLs are not accurate due to the limited density of microsatellite markers. Long-term fine-mapping experiments are needed to refine these loci and investigate causative variants [7]. Furthermore, most of the single nucleotide polymorphisms (SNPs) associated with IMF in genome-wide association studies only explain a small part of the total genetic variance. Studies identifying genetic variation that explains this “missing heritability” of IMF are urgently needed [8].
Since they reside in regulatory elements of the genome, noncoding genomic variants located in intronic regions of protein-coding genes or in intergenic regions may have functional roles in the expression of specific phenotypes or traits. In pigs, long intergenic non-coding RNAs (lincRNAs) have been reported to be associated with permanent molars, adipose and muscle development, and energy metabolism [9,10,11]. Although several lincRNAs have been reported associated with pork and poultry IMF [12,13,14,15], little is known about the mechanism of lincRNA gene regulation in pig IMF.. The objectives of this work were to perform RNA sequencing analysis using an IMF character segregation population which construct using high IMF pigs (Min pig) and low IMF pigs (Large white pigs) crossbred and to analyze the joint or independent effects of lincRNAs on IMF. Moreover, we aimed to identify genetic markers that may be suitable for inclusion in animal genetic improvement programs and provide new targets for the treatment of insulin resistance in humans.
Results
RNA sequencing, data mapping, and transcript identification
RNA sequencing of longissimus dorsi muscle tissue has been done first in our research. After filtering, a total of 579.53 million clean reads (97.22% of the raw data) were obtained. More than 75% of the clean data could be mapped to the reference genome (v11.1 ftp://ftp.ensembl.org/pub/release-102/fasta/sus_scrofa/dna/). A total of 26,276 transcript units were identified, including 4671 lincRNAs. Among these 26,276 units, 59.7% encoded proteins, 3.4% were miscellaneous RNAs, 2.2% were microRNAs (miRNAs), 0.6% were mitochondrial ribosomal RNAs (rRNAs), 0.2% were small nuclear RNAs (snRNAs), and the remaining 33.6% were pseudogenes and processed transcripts. The clean data have been submitted to the Genome Sequence Archive, with the accession number CRA001645.
Differentially expressed genes between high- and low-IMF content pigs
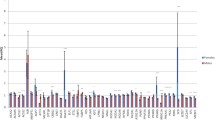
A total of 26 transcripts which have significantly differentially expression (DE,false discovery rate (FDR) < 0.1) between pigs with high and low IMF content were identified using a paired samples model in edgeR [16]. This included 5 novel protein-coding genes, 15 known protein-coding genes and 6 lincRNAs (Table 1 and Fig. 1). Six of the 20 protein-coding genes were upregulated with more than 2-fold changes (FC) in the pigs with low IMF content. And these six genes were N-Acetyl-Alpha-Glucosaminidase (NAGLU), novel gene 1 (Novel1), mitochondrially encoded NADH: ubiquinone oxidoreductase core subunit 6 (ND6), sushi domain containing 3 (SUSD3), novel gene 4 (Novel4), and leucine rich repeat containing 66 (LRRC66). Moreover, two lincRNAs were upregulated in the pigs with high IMF content.
Significantly DE transcripts between high- and low-IMF pigs. N=3 in each group. a Volcano plot of significantly DE transcripts, the significant DE transcripts (FDR < 0.05) were in the color of red, blue lines indicate the threshold of log2 (0.67) and log2 (1.5). b Heat map and cluster represent the 26 DE transcripts differentially regulated in high- and low-IMF pigs, the color of scale bars represent the RPKM of each transcript
Among the 26 DE transcripts, IMF-related LincRNA (IRLnc) and nuclear receptor subfamily 4 group A member 3 (NR4A3), which had similar expression patterns, were located on chromosome 1. Solute carrier family 2 member 1 (SLC20A1), SUSD3 and a novel transcript were located on chromosome 3. Peroxisome proliferator-activated receptor-gamma coactivator-1 (PPARGC1) and LRRC66 were located on chromosome 8. IMF-related LincRNA2 and C2 calcium dependent domain Containing 3 (C2CD3), were located on chromosome 9. Acyl-CoA binding domain containing 7 (ACBD7) and IMF-related LincRNA4 were located on chromosome 10. NAGLU and fatty acid synthase (FASN) were located on chromosome 12. Sulfiredoxin 1 (SRXN1) and IMF-related LincRNA5 were located on chromosome 17. Leptin (LEP) and protein kinase AMP-Activated non-catalytic subunit gamma 2 (PRKAG2) were located on chromosome 18. Other DE transcripts were located on chromosome 2, 6, Mitochondrial and unmapped sequences.
RT-qPCR validation of DE genes
The same pigs with low and high IMF content in RNA-seq analysis were selected for validation by RT-qPCR. According to the RNA-seq abundance, we select 9 DE transcripts for RT-qPCR analysis. As relative quantitation of each transcript between the RT-qPCR and RNA-seq were not in same level, we set the value of qPCR and sequencing in low IMF group to be one. RT-qPCR results showed that 88.89% (8 of 9) of the selected transcripts could be validated in low IMF content vs. high IMF content pigs (Fig. 2). Among the eight validated DE transcripts, there are six known genes which were ACBD7, NR4A3, SRXN1, LEP, FASN, and ND6. One novel gene (novel3) and one lincRNA (IRLnc) could also been validated. Therefore, the sequencing results were reliable and candidate DE mRNAs and lincRNAs could be used for further analysis.
Validation of differentially expressed genes in high - and low - IMF pigs by RT-qPCR. For each gene, the value of qPCR and sequencing in low IMF group were set at 1. N=3 in each group, * represents P< 0.05. ACBD7: Acyl-CoA Binding Domain Containing 7, IRLnc: IMF-related LincRNA, NR4A3: nuclear receptor subfamily 4 group A member 3, SRXN1: Sulfiredoxin 1, LEP: Leptin, FASN: fatty acid synthase, ND6: NADH dehydrogenase subunit 6, novel3: novel protein coding gene 3, C2CD3: C2 Calcium Dependent Domain Containing 3
Identification of sequence homology with 99 vertebrates and query of upstream and downstream genes of IRLnc
Conservation analysis of 100 vertebrate whole genomes showed that there was a 292-bp region within the IRLnc gene (11199-bp) that was conserved between pigs and humans (Fig. 3). To determine whether IRLnc interacts with neighboring genes, we performed a sequence query analysis of the gene in the 500-kb window surrounding IRLnc. Two genes, Sec61 translocon beta subunit (Sec61B) and NR4A3, were found adjacent to IRLnc. We then analyzed the read counts and logFC values of Sec61B and NR4A3 in RNA-seq data and found that Sec61B and NR4A3 had logFC values of 0.03 and 1.94 in pigs with high IMF content compared to those with low IMF content, respectively (FDR = 1 and 0.0.0076, respectively), and this means no expression difference of Sec61B and significant expression differences of NR4A3.
Synteny and sequence conservation of IRLnc PhastCon plot of pig and 99 vertebrates. The grey box shows the region with sequence conservation. The PhastCon plot is relative to loci in the human genome and is derived from 99 vertebrate whole-genome alignments. The consensus logo highlights the 292-bp conserved sequence, which was identified from the 99 vertebrate genome alignments. A score of 4 bits indicates that these bases are perfectly conserved in the 99 vertebrate genomes
Gene expression pattern of Sec61B and NR4A3 in low- and high- IMF pigs
To confirm the differential expression of Sec61B and NR4A3, we validated these findings in a bigger population (five pigs with low IMF content and five with high IMF content). As shown in Fig. 4a, there was no difference in Sec61B expression between the two groups. However, NR4A3 gene expression was significantly different. The expression of Sec61B and NR4A3 in five breeds pigs with different average IMF were also detected to infer the expression of IRLnc and its upstream and downstream genes (Fig. 4b). The results indicated that, the NR4A3 gene expression was significantly high in Laiwu, Mashen, Min and Beijing Black pigs rather than in Large white (P< 0.05). And there are almost none differences of Sec61B expression between these pigs. Thus, we chose NR4A3 for further research.
Gene expression of IRLnc, Sec61B and NR4A3. a The expression of IRLnc, Sec61B and NR4A3 between high- and low-IMF pigs (n=5 in each group), * represents P< 0.05. b The expression of IRLnc, Sec61B and NR4A3 in different breeds (n=5 in each breed), different characters above the bar represent significant differences. Laiwu is a pig breed with extremely high- IMF, Min and Mashen pigs have high IMF, Beijing Black pigs have median IMF, and Large White pigs have median to low IMF
LincRNA-RNA interaction prediction
In order to explore the potential lincRNA-RNA interaction, we calculate the interaction energy of IRLnc and NR4A3. Since IntaRNA software could only analyze sequences less than 2000 bp, we divided the NR4A3 mRNA sequence into three segments (1800 bp each, with 1709 bp; 1621 bp; and 1785 bp effective sequences) for analysis. The nearby segments were designed overlapped to avoid the potential effects of sequence dividing. Six interaction domains with a minimal interaction energy of <− 10 kcal/mol were found in the IRLnc and NR4A3 interaction prediction analysis (Table 2 and Fig. 5). Interestingly, the domain with the lowest minimal interaction energy (− 17.6096 kcal/mol) was located in the 3′UTR region of NR4A3. This result indicated that the conserved sequence of IRLnc may interact with NR4A3 mRNA and directly regulate its expression.
RNA silencing of IRLnc
Three siRNAs targeting IRLnc (si-727, si-2333, and si-2942) were transfected into cells and IRLnc interference efficiency was then tested. The results showed that cells transfected with si-727 and si-2942 had significantly different IRLnc expression levels than cells transfected with the negative control (NC) siRNA (Fig. 6a, P < 0.05), which indicates good knockdown efficiency. Finally, si-727 was selected for subsequent experiments. IRLnc silencing significantly decreased the RNA expression of NR4A3 by approximately 50% (Fig. 6b, P < 0.05). Moreover, IRLnc silencing significantly decreased the protein expression of NR4A3 (Fig. 6c, P < 0.05).
Fold changes of Sec61B and NR4A3. a The transinfected efficiency of three si-IRLnc probes. b Expression of Sec61B and NR4A3 in IRLnc silent cells and normal cells. * represent P< 0.05, n=3 in each group. c Differences of NR4A3 protein expression level in normal and RNA-Silencing cells. * represent P< 0.05, n=3 in each group
Co-localization of IRLnc and NR4A3
In-situ hybridization analysis showed that, in NCs, there was no co-localization of IRLnc and DapB (Fig. 7a). However, co-localization of IRLnc and UBC (Fig. 7b) and IRLnc and NR4A3 (Fig. 7c-e) was observed. Moreover, the results showed that IRLnc and NR4A3 were mainly located at the margin of muscle fibers where intramuscular fat is deposited.
In situ colocalization of IRLnc and NR4A3 mRNA in myoblast and the potential effect pathway of IRLnc on fat deposition (n=3). a-e Representative pseudocolored images of DapB/NR4A3, UBC/NR4A3, IRLnc, NR4A3, and IRLnc/NR4A3 stained with DAPI (nucleus, blue) and for NR4A3 (red), DapB (green), UBC (green), and IRLnc (green), and the color represent co-expression will be orange (red+green). Total magnification of all images is 20 ×. f Potential effect pathway of IRLnc on lipolysis. Red line represents inhibit effect; green line represents promote effect
Discussion
In our research, three pairs of full-sib Large White × Min pigs F2 sows which with extremely different IMF content were selected to do RNA sequencing analysis. Gene annotation and PhastCons analysis were used to mining candidate LincRNAs related to IMF. Co-localization analysis and RNAi-mediated loss of function screens were performed to study the potential interactive mechanism of the candidate LincRNA and its downstream gene.
Investigation of differential expression transcripts
Min is a native Chinese pig breed with an average IMF of 5.22% and Large White is a European pig breed with an average IMF of 2.00%. The offspring of Large White × Min crossbred (F2) population is an ideal IMF separated model to analyze the genetic mechanism of IMF. We used three pairs of full siblings from the F2 population, so that the genetic background of each pair was consistent. The paired samples model in edgeR was also appropriate for our research design. Although thousands of transcripts and lincRNAs were identified in pig genomes, there were only 26 DE transcripts between the groups with high and low IMF content.
Function analysis of DE protein-coding genes
Among these 26 transcripts, 15 protein-coding genes were identified and located to known genes. Among the known genes, ten were associated with fatty acid and myocyte formation or metabolism. One of the SNPs in PPARGC1 (rs8192678[A]) has been reported to be associated with higher triacylglycerol levels (P = 0.005) [17]. FASN and LEP are important factors in fatty acid synthesis [18, 19]. NR4A3 is a regulator of insulin activity in adipocytes [14, 20]. ACBD7 could be involved in energy homeostasis and associated to obesity in humans [21]. Specific knockout of SLC20A1 could significantly decreased hepatic lipogenesis [22]. A SNP in PRKAG2 was associated with plasma free fat acid and glycerol measurements [23]. Adiponectin (ADIPOQ) has the glucose regulation and fatty acid oxidation function [24]. A SNP in Growth arrest and DNA damage-inducible protein GADD45 Alpha (GADD45A) showed significant association patterns for IMF and backfat thickness in Berkshire pigs [25]. ND6 could participated in regulates mitochondrial fatty acid oxidative metabolism, and two rare variants in ND6 were associated with BMI [26, 27]. One gene named LRRC66 was involved in diverse biological processes, including cell adhesion, cellular trafficking, and hormone-receptor interactions [28]. And other genes were mainly associated with diseases such as Mucopolysaccharidosis IIIB [29], stress-induced injury [30], oral-facial-digital syndromes [31], and breast cancer [32]. From the results, intramuscular related genes involved in fat and muscle related function were consistent with previous research, which reported that IMF is regulated through a complex pathway that interacts with muscle, fat, and connective tissue [33, 34].
Our study also identified six DE lincRNAs that may be involved in IMF regulation. These three lincRNAs are located on chromosomes 1, 4 and 10. The lincRNA on chromosome 1 (IRLnc) had not previously been reported associated with IMF in pigs. It displayed a 6.48-fold higher level of expression (logFC = 2.69) in pigs with high IMF content compared to those with low IMF content (FDR = 4.80E-12) and as such, was the locus most significantly associated with IMF content. Therefore, we selected IRLnc for further research.
Potential regulatory effect of IRLnc
To investigate the potential regulatory effect of IRLnc, we first analyzed the IRLnc gene and its surrounding genes. Although all of the genes upstream and downstream of IRLnc are conserved, NR4A3 was the only gene adjacent to IRLnc that displayed differential expression, according to the results of RNA-seq and RT-qPCR analysis. To confirm these results, we detected the expression of IRLnc and NR4A3 in five kinds of purebred pigs. Laiwu pig is famous breed with extremely high- IMF, Min and Mashen pigs have high IMF, Beijing Black pigs have median IMF, and Large White pigs have median to low IMF. And the expression results in purebred pigs consist with the results in crossbred pigs. This observation implied that IRLnc may be a regulatory element for the NR4A3 gene. Thus, we silenced IRLnc in myoblast cells to investigate the influence of IRLnc on NR4A3 RNA and protein expression. Silencing IRLnc resulted in a decrease in NR4A3 expression, thus confirming the speculation that IRLnc may regulate NR4A3.
The results of RNA-RNA interaction prediction were also positive. We found that the 292-bp conserved sequence of IRLnc had a very low energy requirement to interact with the 3′UTR region of NR4A3 mRNA. Therefore, we inferred that IRLnc may regulate the expression of NR4A3 by binding to its regulatory domain. The finding that IRLnc and NR4A3 are co-localized also supports our hypothesis. Together, these results implied that IRLnc may affect NR4A3 expression by directly binding to its regulatory domain.
In previous research, the NR4A family, especially NR4A2 and NR4A3, have been shown to be unnecessary for adipogenesis [35]. Thus, we inferred that NR4A3 may not affect fat deposition through the adipogenesis pathway. Previous studies have shown that NR4A3 gene expression is reduced in skeletal muscles and adipose tissues from multiple rodent models of insulin resistance [14, 20]. Insulin sensitivity is known to be closely related to fat deposition. In the research of Walton et al. (2013), over-expression of NR4A3 induced a decrease in the concentration of circulating catecholamines, leading to poor insulin sensitivity and increased low-density lipoprotein levels [36]. Additionally, poor insulin resistance leads to a decrease in lipolysis [37]. In summary, we propose that IRLnc directly regulates the expression of NR4A3, which then regulates catecholamine catabolism and finally, IMF deposition, by regulating insulin sensitivity (Fig. 7f). A recent study showed that IMF directly modulates muscle insulin sensitivity [38] and we propose that this process may occur through the same pathway. However, the expression regulatory activity of IRLnc and the pathway mediating the effect of IRLnc on IMF require further investigation.
Conclusions
This study performed RNA sequencing analysis using an IMF character segregation population, and provided 26 DE transcripts in high- and low- IMF pigs. This research also provided a global view of the complexity of the mRNA and lincRNA transcriptome in pigs with different IMF content. Using bioinformatic analysis, co-localization analysis and RNAi-mediated loss of function screens, we also explored the potential interactive mechanism of the candidate LincRNA (IRLnc) and its downstream gene (NR4A3). IRLnc that may influence IMF decomposition maybe a potential marker for meat quality selection. Furthermore, as IRLnc could directly regulate the expression of NR4A3 which associated with insulin resistance, we inferred that IRLnc may be a potential therapeutic target for insulin resistance and type 2 diabetes.
Methods
Animals and sample collection
All of the animals used in our research were obtained from the experiment pig farm of Institute of Animal Science, Chinese Academy of Agricultural Sciences (CAAS). Before slaughter, all of the pigs are stunned using 80%-concentrate carbon dioxide for 45 s. When the pigs were confirmed stunned, they were then hoisted on a rail and exsanguinated via carotid artery and the jugular vein. After the blood is gone, pigs were handled on a Stork slaughter process line (Stork B. V, Naarden, Dunth) with standard procedure (removal of hair, eviscerated, cut into two-halves, and so on). In this study, a total number of 46 pigs were used to collect tissues samples, and at least 3 pigs in each group (mostly 5 pigs, as described in each part below) were select to meet calculation power. After experimentation, all of the pigs were sold to the slaughter house.
Six F2 sows (three pairs of full siblings) from a Large White × Min resource population (678 pigs, including 602 F2 individuals, all of the pigs were fed in same diet and raised in same condition. The tissue sampling condition are also uniformed) were used in this study for RNA extraction. In each pair of siblings, there was one high-IMF pig and one low-IMF pig (Table 3). IMF content was measured using an ether extraction method (Soxtec Avanti 2055 Fat Extraction System; Foss Tecator, Hillerød, Denmark).
RNA isolation, sequencing, and mapping
Total RNA was extracted and purified using TRIzol (Invitrogen, Carlsbad, CA, USA) and a RNeasy Mini Kit (Qiagen, Hilden, Germany). A total of 3 μg of RNA per sample was then used for RNA sample preparation. After amplification, PCR products were purified (AMPure XP system; Beckman Coulter, Brea, CA, USA) and library quality was assessed on a Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). Each library was sequenced on a HiSeq 2000 platform (Illumina, San Diego, CA, USA) at the Novogene Bioinformatics Technology Cooperation (Beijing, China). Raw data in fastq format were firstly filtered to clean paired-end data using in-house Perl scripts. A gene model annotation system (Ensembl version 92), the reference genome (Sus scrofa 11.1), and associated files were directly downloaded from the Ensembl website (ftp://ftp.ensembl.org/pub/release-92/fasta/sus_scrofa/dna). An index of the reference genome was constructed using the Bowtie v2.0.6 package [39], with default parameters, and clean reads were aligned to the reference genome using TopHat v2.0.9 [40], with default parameters.
Identification of transcript units
Cufflinks v2.0.2 [41] was used to assemble the aligned reads for each sample. Cuffcompare v2.0.2 [42] was then used to generate intergenic transcripts for each sample assembly. To acquire high-confidence transcripts, two criteria were used to filter the transcripts using in-house Perl scripts: (1) RNA-seq reads must have covered more than 80% of predicted exon nucleotides for a transcript, (2) in at least one sample, there must have been more than three clean reads mapping to the predicted splice structure. Finally, fragments per kilobase of exon, per million fragments mapped values were obtained using Tophat v2.0.9 with --no-novel-juncs and Cufflinks v2.0.2 with -G.
Differential expression analysis
The identification of differentially expressed (DE) transcripts between low-IMF and high-IMF pigs was performed in edgeR using a paired samples model [16]. Significant DE transcripts were determined with the following criteria: false discovery rate (FDR) < 0.1 and log2 fold-change (logFC) value more than 0.58 (log2[1.5, 2]) or less than − 0.58 (log2[0.67, 2]).
Reverse transcription quantitative polymerase chain reaction validation of DE genes
cDNA was synthesized from total RNA using a PrimeScript reverse transcription (RT) reagent kit (Takara Bio Inc., Kusatsu, Japan). Primers were designed for all 26 DE transcripts using Primer 6 software (Table S1). cDNA samples from 10 pigs (five with high IMF content and five with low IMF content) were used as templates for quantitative polymerase chain reaction (qPCR). Reactions were performed on an ABI 7900HT Real-Time System (Applied Biosystems, Foster City, CA, USA) with a 15-μL mixture consisted of 1.5 μL of cDNA, 150 nM of each of the forward and reverse primers, and SYBR® Green PCR Mixture (ABI part number 4472908). Standard PCR cycling conditions were used. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as a control and relative gene expression levels were calculated using the 2−ΔΔCt method [43], where the delta cycle threshold (ΔCt) is the Ct of target gene minus the Ct of GAPDH and ΔΔCt is the ΔCt of the target gene minus the average ΔCt of all individuals. A Student’s t-test was used to analyze the differences in expression between the low- and high-IMF groups.
Identification of sequence homology with 99 vertebrates
NCBI BLASTn was used to identify the sequence homology of selected lincRNAs with 99 vertebrates, including humans. All sequences were retrieved from the University of California Santa Cruz database and the PhastCon conservation plot and consensus logo plot were drawn using in-house R scripts [44].
Expression of IMF-related lincRNA and its upstream and downstream genes
The pigs described above (five low-IMF content and five high IMF content) were selected to detect differences in the expression of IMF-related lincRNA (IRLnc), translocon beta subunit (Sec61B), and nuclear receptor subfamily 4 group A member 3 (NR4A3) using RT-qPCR. Primers for these three transcripts were designed using Primer 6 software (Table S1). Standard PCR cycling conditions were used. The GAPDH gene was used as control and relative gene expression levels were calculated using the 2−ΔΔCt method. A Student’s t-test was used to analyze the differences in expression between the low- and high-IMF groups. Five pigs in each breed (Laiwu, Min, Mashen, Beijing black and Large White) were also selected to detect differences in the expression of IRLnc, Sec61B, and NR4A3 using RT-qPCR. The statistical methods were same as that used in IMF different pigs.
LincRNA-RNA interaction prediction
LincRNA-RNA interactions between the 292-bp conserved sequence of IRLnc and NR4A3 mRNA were predicted using IntaRNA software (server version 4.4.2) [45, 46], with the following parameters: number of (sub) optimal interactions = 5 and min. Number of basepairs in seed = 7.
Cell isolation, tissue culture, and siRNA knockdown of IRLnc
Longissimus dorsi muscle tissues were obtained from one newborn pig. After fractionation and collagenase digestion (12,500 U I/II collagenase/kg of tissue; Sigma, St Louis, MO, USA), primary myoblasts, enriched in bottom stromal vascular fractions, were resuspended and cultured to confluence in Dulbecco’s modified Eagle’s medium supplemented with 10% (vol/vol) fetal bovine serum. After 2 d, cells were incubated in culture medium containing insulin (5 μg/mL) for another 2 d and the culture solution was then changed every 2d.
Three IRLnc-specific siRNAs and one NC siRNA were designed by GenePharma (Shanghai, China; Table S2). When cultured primary myoblasts reached 70–80% confluence, siRNAs (150 nM) were transfected using DharmaFect2 (5 μL/mL), according to the manufacturer’s instructions (Dharmacon, Lafayette, CO, USA). Twelve hours after transfection, cells were induced to differentiate and were then harvested for downstream RNA and protein analysis on the fourth day after differentiation. Total RNA and protein were isolated from the cells using standard procedures. The differences in Sec61B and NR4A3 gene expression between normal and IRLnc-knockdown cells were detected using RT-qPCR. Moreover, the differences in NR4A3 protein between normal and IRLnc-knockdown cells were detected using western blotting, and the signals were normalized to β-actin expression.
In-situ hybridization co-localization of IRLnc and NR4A3
The longissimus dorsi muscles of three 0-d-old piglets were collected for in situ hybridization co-localization analysis. Tissues were freshly harvested, immediately fixed in 4% formalin, processed for paraffin embedding using standard protocols, and then sectioned onto SuperFrost Plus slides (Fisher Scientific, Waltham, MA, USA) at thickness of 5 mm.
Fluorescent probes were designed using IRLnc, NR4A3, UBC, and DapB sequences and synthesized by Advanced Cell Diagnostics (Newark, CA, USA). The NR4A3 probe used the C2 channel (red) and other probes used the C1 channel (green). RNA in-situ hybridization experiments were performed using the Multiplex Fluorescent Reagent Kit V2 kit (Advanced Cell Diagnostics), according to the manufacturer’s protocol. DapB was used as the NC and UBC as the positive control. Fluorescent images were acquired using a TCS SP8 confocal microscope (Leica, Wetzlar, Germany) and IRLnc/NR4A3, IRLnc/UBC, and DapB/NR4A3 co-localization was analyzed using Leica Application Suite X v3.1 software.
Statistical analysis
The data obtained are expressed as mean ± SE, a t-test was used to evaluate the statistical significance of the 2-part comparisons of expression difference. Three replicates in each group were used in vitro. Statistical analysis was carried out using SAS 9.4 statistical software, and the statistical significance was set at P< 0.05.
Availability of data and materials
The sequencing data used in the current study have been submitted to the Genome Sequence Archive, with the accession number CRA001645.
Abbreviations
- ΔΔCt:
-
Delta delta cycle threshold
- DE:
-
Differential expression
- DMEM:
-
Dulbecco’s Modified Eagle’s medium
- FDR:
-
False discovery rate
- GAPDH :
-
Glyceraldehyde-3-phosphate dehydrogenase
- GSA:
-
Genome sequence archive
- GWAS:
-
Genome-wide association study
- IMF:
-
Intramuscular fat
- NC:
-
Negative control
- RT-qPCR:
-
Reverse transcription quantitative polymerase chain reaction
- QTLs:
-
Quantitative trait loci
- Sec61B:
-
Translocon beta subunit
- SNPs:
-
Single nucleotide polymorphisms
- UCSC:
-
University of California Santa Cruz
- CPM:
-
Counts per million
- miRNAs:
-
microRNAs
- NAGLU :
-
N-Acetyl-Alpha-Glucosaminidase
- Novel :
-
Novel gene
- ACBD7 :
-
Acyl-CoA Binding Domain Containing 7
- PPARGC1 :
-
Peroxisome proliferator-activated receptor-gamma coactivator-1
- IRLnc :
-
IMF-related LincRNA
- GADD45A :
-
Growth Arrest And DNA Damage-Inducible Protein GADD45 Alpha
- NR4A3 :
-
Nuclear receptor subfamily 4 group A member 3
- SRXN1 :
-
Sulfiredoxin 1
- snRNAs:
-
Small nuclear RNAs
- rRNAs:
-
Mitochondrial ribosomal RNAs
- LEP :
-
Leptin
- SLC20A1 :
-
Solute Carrier Family 2 Member 1
- FASN :
-
Fatty acid synthase
- PRKAG2 :
-
Protein Kinase AMP-Activated Non-Catalytic Subunit Gamma 2
- ND6 :
-
Mitochondrially Encoded NADH: Ubiquinone Oxidoreductase Core Subunit 6
- ADIPOQ :
-
Adiponectin, SUSD3: Sushi Domain Containing 3
- C2CD3 :
-
C2 Calcium Dependent Domain Containing 3
- LRRC66 :
-
Leucine Rich Repeat Containing 66
References
Madeira MS, Costa P, Alfaia CM, Lopes PA, Bessa RJ, Lemos JP, et al. The increased intramuscular fat promoted by dietary lysine restriction in lean but not in fatty pig genotypes improves pork sensory attributes. J Anim Sci. 2013;91:3177–87.
Bergman BC, Perreault L, Hunerdosse DM, Koehler MC, Samek AM, Eckel RH. Intramuscular lipid metabolism in the insulin resistance of smoking. Diabetes. 2009;58:2220–7.
Valsta LM, Tapanainen H, Mannisto S. Meat fats in nutrition. Meat Sci. 2005;70:525–30.
Rimm EB, Appel LJ, Chiuve SE, Djoussé L, Engler MB, Kris-Etherton PM, et al. Seafood long-chain n-3 polyunsaturated fatty acids and cardiovascular disease: a science advisory from the American Heart Association. Circulation. 2018;138(1):e35–47.
Tocher DR, Betancor MB, Sprague M, Olsen RE, Napier JA. Omega-3 long-chain polyunsaturated fatty acids, EPA and DHA: bridging the gap between supply and demand. Nutrients. 2019;11(1):89.
Hu ZL, Park CA, Wu XL, Reecy JM. Animal QTLdb: an improved database tool for livestock animal QTL/association data dissemination in the post-genome era. Nucleic Acids Res. 2013;41:D871–9.
Xu L, Cole JB, Bickhart DM, Hou Y, Song J, VanRaden PM, Sonstegard TS, et al. Genome wide CNV analysis reveals additional variants associated with milk production traits in Holsteins. BMC Genomics. 2014;15:683.
Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53.
Wang F, Li Y, Wu X, Yang M, Cong W, Fan Z, et al. Transcriptome analysis of coding and long non-coding RNAs highlights the regulatory network of cascade initiation of permanent molars in miniature pigs. BMC Genomics. 2017;18:148.
Xia J, Xin L, Zhu W, Li L, Li C, Wang Y, et al. Characterization of long non-coding RNA transcriptome in high-energy diet induced nonalcoholic steatohepatitis minipigs. Sci Rep. 2016;6:30709.
Zhou ZY, Li A, Wang LG, Irwin DM, Liu YH, et al. DNA methylation signatures of long intergenic noncoding RNAs in porcine adipose and muscle tissues. Sci Rep. 2015;5:15435.
Sun Y, Chen X, Qin J, Liu S, Zhao R, Yu T, et al. Comparative analysis of long noncoding RNAs expressed during intramuscular adipocytes Adipogenesis in fat-type and lean-type pigs. J Agric Food Chem. 2018;66(45):12122–30.
Zou C, Li L, Cheng X, Li C, Fu Y, Fang C, et al. Identification and Functional Analysis of Long Intergenic Non-coding RNAs Underlying Intramuscular Fat Content in Pigs. Front Genet. 2018;9:102.
Huang W, Zhang X, Li A, Xie L, Miao X. Differential regulation of mRNAs and lncRNAs related to lipid metabolism in two pig breeds. Oncotarget. 2017;8:87539–53.
Zhang M, Ma X, Zhai Y, Zhang D, Sui L, Li W, et al. Comprehensive Transcriptome analysis of lncRNAs reveals the role of lncAD in chicken intramuscular and abdominal Adipogenesis. J Agric Food Chem. 2020;68(11):3678–88.
McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–97.
Queiroz EM, Candido AP, Castro IM, Bastos AQ, Machado-Coelho GL, Freitas RN. IGF2, LEPR, POMC, PPARG, and PPARGC1 gene variants are associated with obesity-related risk phenotypes in Brazilian children and adolescents. Braz J Med Biol Res. 2015;48:595–602.
Oh D, Lee Y, La B, Yeo J, Chung E, Kim Y, et al. Fatty acid composition of beef is associated with exonic nucleotide variants of the gene encoding FASN. Mol Biol Rep. 2012;39:4083–90.
Bouafi H, Bencheikh S, Mehdi Krami AL, Morjane I, Charoute H, Rouba H, et al. Prediction and Structural Comparison of Deleterious Coding Nonsynonymous Single Nucleotide Polymorphisms (nsSNPs) in Human LEP Gene Associated with Obesity. Biomed Res Int. 2019;2019:1832084.
Fu Y, Luo L, Luo N, Zhu X, Garvey WT. NR4A orphan nuclear receptors modulate insulin action and the glucose transport system: potential role in insulin resistance. J Biol Chem. 2007;282:31525–33.
Lanfray D, Caron A, Roy MC, Laplante M, Morin F, Leprince J, et al. Involvement of the Acyl-CoA binding domain containing 7 in the control of food intake and energy expenditure in mice. Elife. 2016;5:e11742.
Forand A, Koumakis E, Rousseau A, Sassier Y, Journe C, Merlin JF, et al. Disruption of the phosphate transporter Pit1 in hepatocytes improves glucose metabolism and insulin signaling by modulating the USP7/IRS1 interaction. Cell Rep. 2016;16(10):2736–48.
Kempe-Teufel D, Machicao F, Machann J, Böhm A, Schick F, Fritsche A, et al. Polygenic risk score of lipolysis-increasing alleles determines visceral fat mass and Proinsulin conversion. J Clin Endocrinol Metab. 2019;104(4):1090–8.
Shaaban Z, Khoradmehr A, Amiri-Yekta A, Jafarzadeh Shirazi MR, Tamadon A. Pathophysiologic mechanism of obesity - and chronic inflammation-related genes in etiology of polycystic ovary syndrome. Iran J Basic Med Sci. 2019;22(12):1378–86.
Cho ES, Lee KT, Choi JW, Jeon HJ, Lee SW, Cho YM, et al. Novel SNPs in the growth arrest and DNA damage-inducible protein 45 alpha gene (GADD45A) associated with meat quality traits in Berkshire pigs. Genet Mol Res. 2015;14(3):8581–8.
Kraja AT, Liu C, Fetterman JL, Graff M, Have CT, Gu C, et al. Associations of mitochondrial and nuclear mitochondrial variants and genes with seven metabolic traits. Am J Hum Genet. 2019;104(1):112–38.
Su X, Jin Y, Shen Y, Kim IM, Weintraub NL, Tang Y. RNAase III-Type Enzyme Dicer Regulates Mitochondrial Fatty Acid Oxidative Metabolism in Cardiac Mesenchymal Stem Cells. Int J Mol Sci. 2019;20(22):E5554.
Raj K, Ellinwood NM, Giger U. An exonic insertion in the NAGLU gene causing Mucopolysaccharidosis IIIB in schipperke dogs. Sci Rep. 2020;10(1):3170.
Yang Q, Zhao X, Xing Y, Jiang C, Jiang K, Xu P, et al. A model of mucopolysaccharidosis type IIIB in pigs. Biol Open. 2018;7(10):bio035386.
Kim T, Li D, Terasaka T, Nicholas DA, Knight VS, Yang JJ, Lawson MA. SRXN1 is necessary for resolution of GnRH-induced oxidative stress and induction of gonadotropin gene expression. Endocrinology. 2019;160(11):2543–55.
Bruel AL, Franco B, Duffourd Y, Thevenon J, Jego L, Lopez E, et al. Fifteen years of research on oral-facial-digital syndromes: from 1 to 16 causal genes. J Med Genet. 2017;54(6):371–80.
Rodriguez-Ruiz ME, Buqué A, Hensler M, Chen J, Bloy N, Petroni G, et al. Apoptotic caspases inhibit abscopal responses to radiation and identify a new prognostic biomarker for breast cancer patients. Oncoimmunology. 2019;8(11):e1655964.
Hocquette JF, Gondret F, Baeza E, Medale F, Jurie C, Pethick DW. Intramuscular fat content in meat-producing animals: development, genetic and nutritional control, and identification of putative markers. Animal. 2010;4:303–19.
Li Q, Huang Z, Zhao W, Li M, Li C. Transcriptome analysis reveals long Intergenic non-coding RNAs contributed to intramuscular fat content differences between Yorkshire and Wei pigs. Int J Mol Sci. 2020;21(5):1732.
Au WS, Payne VA, O'Rahilly S, Rochford JJ. The NR4A family of orphan nuclear receptors are not required for adipogenesis. Int J Obes. 2008;32:388–92.
Pearen MA, Goode JM, Fitzsimmons RL, Eriksson NA, Thomas GP, Cowin GJ, et al. Transgenic muscle-specific Nor-1 expression regulates multiple pathways that effect adiposity, metabolism, and endurance. Mol Endocrinol. 2013;27:1897–917.
Kim JY, Nasr A, Tfayli H, Bacha F, Michaliszyn SF, Arslanian S. Increased lipolysis, diminished adipose tissue insulin sensitivity, and impaired beta-cell function relative to adipose tissue insulin sensitivity in obese youth with impaired glucose tolerance. Diabetes. 2017;66:3085–90.
Sachs S, Zarini S, Kahn DE, Harrison KA, Perreault L, Phang T, et al. Intermuscular adipose tissue directly modulates skeletal muscle insulin sensitivity in humans. Am J Physiol Endocrinol Metab. 2019;316:E866–79.
Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25.
Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11.
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5.
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat Protoc. 2012;7:562–58.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)). Method Methods. 2001;25:402–8.
Raney BJ, Dreszer TR, Barber GP, Clawson H, Fujita PA, Wang T, et al. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC genome browser. Bioinformatics. 2014;30(7):1003–5.
Mann M, Wright PR, Backofen R. IntaRNA 20: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 2017;45:W435–9.
Wright PR, Georg J, Mann M, Sorescu DA, Richter AS, Lott S, et al. CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. Nucleic Acids Res. 2014;42:W119–23.
Acknowledgements
We thank the researchers at our laboratories for their dedication and hard work. We thank editage for their professional English language editing services.
Funding
This work was jointly supported by the National Natural Science Foundation of China (Grant No. 31872337, 31501919), China Agriculture Research System (CARS-35), and Agricultural Science and Technology Innovation Project (ASTIP-IAS02). The funding bodies had no role in the design of the study, or collection, analysis and interpretation of data and writing the manuscript.
Author information
Authors and Affiliations
Contributions
LW1 carried out the molecular genetic studies, participated in the sequence alignment and drafted the manuscript. TZ and ZZ participated in the sequence alignment. LW2 and XH participated in the design of the study and performed the statistical analysis. LZ and HY conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All methods and procedures in the study were carried out according to the standard guidelines of Experimental Animals which was established by Ministry of Science and Technology (Beijing, China). The experimental protocols were approved by the Science Research Department of the Institute of Animal Science, Chinese Academy of Agricultural Sciences (CAAS) (Beijing, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
cn- stream genes. Table S2. Primers of DE si-RNA of IRLnc.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, L., Zhou, ZY., Zhang, T. et al. IRLnc: a novel functional noncoding RNA contributes to intramuscular fat deposition. BMC Genomics 22, 95 (2021). https://doi.org/10.1186/s12864-020-07349-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-020-07349-5