Abstract
Background
Weeping lovegrass (Eragrostis curvula [Shrad.] Nees) is a perennial grass found in semi-arid regions that is well adapted for growth in sandy soils and drought conditions. E. curvula constitutes a polymorphic complex that includes cytotypes with different ploidy levels (from 2x to 8x), where most polyploids are facultative apomicts, although both sexual reproduction and full apomixis have been reported in this species. Apomixis is thought to be associated with silencing of the sexual pathway, which would involve epigenetic mechanisms. However, a correlation between small RNAs and apomixis has not yet been conclusively established.
Results
Aiming to contribute to the elucidation of their role in the expression of apomixis, we constructed small RNA libraries from sexual and apomictic E. curvula genotypes via Illumina technology, characterized the small RNA populations, and conducted differential expression analysis by comparing these small RNAs with the E. curvula reference transcriptome. We found that the expression of two genes is repressed in the sexual genotype, which is associated with specific microRNA expression.
Conclusion
Our results support the hypothesis that in E. curvula the expression of apomixis leads to sexual repression.
Similar content being viewed by others
Background
Weeping lovegrass (Eragrostis curvula [Shrad.] Nees) is a perennial grass of the Poaceae family, subfamily Chloridoideae [1]. This native grass of Southern Africa is found in many semi-arid regions of the world since it is able to adapt to sandy soils and drought conditions, thus representing an excellent forage resource for marginal regions [2]. The genus Eragrostis comprises more than 250 species, with E. curvula and E. tef the best studied to date [3,4,5,6]. Eragrostis is characterized by a basic number of x = 10 chromosomes [7]. E. curvula constitutes a complex that includes cytotypes with different ploidy levels (from 2x to 8x). Diploid (2n = 2x = 20) plants are rare and reproduce sexually [8]. By contrast, most E. curvula polyploids are facultative apomicts, although both sexual reproduction and full apomixis have also been reported [9].
In E. curvula, reproduction occurs via pseudogamous diplosporous apomixis [10, 11], where a megasporocyte undergoes two rounds of mitotic division to form a nonreduced tetranucleate embryo sac with an egg, two synergids, and one polar nucleus [12, 13]. Diplospory has been found in species from the Asteraceae, Solanaceae, Rosaceae, Poaceae, and Brassicaceae families, occurring in plants belonging to 68 genera [14,15,16]. Embryo sac development in E. curvula occurs via Eragrostis-type apomixis, a process specific to this grass, which contains only four nonreduced nuclei at maturity [10]. E. curvula embryos are parthenogenetic, but fertilization of the polar nucleus is required for endosperm development.
Apomictic reproduction involves a complex set of mechanisms that result in asexual seed production without meiosis or fertilization; thus, the resulting embryo is genetically identical to its maternal parent [17, 18]. During evolution, apomixis emerged independently multiple times, as evidenced by the variety of apomictic mechanisms described and the many occurrences of apomictic species observed in phylogenetic analyses of angiosperms [19, 20]. Apomixis is inherited as a dominant trait [17]. Although early genetic studies proposed that a single, dominant locus controls apomixis, the developmental stages of apomixis, i.e., meiotic avoidance, parthenogenesis, and fertilization-independent endosperm development, are controlled by independent loci in some species [16, 17, 21].
Some apomixis loci have been localized to low-recombination regions and appear to be associated with heterochromatin and/or substantially repetitive sequences [17]. Although such chromosomal structures and apomixis are thought to be linked, recent evidence suggests that it is more likely that these repetitive chromosomal structures occurred as a consequence of asexual reproduction and suppressed recombination, which may have evolved to maintain the genic elements required for apomixis [17].
Assuming that apomixis is a consequence of spatial and temporal changes in expression of sexual pathway-related genes [21], differential expression analysis between stages and/or tissues of sexual and apomictic genotypes represents a useful tool for unraveling the transcriptional pathways involved in this reproductive mode. Stress-like genes are expressed in aposporous initial cells in Brachiaria brizantha [22] and Hieracium praealtum [23], suggesting these genes play a role in the induction of apospory. In B. brizantha, these genes encode proteins including a helicase and a MADS-box transcription factor [22, 24].
Comparative analysis between apomictic and sexual Boechera species revealed a global decrease in gene expression within the apomict versus sexual species during the early stages of development [25]. Comparative gene expression studies of diplosporous species should focus on identifying genes involved in the avoidance of meiosis by the megaspore mother cell. Unlike the case in aposporic species, the frequency of apomixis in E. curvula, as evidenced by the proportion of sexual-to-apomictic embryo sacs, increases under stress conditions, such as in vitro culture, polyploidization [4], and water deprivation [26]. Several genes are thought to be involved in apomixis in this species, including methyltransferases, kinases and transposon sequences [3, 27,28,29], but conclusive evidence for their roles in this process is currently lacking.
The involvement of epigenetic mechanisms in the regulation and/or expression of apomixis also has been studied in various apomictic species. Apomixis is thought to result from epigenetic deregulation of the sexual pathway, thus accounting for the facultative nature of apomixis. The increase of sexual embryo sacs in E. curvula under different stress conditions shows that sexual and apomictic pathways coexist in facultative apomicts and that stress deregulates the silencing of the sexual one, proving the epigenetic nature of this silencing [26]. Small RNAs (sRNAs) play roles in this epigenetic mechanism, as they are huge contributors to the phenotypic plasticity of plants and are involved in plant development, reproduction, and genome reprogramming [30].
The major classes of sRNAs in plants include microRNAs (miRNAs), hairpin-derived small-interfering RNAs (hp-siRNAs), natural antisense siRNAs (natsiRNAs), secondary siRNAs, and heterochromatic siRNAs (hetsiRNAs). miRNAs are transcribed from single-stranded hairpin-folded RNA molecules known as long primary microRNAs (pri-miRNAs) by RNA Polymerase II [30]. The enzyme Dicer-like 1 initially produces small stem-loop precursor microRNAs (pre-miRNAs) from pri-miRNA, and mature miRNA duplexes consisting of the active miRNA strand and its complementary strand (typically 20 to 22 nucleotides long) are subsequently produced [30]. One strand from the initial duplex associates with an Argonaute (AGO) protein and hybridizes with target RNAs, a process guided by Watson-Crick pairing [30]. AGO-miRNA complexes are involved in post-transcriptional gene silencing via messenger RNA (mRNA) cleavage or translational repression [31]. Most miRNA genes are species- or family-specific, suggesting that they evolve rapidly and have high turnover rates [32].
AGO genes in sexual plants influence the number of cells that have the capacity to initiate embryo sac development [33, 34]. For example, the phenotype of maize ago104 mutants mimics diplospory, with diploid gametes originating from a single megaspore mother cell per ovule undergoing mitosis [34]. The repression of the somatic fate of germ cells might result from the accumulation of AGO104 protein surrounding the megaspore mother cell [34]. In E. curvula, in situ hybridization studies suggest that EcAGO104 could have a similar function as maize AGO104, as evidenced by its differential regulation during male and female reproductive developments between apomictic and sexual plants [29].
Several studies have focused on the expression of sRNAs in apomict and sexual plants, but no correlation between sRNAs and apomixis has thus far been conclusively established [35, 36]. The use of various types of differential gene expression analyses of small and large RNAs between apomictic and sexual individuals of a single species represents a promising approach for investigating the regulation of apomixis. Here, we developed sRNA libraries from apomictic and sexual E. curvula genotypes, providing a valuable tool for studying the roles of these molecules in the expression of apomixis. We identified a set of genes that were repressed in the sexual genotypes via a compatible miRNA-mRNA interaction, supporting the hypothesis that apomixis in E. curvula leads to sexual repression.
Results
Characterization of the sRNA library profiles
To elucidate the epigenetic mechanisms involved in the regulation of reproductive behavior in E. curvula, we developed four sRNA libraries via Illumina sequencing technology. The sRNA fraction was obtained from spikelets at all developmental stages from two E. curvula genotypes with contrasting reproductive modes: the apomictic cultivar ‘Tanganyika’ and the sexual cultivar ‘OTA-S’. For sequencing purposes, two biological replicates of each genotype were used and resulted in four libraries, hereafter referred to as T3P1 and T3P2 (from Tanganyika inflorescences) and O2P1 and O2P2 (from OTA inflorescences). Near ~ 40 million clean reads were generated per library, with an average length of ~ 23 bp and two peaks positioned at 21 and 24 bp, as expected (Table 1; Fig. 1). The quality scores were classified as very good (green); the percentage of adaptors, N content, GC content, and level of sequence duplication indicated that the four libraries were suitable for analysis (Additional file 1: Figure S1 and Additional file 2: Figure S2). The reads were deposited in the Sequence Reads Archive (SRA) database at NCBI as BioProject PRJNA378998 “Eragrostis curvula sRNA raw sequence reads” including Biosamples SAMN06564601 (biological replicates of reads from Tanganyika) and SAMN06564600 (biological replicates of reads from OTA-S). Sequence tags were labeled with the name of the library and the read count of each individual sequence. Statistics of the libraries depuration are shown in Tables 1 and 2.
Identification of conserved miRNAs
The sequence tags were aligned against miRBase 22.0, revealing that ~ 0.4% of each library corresponded to conserved miRNAs (Table 3a, b), for a total of 469,504 sequence tags. We analyzed the miRNAs by family, finding that ~ 75% of the sequence tags were distributed among 74 miRNA families (Additional file 4: Table S1). The greatest number of miRNAs belonged to the miR2275 and miR156 families, followed by miR396, miR827, miR169, miR5072, miR-8175, and MIR894 (Additional file 4: Table S1). We focused on the most highly conserved families and designed primers to validate their presence in biological samples (Table 4). qRT-PCR analysis of E. curvula miRNAs revealed miRNAs matching ata-miR2275a-3p, zma-miR2275c-3p, gma-miR156d, stu-miR156d-3p, ata-miR396b-5p, ssp-miR827, ath-miR-8175, and ppt-MIR894, which we named ecu-miR2275a, ecu-miR2275b, ecu-miR156a, ecu-miR156b, ecu-miR396, ecu-miR827, ecu-miR-8175, and ecu-miR894, respectively (Fig. 2). qRT-PCR confirmed that there were no significant differences in the expression levels of these miRNAs between genotypes, as suggested by the RT-PCR (Fig. 2).
Specific amplification of conserved miRNAs from E. curvula. For clarity, a single run per genotype is shown. The miRNA assayed is listed above each amplification band. O, cDNA from genotype OTA-2; T, cDNA from genotype Tanganyika; (−), negative control; M, molecular weight markers. The band at the bottom is 100 bp
Identification of miRNA targets
Given the lack of information about E. curvula miRNAs and its genome, which is restricted to the sexual diploid Victoria [5], we aimed at identifying the miRNAs based on their possible targets.
A huge amount of possible targets were retrieved in the interface psRNATarget (http://plantgrn.noble.org/psRNATarget/) [37] output. Due to our inability to validate such a large set of data, we selected several miRNAs and their targets, focusing on the possible differential representation of miRNAs and/or transcripts according to the reproductive mode of the plant and the function of the encoded protein.
Two novel miRNA, GAACTGTTAGAGTTTGCCGCG and TATATTTTGAAACAGAGGGAG that shares partial identity with osa-miR812k and hvu-miR5049b, respectively and thus were named ecu-miR821 and ecu-miR5049, and a third one, ecu-miR8175, were found to be in silico differentially expressed in the sexual genotype OTA-S (logFC > |2|; p value < 0.01) and was further validated on cDNA samples by qRT-PCR (Fig. 3a). Our interest was focused on these miRNAs since the first one targets MADS-box transcription factor 6 (encoded by isotig28724) that was expressed in the apomictic genotype and the second one targets an uncharacterized transcript coded by isotig46613, only detected in cDNA obtained from OTA-2 (Fig. 3b), suggesting that the regulation of the expression of this mRNA could not be attributed exclusively to this miRNA. The third one, ecu-miR8175, targets isotig39554 (GFVM01039049.1), encoding a transposable element protein, expressed exclusively in the apomictic genotype Tanganyika (Fig. 3b).
We searched for targets of the conserved miRNAs and analyzed a subset of mRNA targets. Among the conserved miRNAs that targeted E. curvula transcripts, the greatest numbers belonged to the miR156 and miR8175 families. Two members of the miR156 family, ecu-mir156a and ecu-mir156b, were identified in our libraries, which share identity with gma-miR156d and stu-miR156d-3p, respectively (Table 4). ecu-mir156a, which targets isotig18002, isotig37981, and isotig40670, encodes a squamosa promoter-binding-like (SPL) protein. These targets were successfully amplified from OTA-2 and Tanganyka genomic DNA (Fig. 4). However, no amplification product was obtained when we assayed biological replicates of cDNAs from both genotypes, which is in agreement with the notion that ecu-miR156a (Fig. 2) plays a role in regulating mRNAs predicted to be isotig18002, isotig37981, and isotig40670.
Other targets of the conserved miRNAs identified through the interface psRNATarget include the following: ecu-miR396 targets isotig19537 (encoding a growth-regulating factor family protein); ecu-miR827 targets isotig22795 (encoding an SPX domain-containing membrane protein); ecu-miR2275a targets isotig16120 (encoding a membrane-anchored ubiquitin-fold protein); ecu-miR2275b targets isotig35874 (encoding a 3-phosphoinositide-dependent protein kinase). No mRNA targets were identified for ecu-mir894.
Discussion
Apomixis has emerged independently multiple times during angiosperm evolution, resulting in various apomictic mechanisms [19, 20]. These mechanisms might involve epigenetic silencing of the sexual pathway or, as recently reviewed by [38], of the apomictic one, if the apomictic state is considered to be the baseline, with its silencing having allowed the development of sexuality. In both cases, the epigenetic silencing could be accounted for by the facultative nature of apomixis. Since miRNAs contribute to the phenotypic plasticity of plants and are involved in development, reproduction, and genome reprogramming [30], they represent exciting candidates for studying the modulation of the expression of apomixis in E. curvula. The existence of natural sexual and apomictic tetraploid genotypes makes this species an invaluable model system for studying apomixis, particularly considering the Eragrostis-type of diplosporous apomixis described in E. curvula [13]. Elucidating the mechanisms underlying apomixis in E. curvula would have a strong impact on agriculture at the global level, as it would provide molecular tools to facilitate the transfer of apomixis to species of agronomic interest. The scientific interest in E. curvula prompted us to provide EST [39] and full transcriptome data [3] for this species to the scientific community.
The possible link between sRNAs and apomixis remains unclear. Therefore, differential expression analysis combining small and large RNAs from E. curvula genotypes with contrasting reproductive modes represents a valuable approach for investigating the mechanism regulating apomixis. In the present study, we produced sRNA libraries from the spikelets of apomictic and sexual genotypes of E. curvula via Illumina sequencing technology, which met the quality criterion needed to be used for sRNA characterization in this species. To the best of our knowledge, this is the first available set of sRNA data for this species, which has the added benefit of being obtained from genotypes with contrasting reproductive modes. This dataset, combined with previous transcriptomic data, allowed us to identify conserved miRNAs and their targets.
As expected, when we compared the sequences to those in miRBase, a small proportion (~ 0.4%) of the sequence tags showed identity with miRNAs from the database and were further classified as conserved miRNAs. A total of 75 miRNA families covered 75% of the matching sequence tags, whereas other miRNA families were not as well represented in E. curvula. Moreover, nearly 10% of the miRNAs from miRBase were detected in our libraries, suggesting that non-conserved miRNAs could play roles in the regulation of translation in E. curvula. Most of the miRNAs belonged to the miR2275 family, which trigger the biogenesis of 24-nucleotide phased sRNAs [40], as well as the miR156 family; these miRNAs are mainly expressed during the early stages of shoot development and are involved in repressing the transition from the juvenile to the adult phase of vegetative development by inhibiting the cleavage of their target SPL protein [41]. At the stages included in this analysis, ecu-miR156a was expressed in both E. curvula genotypes. Given that SPL genes were amplified from genomic DNA, the most plausible explanation is that ecu-miR156a is repressing the expression of the mRNA encoding SPL. Interestingly, seven additional transcripts encoding SPL isoforms were identified in the reference transcriptome, but they are not targeted by miR156, as revealed by analysis using the psRNATarget interface.
Based on its similarity to the transposable element protein retrotrans_gag (ABA97988.1), isotig39554 (GFVM01039049.1) is a putative transposable element; its expression was repressed in the sexual genotype by ecu-miR8175 overexpression, whereas it was expressed in the apomictic genotype. Ecu-miR8175 expression is modulated under various conditions [42,43,44], but this is the first time that a link was proposed between the differential expression of this miRNA and the reproductive mode. Previously reported targets for miR8175 include an aquaporin-like superfamily gene in barley [42], an ABA-dependent kinase gene in Arabidopsis thaliana [45], and several unrelated genes in Jatropha curcas [46], suggesting that even when a miRNA is conserved across species, its roles can vary.
Conclusions
In the present study, we explored the role of miRNA-mRNA interactions in apomixis in the grass E. curvula using a combination of previously acquired transcriptomic data [3] and newly developed small RNA libraries. Supporting the hypothesis that apomixis results from the deregulation of the sexual pathway, we found that two genes, a MADS-box transcription factor gene and a transposon, were specifically repressed in the sexual genotype, most likely due to interactions with miRNAs. These results strongly suggest that miRNA is involved in the expression of apomixis in E. curvula via its repressive effects on gene expression. Further studies are needed to identify the link among these genes.
Methods
Plant materials
Two tetraploid accessions of Eragrostis curvula (weeping lovegrass) provided by the United States Department of Agriculture (USDA) were used in this work: the diplosporous apomictic genotype ‘Tanganyika’ (USDA: PI234217) (2n = 4× = 40) and the sexual genotype ‘OTA-S’ (USDA: PI574506) (2n = 4× = 40). Both genotypes were grown in a greenhouse under natural light conditions at 25 °C.
RNA extraction and sequencing
Four RNA samples were prepared: O2P1 and O2P2 were biological replicates derived from spikelets with basal flowers at the beginning of anthesis from two plants of the sexual genotype OTA-S; T3P1 and T3P2 were biological replicates obtained from spikelets of two plants of the apomictic genotype Tanganyika. For both genotypes spikelets with basal flowers at the beginning of anthesis were collected for RNA extraction based on the reproductive calendar showed in Selva et al., [29] where all developmental stages are represented, from early archespores to mature female gametophytes. The development of the E. curvula spikelet is typically heterochronic. Basal flowers within a spikelet are more developed than flowers at the top of the spikelet. In sexual genotype, from the top to the base we found flowers containing ovules at archesporial cell, megaspore mother cell, embryo sacs with bi, tetra and octanucleate stage and mature embryo sacs with proliferating antipodal cells. Apomictic genotype shows archesporial cell, megaspore mother cell, megaspore mother cell elongated, bi and tetranucleate stage and mature embryo sac without antipodal cells.
For each sample, 30 mg of fresh tissue was ground to a fine powder in liquid nitrogen. Total RNA was extracted from the plant tissue as two fractions, small and large RNA, including RNA sequences smaller and larger than 200 bp, respectively, using a commercial RNA purification kit (Macherey-Nagel) according to the manufacturer’s instructions. The RNA was resuspended in diethylpyrocarbonate (DEPC)-treated water. The large RNA fraction was used for cDNA library construction [3], and the sRNA fraction was quantified by UV spectrometry (260 nm). The amount of sRNA ranged from 0.8 to 1.4 μg. sRNA library preparation and sequencing were conducted by GenXPro GmbH (Germany). Briefly, RNA was successively ligated to modified 3′ and 5′ adapters (TrueQuant RNA adapters, GenXPro), followed by reverse transcription and PCR amplification using specific primers. The amplified libraries were size-selected by polyacrylamide gel electrophoresis. The miRNA population, containing flanking p5 and p7 adapters, was sequenced on a HiSeq 2000 (Illumina).
Bioinformatic analysis
After trimming the reads for adaptors and low-quality sequences as well as sequences shorter than 17 nt and longer than 33 nt, the occurrence of each unique sequence read was counted. The four individual libraries were separately collapsed by aligned using Bowtie (1.1.2) under the following parameters: --wrapper basic-0 -f -v 0 -a --best --strata -S --sam. Following the statistical criteria generally accepted for sRNA library analysis, sequence tags with counts < 3 were discarded. The remaining sequence tags were compared with the sequences of noncoding RNAs families (rRNA, tRNA, snRNA, and snoRNA) via alignment using Bowtie against the Rfam database (version 12.2) to identify degradation fragments of noncoding RNAs. Sequence tags sharing identity with mitochondrial and chloroplastic DNA sequences were also discarded.
Identification of conserved miRNAs
To identify and annotate conserved miRNAs, the four sequence datasets obtained after the bioinformatics analysis described above were mapped onto the miRNA precursors and mature miRNAs from miRBase (release 22, http://www.mirbase.org/) using local BLAST without gaps and a maximum number of mismatches of 2, excluding hits with coverage < 90%. Unique reads that mapped to known miRNAs were classified as conserved miRNAs.
Identification of novel miRNAs and mRNA targets
Reads were classified as novel miRNAs based on the identification of their possible targets. The sequence tags from the four libraries were subjected to BLAST analysis against the reference transcriptome, allowing no mismatches and no gaps. Sequence tags that matched each transcript were graphed and manually analyzed. When a transcript showed reads targeting across its entire length, the reads were considered to be degradation products and were discarded as possible novel miRNAs. The remaining sequence tags were used as input for further analysis through the interface psRNATarget (http://plantgrn.noble.org/psRNATarget/) [37] with the following parameters: 1) maximum expectation: 2.5 (range: 0–5.0); 2) length for complementary scoring (hsp size): 20 (range: 15–30 bp); 3) target accessibility – allowed maximum energy to un-pair the target site (UPE): 20 (range: 0–100); 4) seed region: 2–13; 5) mismatches allowed in the seed region: 0; 6) flanking length around target site for target accessibility analysis: 17 bp upstream/13 bp downstream; 7) range of central mismatch leading to translational inhibition: 9–11 nt; and 8) bulges on the target sequence in the alignment with miRNA not allowed. Previous trimming of degradation products reduced the number of possible miRNA candidates, thereby increasing the accuracy of the predicted data.
Quantitative RT-PCR (qRT-PCR) and stem-loop qRT-PCR
For the validation assays, total RNA was isolated from Tanganyika and OTA-S spikelets collected under the same criteria used to obtain the samples for sequencing. However, an alternative, two-phased protocol was used for RNA isolation. The first phase involved a modified cetyltrimethylammonium bromide (CTAB)-based protocol [47]. The tissues (~ 35 mg) from two biological replicates per sample were homogenized in liquid nitrogen and combined with CTAB extraction buffer (100 mM Tris HCl, pH 8, 1.4 M NaCl, 20 mM EDTA pH 8, 2% [w/v] CTAB, and 0.5% β-mercaptoethanol [v/v]). After incubation at 65 °C for 10 min, chloroform was added and the sample was centrifuged at 12,000 x g (10 min). RNA was precipitated from the aqueous phase overnight on ice using 3 M LiCl, in the second phase, samples obtained in phase I were mixed with TRIzol™ Reagent (Invitrogen). Following the addition of chloroform, the homogenate was separated into three phases via centrifugation. RNA was precipitated from the upper layer with isopropanol and washed with ethanol. The samples were resuspended in 20–25 μl of DEPC water and stored at − 80 °C. The RNA was quantified using a DeNovix spectrophotometer and its integrity checked by agarose gel electrophoreses.
The validation of the conserved miRNAs was carried out as follows: cDNA was synthetized from 3 μg of total DNA-free RNA per sample using a Mir-X miRNA First-Strand Synthesis kit (Clontech Laboratories, Inc.) following the manufacturer’s recommendations. Basically, in a one-step reaction, RNA was polyadenylated and reverse transcribed using poly(A) polymerase and SMART® MMLV. The cDNA working solutions were obtained from stocks via 10-fold dilution. To measure conserved miRNA expression, with the predicted conserved miRNAs used as forward primers and the reverse primers provided in the Mir-X miRNA First-Strand Synthesis kit (Table 5). RT-PCR was conducted in a 25-μL reaction volume containing 1.5 μl cDNA template, 0.25 μM of each primer, 0.8 mM dNTPs, 1.5 mM MgCl2, 1X Taq buffer, and 1 U Taq polymerase (Invitrogen, Brazil) using a MyCycler Thermal Cycler (Bio-Rad, Hercules, CA, USA). The cycling conditions were as follows: 1) denaturation step, 2 min at 94 °C; 2) 38 cycles of 94 °C for 15 s, 20 s at the optimal annealing temperature for each primer pair, and 72 °C for 30 s; and 3) extension step, 72 °C for 3 min. The amplification products were separated by 2% (w/v) agarose gel electrophoresis, visualized after staining with ethidium bromide under UV light, and imaged with a Kodak Easy share Z7590 zoom digital camera. Quantitative reverse-transcription PCR (qRT-PCR) was conducted in reactions including 50 pmol of forward and reverse primers, 5 μl of 100-fold diluted cDNA, and 10 μl of Real Mix (Biodynamics, Buenos Aires, Argentina). Amplifications were carried out in a Rotor Gene 6000 (Corbett Research, Sydney, Australia). Cycling consisted of 94 °C for 2 min, followed by 45 cycles at 94 °C for 15 s, 20 s at the optimal annealing temperature for each primer pair, and 72 °C for 30 s. The specificity of the amplification products was verified by melting curve analysis performed using 1-s cycles from 72 to 95 °C, increasing the temperature by 0.5 °C per cycle. Actin housekeeping gene was amplified using E. curvula-based primers (F: 5′- AATGAGCTCCGTGTAGCACCAGAA-3′; R: 5′-ACATGGCTGGAACACTGAAGGTCT-3′) as described previously [27].
Novel miRNA validation was conducted via stem-loop validation assays [48]. RNA was extracted using a NucleoSpin® miRNA kit (Macherey-Nagel, Düren, Germany). The ImProm-II™ reverse transcription system (Promega) was used to reverse transcribe 20 ng of the small fraction of RNA from OTA-S and Tanganyika spikelets using 50 nM stem-loop RT primer, designed as described in [48] (Table 5). Each 20-μl reaction was incubated in a BioRad Thermocycler for 5 min at 25 °C, 60 min at 42 °C, 15 min at 70 °C (to inactivate the reverse transcriptase), and held at 4 °C. The reaction mixture for qRT-PCR consisted of 0.5 μM of a forward primer specific for each miRNA, 0.5 μM of universal reverse stem-loop [48], 1 μl of RT product, and 7.5 μl of 2x Real Mix (Biodynamics, Buenos Aires, Argentina). The cycling conditions were similar to those used for cDNA amplification (95 °C for 5 min, cycle 95 °C for 5 s, 60 °C for 10 s, and 72 °C for 1 s). A melting curve was produced at the end of each reaction to check the specificity of amplification (ramp up from 65 to 95 °C by 0.2 °C per step, waiting 1 s after each step). The qRT-PCR was performed in technical triplicates of two biological samples from OTA-S and Tanganyika spikelets. The 2-ΔΔCt method [49] was used to normalize transcript levels between cDNA samples. Amplification efficiency was calculated with Rotor-Gene 6000 Series Software 1.7 for the samples and the corresponding internal control. Experiments were conducted on technical triplicates of three biological samples. Differences between mean values were evaluated by Student’s t-tests. P values< 0.05 were considered significant.
Validation assays of mRNA targets were conducted, using technical triplicates of two biological replicates. To carry out this purpose, specific primers were designed using Primer3 (http://bioinfo.ut.ee/primer3), synthesized by Macrogen (Seoul, Korea), and assayed using cDNA samples from plants with sexual and apomictic genotypes (Table 5).
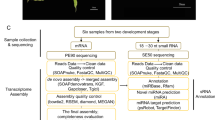
A flowchart diagram was done to illustrate the criteria followed in the methodology of the manuscript (Additional file 3: Figure S3).
Differential expression analyses
Differential expression between two conditions was analyzed using the EdgeR package [50], and P-values adjusted using the Benjamini and Hochberg method [51], to control the false discovery rate. The corrected P-value of 0.01 and log2 (fold-change) of 2 were set as the threshold for significant differential expression.
Availability of data and materials
The datasets generated for this study can be found in NCBI, BioProject PRJNA378998 “Eragrostis curvula sRNA raw sequence reads”, including Biosamples SAMN06564601 (biological replicates of reads from Tanganyika) and SAMN06564600 (biological replicates of reads from OTA-S).
Abbreviations
- AGO:
-
Argonaute
- EST:
-
Expressed sequence tag
- hetsiRNA:
-
heterochromatic siRNA
- hp-siRNA:
-
hairpin-derived siRNA
- LogFC:
-
Logarithm fold change
- miRNA:
-
microRNA
- mRNA:
-
messenger RNA
- natsiRNA:
-
natural antisense siRNA
- O2P1 and O2P2:
-
RNA sampled from OTA, plants 1 and 2, respectively
- pre-miRNAs:
-
Stem-loop precursor microRNAs
- pri-miRNA:
-
primary microRNA
- qRT-PCR:
-
Real-time reverse transcription-PCR
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- siRNA:
-
small-interfering RNA
- SPL:
-
Squamosa-promoter binding-like protein
- sRNA:
-
small RNA
- T3P1 and T3P2:
-
RNA sampled from Tanganyika, plants 1 and 2, respectively
References
Watson L, Dallwitz MJ. The grass genera of the world. CAB international: Oxfordshire; 1992.
Covas G, Cairnie A. Introducción del pasto lloron en la Argentina. In: Fernandez O, Brevedan RE, Gargajo A, editors. El Pasto lloron: su biología y manejo. Bahía Blanca: CERZOS; 1991. p. 1–6.
Garbus I, Romero JR, Selva JP, Pasten MC, Chinestra C, Carballo J, et al. De novo transcriptome sequencing and assembly from apomictic and sexual Eragrostis curvula genotypes. PLoS One. 2017;12:e0185595.
Zappacosta DC, Ochogavía AC, Rodrigo JM, Romero JR, Meier MS, Garbus I, et al. Increased apomixis expression concurrent with genetic and epigenetic variation in a newly synthesized Eragrostis curvula polyploid. Sci Rep. 2014;4:4423.
Carballo J, Santos BACM, Zappacosta D, Garbus I, Selva JP, Gallo C, Díaz A, Albertini E, Caccamo M, Echenique V. A high-quality genome of Eragrostis curvula grass provides insights into Poaceae evolution and supports new strategies to enhance forage quality. Sci Rep. 2019;9(1):10250.
Assefa K, Cannarozzi G, Girmam D, Kamiesm R, Chanyalewm S, Plaza-Wüthrich S, et al. Genetic diversity in tef [Eragrostis tef (Zucc.) trotter]. Front Plant Sci. 2015;26:177.
Streetman LJ. Reproduction of the Lovegrasses, the genus Eragrostis-IE chloromelas Steud. E. curvula (Schrad.) Nees, E. lehmanniana Nees and E. superba Peyr. Wrightia. 1963;3:41–51.
Voigt P. Discovery of sexuality in Eragrostis curvula (Schrad.) Nees. Crop Sci. 1971;11:424–5.
Voigt P, Bashaw E. Facultative apomixis in Eragrostis curvula. Crop Sci. 1976;16:803–5.
Crane C. Classification of apomictic mechanisms. In: Savidan Y, Carman JG, Dresselhaus T, editors. Flowering of Apomixis: from mechanisms to genetic engineering. Mexico City: CIMMYT; 2001. p. 168–211.
Voigt P, Rethman N, Poverene M. Lovegrasses. In: Moser LE, Burson BL, LE, editors. Warm-season (C4) grass. Madison: American Society of Agronomy; 2004. p. 1027–56.
Asker S E, Jerling L. Apomixis in Plants. Boca Raton: CRC Press, Inc.; 1992.
Meier M, Zappacosta D, Selva JP, Pessino S, Echenique V. Evaluation of different methods for assessing the reproductive mode of weeping lovegrass plants, Eragrostis curvula (Schrad.) Nees. Aust J Bot. 2011;59:253–61.
Hojsgaard D, Klatt S, Baier R, Carman JG, Hörandl E. Taxonomy and biogeography of apomixis in angiosperms and associated biodiversity characteristics. Crit Rev Plant Sci. 2014;33:414–27.
Nogler G. Gametophytic apomixis. In: Embryology of Angiosperms, edited by B. M. Johri. Springer, Berlin; 1984. 475–518.
Barcaccia G, Albertini E. Apomixis in plant reproduction: a novel perspective on an old dilemma. Plant Reprod. 2013;26:159–79.
Hand ML, Koltunow AM. The genetic control of apomixis: asexual seed formation. Genetics. 2014;197:441–50.
Brukhin V. Molecular and genetic regulation of apomixis. Russ J Genet. 2017;53:943–64.
Carman JG. Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol J Linn Soc. 1997;61:51–94.
Van Dijk P, Vijverberg K. The significance of apomixis in the evolution of the angiosperms: a reappraisal. In: Bakker F, Chatrou L, Gravendeel B, Pelser PB, editors. eds Plant species-level systematics: new perspectives on pattern and process. Ruggell: Gantner Verlag; 2005. p. 101–16.
Grossniklaus U. From sexuality to apomixis: molecular and genetic approaches. In: Savidan Y, Carman JG, Dresselhaus T, editors. Eds The Flowering of Apomixis: from Mechanisms to Genetic Engineering. Mexico: D.F. CIMMYT; 2001. p. 168–211.
Silveira ED, Guimaraes LA, de Alencar Dusi DM, Da Silva FR, Martins NF, do Carmo Costa MM, et al. Expressed sequence-tag analysis of ovaries of Brachiaria brizantha reveals genes associated with the early steps of embryo sac differentiation of apomictic plants. Plant Cell Rep. 2012;31(2):403–16.
Okada T, Hu Y, Tucker MR, Taylor JM, Johnson SD, Spriggs A, et al. Enlarging cells initiating apomixis in Hieracium praealtum transition to an embryo sac program prior to entering mitosis. Plant Physiol. 2013:113.
Guimarães DA, Dusi DMA, Masiero S, Resentini S, Gomes AM, Silveira D, et al. BbrizAGL6 is differentially expressed during embryo sacformation of apomictic and sexual Brachiaria brizantha plants. Plant Mol Biol Rep. 2013;31:1397–406.
Sharbel TF, Voigt ML, Corral JM, Galla G, Kumlehn J, Klukas C, et al. Apomictic and sexual ovules of Boechera display heterochronic global gene expression patterns. Plant Cell. 2010;22:655–71.
Rodrigo JM, Zappacosta DC, Selva JP, Garbus I, Albertini E, Echenique V. Apomixis frequency under stress conditions in weeping lovegrass (Eragrostis curvula). PLoS One. 2017;12(4):e0175852.
Selva JP, Pessino SC, Meier MS, Echenique VC. Identification of candidate genes related to polyploidy and/or apomixis in Eragrostis curvula. Am J Plant Sci. 2012;19:403.
Romero J, Selva JP, Pessino S, Echenique VC, Garbus I. Repetitive sequences in Eragrostis curvula. cDNA EST libraries obtained from genotypes with different ploidy. Biol Plant. 2016;60:55–67.
Selva JP, Siena L, Rodrigo JM, Garbus I, Zappacosta D, Romero JR, et al. Temporal and spatial expression of genes involved in DNA methylation during reproductive development of sexual and apomictic Eragrostis curvula. Sci Rep. 2017;7:15092.
Borges F, Martienssen RA. The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol. 2015;16:727–41.
Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet. 2013;14:447–59.
Cuperus JT, Fahlgren N, Carrington JC. Evolution and functional diversification of MIRNA genes. Plant Cell. 2011;23:431–42.
Olmedo-Monfil V, Durán-Figueroa N, Arteaga-Vázquez M, Demesa-Arévalo E, Autran D, Grimanelli D, et al. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature. 2010;464:628.
Singh M, Goel S, Meeley RB, Dantec C, Parrinello H, Michaud C, et al. Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell. 2011. https://doi.org/10.1105/tpc.110.079020.
Amiteye S, Corral JM, Vogel H, Sharbel TF. Analysis of conserved microRNAs in floral tissues of sexual and apomictic Boechera species. BMC Genomics. 2011;12:500.
Galla G, Volpato M, Sharbel TF, Barcaccia G. Computational identification of conserved microRNAs and their putative targets in the Hypericum perforatum L. flower transcriptome. Plant Rep. 2013;26:209–29.
Dehury B, Panda D, Sahu J, Sahu M, Sarma K, Barooah M, et al. In silico identification and characterization of conserved miRNAs and their target genes in sweet potato (Ipomoea batatas L.) expressed sequence tags (ESTs). Plant Signal Behav. 2013;8(12):e26543.
Albertini E, Barcaccia G, Pupilli F, Carman J. Did apomixis evolve from sex or was it the other way around? J Exp Botany. 2019; in press.
Cervigni GD, Paniego N, Díaz M, Selva JP, Zappacosta D, Zanazzi D, et al. Expressed sequence tag analysis and development of gene associated markers in a near-isogenic plant system of Eragrostis curvula. Plant Mol Biol. 2008;67:1–10.
Song X, Li P, Zhai J, Zhou M, Ma L, Liu B, et al. Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J. 2012;69:462–74.
Xu M, Hu T, Zhao J, Park M-Y, Earley KW, Wu GW, et al. Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet. 2016;12:e1006263.
Wu L, Yu J, Shen Q, Huang L, Wu D, Zhang G. Identification of microRNAs in response to aluminum stress in the roots of Tibetan wild barley and cultivated barley. BMC Genomics. 2018;19:560.
Ghosh S, Singh K, Shaw AK, Azahar I, Adhikari S, Ghosh U, et al. Insights into the miRNA-mediated response of maize leaf to arsenate stress. Environ Exper Bot. 2017;137:96–109.
Sarkar D, Maji RK, Dey S, Sarkar A, Ghosh Z, Kundu P. Integrated miRNA and mRNA expression profiling reveals the response regulators of a susceptible tomato cultivar to early blight disease. DNA Res. 2017;24:235–50.
Liu Y, El-Kassaby YA. Regulatory crosstalk between microRNAs and hormone signalling cascades controls the variation on seed dormancy phenotype at Arabidopsis thaliana seed set. Plant Cell Rep. 2017;36:705–17.
Prakash VN, Jadeja VJ. Identification and characterization of miRNA transcriptome in Jatropha curcas by high-throughput sequencing. Plant Gene. 2018;15:44–50.
Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Report. 1993;11:113–6.
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, et al. Real-time quantification of microRNAs by stem–loop RT–PCR. Nucleic Acids Res. 2005;33:e179. https://doi.org/10.1093/nar/gni178.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25:402–8.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:1.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300.
Acknowledgements
Not applicable.
Funding
This work was granted by the Agencia Nacional de Promoción CientÍfica y Tecnológica (ANPCyT, PICT 2013–2029 to I.G. and PICT Raíces 2014–1243 to V.E), CONICET (PIP 11220110100767 to V.E and 11220150100963 to I.G) and the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie Grant Agreement No 645674 to E.A. Roles of the funding bodies: PIP 11220110100767: study design and sample collection; libraries construction and sequencing. PICT 2013–2029, PIP 11220150100963 and PICT Raíces 2014–1243: molecular data acquisition, analysis, and interpretation; molecular biology supplies. European Union’s Horizon 2020: writing and publishing founding.
Author information
Authors and Affiliations
Contributions
Conception: IG, VE; Experimental design: IG; Materials: JPS, VE; Experimental procedures: MCP, JPS, AB; Bioinformatic data processing: IG, JC; Analysis and interpretation: IG; Writing: IG, VE; Critical review: EA, VCE. All the authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1:
Figure S1. Quality scores across all bases as a function of their positions in the reads for O2P1, O2P2, T3P1, and T3P2. The graphs are the output of the FastQC (0.11.5) program.
Additional file 2: Figure S2.
Other quality parameters of the small RNA libraries sequenced on the Illumina platform. a) Presence of adaptors against the position in the read; b) N content against the position in the read (%); c) theoretical (blue line) and actual (red line) GC distribution across all sequences (%); d) sequence duplication level. For clarity, only the graphs obtained for the O2P1 library are shown as representatives of the other three libraries. The graphs are the output of FastQC.
Additional file 3:
Figure S3. A flowchart diagram. The criteria followed to analyze is the data is shown.
Additional file 4:
Table S1. Major conserved miRNA families identified in small RNA libraries O2P1, O2P1, T3P1, and T3P2. The last file shows the total number of miRNAs identified from these families and the percentage of miRNAs from these families compared to the total number of miRNAs characterized.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Garbus, I., Selva, J.P., Pasten, M.C. et al. Characterization and discovery of miRNA and miRNA targets from apomictic and sexual genotypes of Eragrostis curvula. BMC Genomics 20, 839 (2019). https://doi.org/10.1186/s12864-019-6169-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-019-6169-0