Abstract
Background
In the pig production industry, artificial insemination (AI) plays an important role in enlarging the beneficial impact of elite boars. Understanding the genetic architecture and detecting genetic markers associated with semen traits can help in improving genetic selection for such traits and accelerate genetic progress. In this study, we utilized a weighted single-step genome-wide association study (wssGWAS) procedure to detect genetic regions and further candidate genes associated with semen traits in a Duroc boar population. Overall, the full pedigree consists of 5284 pigs (12 generations), of which 2693 boars have semen data (143,113 ejaculations) and 1733 pigs were genotyped with 50 K single nucleotide polymorphism (SNP) array.
Results
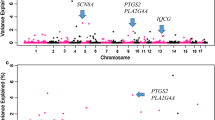
Results show that the most significant genetic regions (0.4 Mb windows) explained approximately 2%~ 6% of the total genetic variances for the studied traits. Totally, the identified significant windows (windows explaining more than 1% of total genetic variances) explained 28.29, 35.31, 41.98, and 20.60% of genetic variances (not phenotypic variance) for number of sperm cells, sperm motility, sperm progressive motility, and total morphological abnormalities, respectively. Several genes that have been previously reported to be associated with mammal spermiogenesis, testes functioning, and male fertility were detected and treated as candidate genes for the traits of interest: Number of sperm cells, TDRD5, QSOX1, BLK, TIMP3, THRA, CSF3, and ZPBP1; Sperm motility, PPP2R2B, NEK2, NDRG, ADAM7, SKP2, and RNASET2; Sperm progressive motility, SH2B1, BLK, LAMB1, VPS4A, SPAG9, LCN2, and DNM1; Total morphological abnormalities, GHR, SELENOP, SLC16A5, SLC9A3R1, and DNAI2.
Conclusions
In conclusion, candidate genes associated with Duroc boars’ semen traits, including the number of sperm cells, sperm motility, sperm progressive motility, and total morphological abnormalities, were identified using wssGWAS. KEGG and GO enrichment analysis indicate that the identified candidate genes were enriched in biological processes and functional terms may be involved into spermiogenesis, testes functioning, and male fertility.
Similar content being viewed by others
Background
In the pig production industry, artificial insemination (AI) plays an important role in enlarging the beneficial impact of elite boars. Sperm quality, affected by genetic and environmental factors simultaneously and with moderate to low heritability [1, 2], is essential for guaranteeing the success of AI. For both academic researchers and AI station managers, understanding the genetic background and detecting genetic markers associated with semen traits can help in improving genetic selection for such traits and accelerate genetic progress. Biologically, the process of spermatogenesis, which mainly contains three steps including mitotic phase, meiotic phase, and spermiogenesis, involves complex coordination among many genes and different cell types and occurs in the seminiferous tubules of the testes [3]. Mutations in related genes and status changes in corresponding cells will potentially affect the sperm quality and male fertility. From the aspect of genetic control and animal breeding, detecting candidate genes responsible to sperm quality and fertility is an essential step.
With the fast development of sequencing technology and commercially availability of dense marker panels, researchers can precisely identify quantitative trait loci (QTL) by searching for association between genetic markers and phenotypic records, which is known as genome-wide association study (GWAS) [4]. Though many challenges, such as the need for efficient study design especially for replication efforts and technologies for capturing genetic variation, the missing heritability problem, reducing the bias introduced into a dataset, and utilizing of new resources available, remained to be addressed [5], GWAS has been successfully implemented in detecting genetic risk factors for human diseases and mapping QTL for economically important traits in both animal and plant breeding populations. Among the established GWAS approaches, the weighted single step GWAS (wssGWAS) [6] is preferable for association study in domesticated animals, for which large numbers of individuals are usually phenotyped but less genotyped. Technically, the wssGWAS calculates genomic estimated breeding values (GEBVs) of individuals by solving mixed model equations using H (a blend of pedigree derived relationship matrix A and weighted genomic relationship matrix Gw, which is constructed in the manner of weighting SNPs according to their genetic variances, [7, 8]) as relatedness matrix, and converts the GEBVs into marker effects in the genotyped subpopulation based on the underlying equivalency between marker effect models and breeding value models. Genetic variance of certain chromosome window is subsequently calculated as the variation of the genetic values possessed by SNPs located in that window (i.e., Var(Zwindowgwindow), Zwindow and gwindow represent genotypes and marker effects of SNPs located in the window, respectively).
In several previous studies related to boar semen traits, associated genetic markers or candidate genes have been reported for both sperm quantity (i.e. number of sperm cells per ejaculation) and quality (i.e. sperm motility and morphological abnormalities) traits, [9,10,11,12,13]. However, the genetic background of breeds or boar populations, number of boars used for association study, and density of genetic marker panels varied a lot among the previous studies. The Duroc boars have been used as terminal sire in most of the modern three-way crossbreeding pig production system. While due to the relatively small numbers of Duroc boars in one herd and difficulty in phenotypic data collecting, seldom association studies have been conducted. In this study, we utilized wssGWAS to detect genetic regions and further candidate genes associated with semen traits in a Duroc boar population. In order to further understand the genetic control of semen traits, post GWAS analyses, KEGG and GO enrichment were performed to find biological processes and functional terms in which the candidate genes were involved.
Results
In this study, we identified genomic regions associated with semen traits in a Duroc boar population via wssGWAS [6]. In order to investigate the genetic background of the semen traits under consideration, we estimated the heritabilities of these traits by fitting model (1) using the pedigree derived numerator relationship matrix. Variance component and heritability estimations are presented in Table 1. The estimated heritabilities were 0.18, 0.16, 0.16, and 0.26 for the number of sperm cells, sperm motility, sperm progressive motility, and total morphological abnormalities, respectively. The heritability ranged from moderate to low, which were at the similar magnitude compared to Marques et al., (2018, 2017) while higher than that of Wolf (2010), in which relatively smaller Large White and Landrace boar populations were used for heritability estimation.
Figure 1 shows the proportion of variances explained by each 0.4 Mb windows for the traits under study. Table 2 shows the first three most important QTL regions and the candidate genes, which have been previously reported to be associated with mammal spermiogenesis, testes functioning, and male fertility, for the four traits. Overall, the first three QTL regions together explained about 5.12~11.97% of the genetic variances of the semen traits under study (Table 2). For each trait, the most important windows explained approximately 2%~ 6% of the total genetic variances (Table 2). Furthermore, the identified windows (windows that explained more than 1% of total genetic variances) explained totally 28.29, 35.31, 41.98, and 20.60% of genetic variances (not phenotypic variance) for number of sperm cells, sperm motility, sperm progressive motility, and total morphological abnormalities, respectively (Fig. 1 and Additional file 1: Table S1, Additional file 2: Table S2, Additional file 3: Table S3 and Additional file 4: Table S4). From the aspect of pig breeding, the relatively high proportions of genetic variances explained by the identified QTL regions may imply the possibility of integrating these QTL mapping results into estimating of genetic breeding values of semen traits [14, 15].
Table 3 shows the GO terms and KEGG pathways in which the identified genes enriched. Four GO terms and one KEGG pathway were enriched for the number of sperm cells. The enriched GO terms are involved in positive regulation of myeloid cell differentiation (GO:0045639), steroid hormone mediated signaling pathway (GO:0043401), granulocyte colony-stimulating factor receptor binding (GO:0005130), and enzyme regulator activity (GO:0030234). The enriched KEGG pathway is involved in thyroid hormone signaling (ssc04919). For sperm motility, three GO terms were enriched for the candidate genes, while no KEGG pathway was targeted. The GO terms play roles in cellular calcium ion homeostasis (GO:0006874), protein phosphorylation (GO:0006468), and protein kinase activity (GO:0004672). For progressive motility, seven GO terms and two KEGG pathways were enriched for the identified candidate genes. These GO terms and KEGG pathways are involved in nucleotide metabolic and biosynthesis processes. Three GO terms were enriched for the total morphological abnormalities, while no KEGG pathway was targeted. These GO terms are related to transition between slow and fast fiber (GO:0014886), sensory perception of sound (GO:0007605), endochondral ossification (GO:0001958), cullin family protein binding (GO:0097602), RNA polymerase II transcription factor activity, and sequence-specific DNA binding (GO:0000981).
Discussion
In this study, we conducted association study for boar semen traits in a large Duroc population. To our knowledge, this is the first time that genome-wide association study results are reported in such a large Duroc boar population. Based on the literature (cited above), some of the identified candidate genes have been previously reported to be involved in mammal spermiogenesis, testes functioning, and male fertility. Among the candidate genes identified in this study, DNAI2 was previously reported to be associated with sperm motility of large white boars [9], while no more genes overlapped with that identified in large white and landrace boars. Similarly, in the study conducted by Marques et al., [9], where wssGWAS was also used, no overlapping regions were identified between large white and landrace boars. However, gene network analysis revealed that genes identified in different lines were involved in the same biological processes [9]. Therefore, it is hard to draw the conclusion that the identified candidate genes in this study are specific to Duroc line. Nevertheness, functional validation experiments of homologous genes in human and model organisms (literature mentioned in the following paragraphs) in the literature provided evidence of role of the identified genes in semen traits. Considering genetic improvement of semen traits, the identified candidate genes may provide knowledge for developing genomic prediction models based on the trait-specific genomic best linear unbiased prediction (GBLUP) [15] by assigning higher genetic variances to genetic markers located in these regions.
For number of sperm cells, 18 relevant QTL regions located on SSC2 (SSC: Sus scrofa chromosome), 3, 5, 9, 10, 12, 14, 15, 16, and 17 were identified. Sixty-five genes were annotated in these genomic regions according to the National Center for Biotechnology Information (NCBI) (Additional file 1: Table S1). Among the identified genes, some has been previously reported to be associated with sperm biological function. The Tudor domain-containing protein 5 (TDRD5) and quiescin Q6/sulfhydryl oxidase 1 (QSOX1), located in the region of 121.15–121.95 Mb on SSC9, were considered as candidate gene for the number of sperm cells (Table 2). TDRDs are an evolutionarily conserved family of proteins involved in germ cell development, and TDRD5 is required for retrotransposon silencing, chromatoid body assembly, and spermiogenesis in mice [16]. The QSOX family was reported to have biological functions in male germ cell development [17]. BLK gene, located in the region of 14.49–15.29 Mb on SSC14, is an Src-related tyrosine kinase in the events leading to proper sperm formation. Previous study reported that BLK is involved in sperm function and integrity [18]. The members of tissue inhibitor of metalloproteinase (TIMP), among which TIMP3 was identified in the region of 11.71–12.51 Mb on SSC5, were reported to exist in human seminal plasma as complexes and may play roles in sperm function [19]. The thyroid hormone receptor alpha (THRA), identified in the region of 22.19–22.56 Mb on SSC12, is responsible for sertoli cell development, sperm production, and testis weight [20] by regulating the function of triiodothyronine (T3), which played a key role in Sertoli cell development [21]. Colony stimulating factor 3 (CSF3), another gene located in the region of 22.19–22.56 Mb on SSC12, is involved in reactive oxygen species (ROS) of sperm cells and associated with abnormalities in sperm histone transition [22]. A third gene located in the region of 22.19–22.56 Mb on SSC12, Zona pellucida binding protein 1 (ZPBP1), a spermatid and spermatozoon protein, is responsible for sperm morphology. Lin et al., (2007) reported that male mice lacking ZPBP1 were sterile, with abnormal round-headed sperm morphology and no forward sperm motility.
For sperm motility, 20 relevant QTL regions located on SSC1, 2, 3, 4, 7, 8, 9, 12, 13, 14, 15, 16, and 17 were identified. Fifty-nine genes were annotated in these genomic regions (Additional file 2: Table S2). Within the window explained the largest proportion of genetic variance of sperm motility (1.29–2.09 Mb on SSC13), six genes were targeted, while none of them have ever been previously reported to be associated with semen traits. The protein phosphatase 2 regulatory subunit B beta gene (PPP2R2B), located in the region of 147.71–148.51 Mb on SSC2, was indicated to be associated with biology of sperm in a Co-Expression Network Analysis conducted in Bos taurus and Sus scrofa [23]. For the NIMA related kinase 2 (NEK2) gene, located in the region of 131.55–132.35 Mb on SSC9 and showed biased expression in reproductive tissues (testis of human and ovary of swine), several studies have demonstrated its role in the process of meiotic cell cycle and the first meiotic division in mouse spermatocytes [24,25,26,27]. Ndrp3, a member of the N-myc downstream regulated gene family (NDRG), is enhanced expressed specifically in germ cells and involved in regulation of the male meiosis in mouse [18]. The ADAM metallopeptidase domain 7 (ADAM7), which located in the region of 8.49–8.86 Mb on SSC14, is associated with epididymosomes, integrated into sperm plasma membrane, and related to fertility and sperm integrity [28, 29]. S-phase kinase associated protein 2 (SKP2) was identified in the region of 21.48–21.86 Mb on SSC16. The Skp2 protein is the receptor subunit of an SCF-type ubiquitin ligase and is a major regulator of the progression of cells into S phase of the cell cycle, Skp2-deficient mice shown reduction in the number of mature gametes and thus decreased fertility [30]. Moreover, Skp2 was reported to be involved in the manipulation of boar sperm production [31]. The ribonuclease T2 (RNASET2) gene, located in the region of 1.77–2.16 Mb on SSC1, have been reported to be involved in sperm motility impairment [32] and interacted with AKAP4 in human sperm tail and subsequently reduced sperm motility by suppressing PKA/PI3K/calcium signaling pathways [33].
For progressive motility, 24 relevant QTL regions located on SSC1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, and 17 were identified (Additional file 3: Table S3). Seventy-three genes were annotated in these regions. Within the first three genomic regions that explained the largest proportions of genetic variance, 11 genes were identified but none of them have been previously reported to be directly associated with semen traits in the literatures. However, in the region of 18.51–18.87 Mb on SSC3, SH2B adaptor protein 1 (SH2B1) was identified. Previous study in mice have shown that expression of Sh2b1 on the mRNA level in obesity mice was decreased, which resulted in significant reduction in male fertility and exacerbated reproductive toxicity and germ cell mutagenicity [34]. Src family tyrosine kinase (BLK), also identified for the number of sperm cells, is an Src-related tyrosine kinases in the events leading to proper sperm formation, was reported to potentially involved in sperm function and integrity [18]. Laminin subunit beta 1 (LAMB1) gene, which directs the expression of beta-galactosidase during development of the mouse testis and ovary [35], was identified in the region of 107.72–108.07 Mb on SSC9. Acuolar protein sorting 4 homolog A (VPS4A) gene, located in the region of 17.44–17.8 Mb on SSC6, was previously reported to be involved in human sperm acrosome reaction [36]. Sperm associated antigen 9 (SPAG9) gene, which codes an acrosomal molecule that plays role in acrosome reaction and sperm-egg binding [37], was identified in the region of 27.39–27.78 Mb on SSC12. Lipocalin 2 (LCN2) gene, located in the region of 268.37–268.73 Mb on SSC1, is involved in sperm maturation and fertilization [38]. Dynamin 1 (DNM1), another gene located in the same region on SSC1, acts as a key mediator of sperm acrosome formation and function, is essential for mammalian spermatogenesis [39].
For total morphological abnormalities, 16 relevant QTL regions located on SSC1, 3, 4, 8, 9, 10, 11, 12, 13, 15, 16, and 17 were identified (Additional file 4: Table S4). Sixty-seven genes were annotated in these regions according to NCBI. The growth hormone receptor (GHR) gene, which shows significant association with embryos survival rate and involved in fertilization rate of dairy cattle [40], was identified in the region of 26.76–27.56 Mb on SSC16 (Table 2). Selenoprotein P (SELENOP), another gene located in the same region, is abundantly expressed in the testis and affected the sperm quality of rats [41]. Solute carrier family 16 member 5 (SLC16A5) gene, for which a protein-coding variation is associated with cisplatin-induced ototoxic effects and involved in human germ cell testicular cancer [42], was identified in the region of 6.20–7.00 Mb on SSC12. SLC9A3 regulator 1 (SLC9A3R1), another gene located in the same region, is important for mouse sperm capacitation through functional interactions with SLC26A3, SLC26A6, and CFTR [43]. Dynein axonemal intermediate chain 2 (DNAI2) gene, of which mutations may resulted in defects of sperm flagella in human [9, 44, 45], was identified on SSC12.
Conclusions
In conclusion, we identified candidate genes associated with the number of sperm cells, sperm motility, sperm progressive motility, and total morphological abnormalities in a Duroc boar population using wssGWAS. Functional validation experiments of homologous genes in human and model organisms (literature mentioned in the Discussion section) in the literature provided evidence of role of the identified genes in semen traits. The identified candidate genes may provide knowledge for developing genomic evaluation models by assigning higher weights to genetic markers located in these regions.
Methods
Phenotypes, genotypes, and pedigree
The studied Duroc boar population was a mixed herd consisted of boars from pure Danish lines, pure American lines, and progenies from the crosses between Danish and American Duroc lines. For more detailed description of the conditions where the boars were raised, please refer to our recently published paper [46]. Overall, 5284 pigs (12 generations) were included in the pedigree, of which 2693 boars with semen trait records. Four traits, total sperm morphological abnormalities (ABN), number of sperm cells (Ncells), sperm motility (MOT, defined as proportion of moving sperms), and sperm progressive motility (PMOT, defined as proportion of sperms moving in strait line) were recorded in the years from 2015 to 2018. The fresh semen was evaluated immediately after ejaculate using the UltiMateTM CASA system (Hamilton Thorne Inc., Beverly, MA, USA). Number of sperm cells was calculated as the product of the semen volume (mL) and concentration (106 mL− 1). For the traits of total morphological abnormalities, number of sperm cells, and motility, 143,113 individual ejaculates (53 ejaculates per boar on average) of 2693 boars were used for the association study (Table 4). For the trait progressive motility, 29,526 observations of 1304 out of the 2693 boars were used for association study (Table 4).
Total DNA of 1733 pigs were extracted (semen of boars and ear tissue samples of sows) using the genome extraction kits produced by Wuhan NanoMagBio Technology Co., Ltd. Those pigs were genotyped using the GGP 50 k SNP array (GeneSeek, US), which contains 50,703 SNPs. Overall, 1627 out of the genotyped pigs were related to boars with phenotypic records through pedigree and used for further analysis. SNPs, for which the Sscrofa10.2 physical positions were provided by the SNP array producer, were remapped to Sscrofa11.1 using the genome remapping procedure available in NCBI (https://www.ncbi.nlm.nih.gov/). SNPs with unknown positions (9405 SNPs) and that unable to map to Sscrofa11.1 (1842 SNPs) were removed from the dataset. Moreover, SNPs on sex chromosomes (2469 SNPs) were excluded to avoid sex bias in constructing genomic relationship matrix and calculating genetic breeding values (1231 boars and 392 sows were included in the genotype file and used for constructing the genomic relationship matrix). Autosome SNPs were filtered via plink [47] using the following criteria: individual call rate ≥90% (4 pigs removed); SNP call rate ≥90% (673 SNPs removed); minor allele frequency ≥0.01 (5345 SNPs removed); Hardy–Weinberg equilibrium p-value ≥10−6 (2680 SNPs removed). After quality control, 1623 pigs, among which 1231 had semen evaluation data, and 28,289 SNPs remained for further analysis. Missing genotypes were finally imputed using Beagle software (version 4.1) [48, 49].
Statistical model
The weighted single step GWAS procedure [6, 50] was used for association study. Briefly, genomic estimated breeding values (GEBVs) were estimated via a weighted single step GBLUP and marker effects were calculated from the GEBVs. QTL regions were selected based on the proportion of genetic variance explained by certain chromosome segments.
The following repeatability model was used in this study.
where y denoted the response; X, Z, and W denoted the design matrices; b was fixed effects (overall mean and year-season of ejaculation); \( \mathbf{a}\sim \mathrm{N}\left(0,\mathbf{H}{\sigma}_a^2\right) \) was a vector of genetic values; \( \mathbf{p}\sim \mathrm{N}\left(0,\mathbf{I}{\sigma}_p^2\right) \) was a vector of random permanent boar effect; covariates Age and Intv denoted the age of the boars at months when ejaculating and collection interval (days), respectively. \( \mathbf{e}\sim \mathrm{N}\left(0,\mathbf{I}{\sigma}_e^2\right) \) denoted the residuals. I denoted the identity matrix. The inverse of H matrix, a blend of pedigree and genetic marker derived matrices [7, 8], was calculated as
where A denoted the pedigree derived relationship matrix; A22 was a sub matrix of A corresponding to the genotyped individuals; Gω = 0.9G + 0.1A22, these weights were used to scale the genomic information to be compatible with the pedigree information and to control bias [51, 52]; \( \boldsymbol{G}=\frac{\boldsymbol{Z}\boldsymbol{D}{\boldsymbol{Z}}^{\prime }}{\sum_{i=1}^m2{p}_i\left(1-{p}_i\right)} \) was the genomic relationship matrix [53], where Z was a matrix of genotypes (with 0-2p, 1-2p, and 2-2p represented genotypes AA, Aa, and aa, respectively; p denoted the minor allele frequency (MAF)), D denoted a diagonal matrix contained the SNP weights, pi denoted MAF of the ith SNP, and m denoted the number of SNPs.
Variance components and further heritability of the studied traits were obtained via average information restricted maximum likelihood (AI-REML) [54] procedure using pedigree. Marker effects and further weights for constructing G were calculated in an iterative way used by Wang et al., [6, 50]. An iteration procedure with the following steps was used for association study.
-
Step1: Initialization, let t = 1, D(t) = I, G(t) = λZD(t)Z′, and \( \uplambda =\frac{1}{\sum_{i=1}^m2{p}_i\left(1-{p}_i\right)} \);
-
Step2: GEBV estimation, calculate GEBVs (\( \hat{\boldsymbol{a}} \)) via ssGBLUP with \( {\mathbf{H}}^{-1}={\boldsymbol{A}}^{-1}+\left[\begin{array}{cc}\mathbf{0}& \mathbf{0}\\ {}\mathbf{0}& {\left(0.9{\boldsymbol{G}}_{(t)}+0.1{\boldsymbol{A}}_{22}\right)}^{-\mathbf{1}}-{\boldsymbol{A}}_{22}^{-1}\end{array}\right] \);
-
Step3: Marker effects calculation, obtain marker effects via \( {\hat{\boldsymbol{g}}}_{(t)}=\uplambda {\boldsymbol{D}}_{(t)}{\boldsymbol{Z}}^{\prime }{\boldsymbol{G}}_{(t)}^{-1}\hat{\boldsymbol{a}} \);
-
Step4: SNP weights calculation, get SNP weights for the next iteration via \( {d}_{i\left(t+1\right)}={\hat{g}}_{i(t)}^22{p}_i\left(1-{p}_i\right) \);
-
Step5: SNP weights normalization, rescale the weights to keep the total genetic variance constant via \( {\boldsymbol{D}}_{\left(t+1\right)}=\frac{tr\left({\boldsymbol{D}}_{(1)}\right)}{tr\left({\boldsymbol{D}}_{\left(t+1\right)}\right)}{\boldsymbol{D}}_{\left(t+1\right)} \);
-
Step 6: Weighted G construction, calculate G for the next iteration via G(t + 1) = λZD(t + 1)Z′;
-
Step7: Let t = t + 1 and loop to step 2.
The procedure was run for three iterations, as used in Wang et al., [50] and Marques et al., [9], and marker effects obtained from the third iteration were used for calculating proportions of genetic variances explained by subsets of consecutive SNPs. SNPs located within 0.4 Mb (average haploblock length of the commercial pig lines including Duroc [55]) of unoverlapping windows were grouped into subsets and the genetic variances explained by those windows were obtained via
The iteration steps mentioned above were run with BLUF90 software families [51] in a iterative way, AIREMLF90 for variance components estimation, BLUPF90 for GEBVs calculation, and postGSf90 [56] for marker effects converting, respectively.
Candidate genes detection and functional enrichment analysis
QTL regions were selected according to genetic variance of chromosome windows. Windows explained higher than 1% genetic variances were selected as candidate QTL regions, within which candidate genes were searched. The threshold of 1% was set based on literature [9, 57,58,59] and the expected average genetic variance explained by single chromosome segment (0.027, 100% divided by 3707 windows). Consecutive windows with midpoints less than 0.4 Mb apart were treated as overlapped and merged. The first three windows that explained the largest amount of genetic variances were further extended to 0.4 Mb flanking regions of the midpoints on both upstream and downstream.
Candidate genes on “National Center for Biotechnology Information” (NCBI, http://www.ncbi.nlm.nih.gov) were searched in the selected QTL regions. Kyoto Encyclopedia of Genes and Genomes (KEGG) [60] and Gene Ontology (GO) [61] enrichment analyses were performed based on candidate gens for each trait via the Database for Annotation, Visualization, and Integrated Discovery (DAVID, Version 6.8, https://david.ncifcrf.gov/) [62].
Availability of data and materials
The datasets generated and/or analyzed during the current study are not publicly available since the studied population is consisted of the nucleus herd of Guangxi Yangxiang Agriculture and Husbandry Co.,LTD, but are available from the corresponding author on reasonable request.
Abbreviations
- ABN:
-
Total sperm morphological abnormalities
- AI:
-
Artificial insemination
- AI-REML:
-
Average information restricted maximum likelihood
- Co., Ltd.:
-
Company limited
- DAVID:
-
The database for annotation, visualization, and Integrated discovery
- GBLUP:
-
Genomic best linear unbiased prediction
- GEBVs:
-
Genomic estimated breeding values
- GO:
-
Gene ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- MAF:
-
Minor allele frequency
- MOT:
-
Sperm motility
- NCBI:
-
National Center for Biotechnology Information
- Ncells:
-
Number of sperm cells
- PMOT:
-
Sperm progressive motility
- QTL:
-
Quantitative trait loci
- SNP:
-
Single nucleotide polymorphism
- SSC:
-
Sus scrofa chromosome
- SYSU:
-
Sun Yat-Sen University
- wssGWAS:
-
Weighted single-step GWAS
References
Wolf J. Heritabilities and genetic correlations for litter size and semen traits in Czech large white and landrace pigs. J Anim Sci. 2010;9:2893–903.
Marques DBD, Lopes MS, Broekhuijse MLWJ, Guimarães SEF, Knol EF, Bastiaansen JWM, et al. Genetic parameters for semen quality and quantity traits in five pig lines. J Anim Sci. 2017;10:4251–9.
Chu DS, Shakes DC. Spermatogenesis. Adv Exp Med Biol. 2013;757:171–203.
Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108.
De R, Bush WS, Moore JH. Bioinformatics challenges in genome-wide association studies (GWAS). Methods Mol Biol. 2014;1168:63–81.
Wang H, Misztal I, Aguilar I, Legarra A, Muir WM. Genome-wide association mapping including phenotypes from relatives without genotypes. Genet Res. 2012;94:73–83.
Christensen OF, Lund MS. Genomic prediction when some animals are not genotyped. Genet Sel Evol. 2010;42. https://doi.org/10.1186/1297-9686-42-2.
Legarra A, Aguilar I, Misztal I. A relationship matrix including full pedigree and genomic information. J Dairy Sci. 2009;92:4656–63.
Marques DBDD, Bastiaansen JWMM, Broekhuijse MLWJWJ, Lopes MS, Knol EF, Harlizius B, et al. Weighted single-step GWAS and gene network analysis reveal new candidate genes for semen traits in pigs. Genet Sel Evol. 2018;50:1–14.
Kaewmala K, Uddin MJ, Cinar MU, Grosse-Brinkhaus C, Jonas E, Tesfaye D, et al. Association study and expression analysis of CD9 as candidate gene for boar sperm quality and fertility traits. Anim Reprod Sci. 2011;125:170–9.
Kaewmala K, Uddin MJ, Cinar MU, Grosse-Brinkhaus C, Jonas E, Tesfaye D, et al. Investigation into association and expression of PLCz and COX-2 as candidate genes for boar sperm quality and fertility. Reprod Domest Anim. 2012;47:213–23.
Gunawan A, Cinar MU, Uddin MJ, Kaewmala K, Tesfaye D, Phatsara C, et al. Investigation on association and expression of ESR2 as a candidate gene for boar sperm quality and fertility. Reprod Domest Anim. 2012;47:782–90.
Diniz DB, Lopes MS, Broekhuijse MLWJ, Lopes PS, Harlizius B, Guimaraes SEF, et al. A genome-wide association study reveals a novel candidate gene for sperm motility in pigs. Anim Reprod Sci. 2014;151:201–7.
Zhang Z, Erbe M, He J, Ober U, Gao N, Zhang H, et al. Accuracy of whole genome prediction using a genetic architecture enhanced variance-covariance matrix. G3 (Bethesda). 2015;5:615–27.
Zhang Z, Ober U, Erbe M, Zhang H, Gao N, He J, et al. Improving the accuracy of whole genome prediction for complex traits using the results of genome wide association studies. PLoS One. 2014;9:e93017.
Yabuta Y, Ohta H, Abe T, Kurimoto K, Chuma S, Saitou M. TDRD5 is required for retrotransposon silencing, chromatoid body assembly, and spermiogenesis in mice. J Cell Biol. 2011;5:781–95.
Wang T-E, Li S-H, Minabe S, Anderson AL, Dun MD, Maeda K-I, et al. Mouse quiescin sulfhydryl oxidases exhibit distinct epididymal luminal distribution with segment-specific sperm surface associations†. Biol Reprod. 2018;5:1022–33.
Goupil S, La Salle S, Trasler JM, Bordeleau LJ, Leclerc P. Developmental expression of Src-related tyrosine kinases in the mouse testis. J Androl. 2011;32:95-110.
Shimokawa K, Katayama M, Matsuda Y, Takahashi H, Hara I, Sato H. Complexes of gelatinases and tissue inhibitor of metalloproteinases in human seminal plasma. J Androl. 2003;1:73–7.
Holsberger DR, Kiesewetter SE, Cooke PS. Regulation of neonatal Sertoli cell development by thyroid hormone receptor α11. Biol Reprod. 2005;73:396–403.
Cooke PS, Hess RA, Porcelli J, Meisami E. Increased sperm production in adult rats after transient neonatal hypothyroidism. Endocrinology. 1991;129:244–8.
Jiang L, Zheng T, Huang J, Mo J, Zhou H, Liu M, et al. Association of semen cytokines with reactive oxygen species and histone transition abnormalities. J Assist Reprod Genet. 2016;9:1239–46.
Ahmed Khan F, Liu H, Zhou H, Wang K, Qamar MTU, Pandupuspitasari NS, et al. Analysis of Bos taurus and Sus scrofa X and Y chromosome transcriptome highlights reproductive driver genes. Oncotarget. 2017;33:54416–33.
Di Agostino S, Fedele M, Chieffi P, Fusco A, Rossi P, Geremia R, et al. Phosphorylation of high-mobility group protein A2 by Nek2 kinase during the first meiotic division in mouse spermatocytes. Mol Biol Cell. 2004;15:1224–32.
Di Agostino S, Rossi P, Geremia R, Sette C. The MAPK pathway triggers activation of Nek2 during chromosome condensation in mouse spermatocytes. Development. 2002;129:1715–27.
Rhee K, Wolgemuth DJ. Tcp10 promoter-directed expression of the Nek2 gene in mouse meiotic spermatocytes. Mol Cells. 2002;13:85–90.
Rhee K, Wolgemuth DJ. The NIMA-related kinase 2, Nek2, is expressed in specific stages of the meiotic cell cycle and associates with meiotic chromosomes. Development. 1997;124:2167–77.
Choi H, Han C, Jin S, Kwon JT, Kim J, Jeong J, et al. Reduced fertility and altered Epididymal and sperm integrity in mice lacking ADAM7. Biol Reprod. 2015;93:70.
Oh JS, Han C, Cho C. ADAM7 is associated with epididymosomes and integrated into sperm plasma membrane. Mol Cells. 2009;28:441–6.
Fotovati A, Nakayama K, Nakayama KI. Impaired germ cell development due to compromised cell cycle progression in Skp2-deficient mice. Cell Div. 2006;1:4.
Jiao ZJ, Yi W, Rong YW, Kee JD, Zhong WX. MicroRNA-1285 regulates 17beta-estradiol-inhibited immature boar Sertoli cell proliferation via adenosine monophosphate-activated protein kinase activation. Endocrinology. 2015;156:4059–70.
Liu Y, Chen G, Lu L, Sun H, Guo Q, Xue K, et al. RNASET2 in human spermatozoa and seminal plasma: a novel relevant indicator for asthenozoospermia. Andrology. 2013;1:75–84.
Xu Y, Fan Y, Fan W, Jing J, Xue K, Zhang X, et al. RNASET2 impairs the sperm motility via PKA/PI3K/calcium signal pathways. Reproduction. 2018;155:383–92.
Ghanayem BI, Bai R, Kissling GE, Travlos G, Hoffler U. Diet-induced obesity in male mice is associated with reduced fertility and potentiation of acrylamide-induced reproductive toxicity. Biol Reprod. 2010;82:96–104.
Li C, Gudas LJ. Sequences 5′ of the basement membrane laminin beta 1 chain gene (LAMB1) direct the expression of beta-galactosidase during development of the mouse testis and ovary. Differentiation. 1997;62:129–37.
Pocognoni CA, Berberian MV, Mayorga LS. ESCRT (endosomal sorting complex required for transport) machinery is essential for Acrosomal exocytosis in human sperm. Biol Reprod. 2015;93:124.
Jagadish N, Rana R, Mishra D, Garg M, Chaurasiya D, Hasegawa A, et al. Immunogenicity and contraceptive potential of recombinant human sperm associated antigen (SPAG9). J Reprod Immunol. 2005;67:69–76.
Watanabe H, Takeo T, Tojo H, Sakoh K, Berger T, Nakagata N, et al. Lipocalin 2 binds to membrane phosphatidylethanolamine to induce lipid raft movement in a PKA-dependent manner and modulates sperm maturation. Development. 2014;141:2157–64.
Redgrove KA, Bernstein IR, Pye VJ, Mihalas BP, Sutherland JM, Nixon B, et al. Dynamin 2 is essential for mammalian spermatogenesis. Sci Rep. 2016;6:35084.
Khatib H, Huang W, Wang X, Tran AH, Bindrim AB, Schutzkus V, et al. Single gene and gene interaction effects on fertilization and embryonic survival rates in cattle. J Dairy Sci. 2009;92:2238–47.
Zhou J-C, Zheng S, Mo J, Liang X, Xu Y, Zhang H, et al. Dietary selenium deficiency or excess reduces sperm quality and testicular mRNA abundance of nuclear glutathione peroxidase 4 in rats. J Nutr. 2017;147:1947–53.
Drogemoller BI, Monzon JG, Bhavsar AP, Borrie AE, Brooks B, Wright GEB, et al. Association between SLC16A5 genetic variation and cisplatin-induced ototoxic effects in adult patients with testicular cancer. JAMA Oncol. 2017;3:1558–62.
Chavez JC, Hernandez-Gonzalez EO, Wertheimer E, Visconti PE, Darszon A, Trevino CL. Participation of the cl−/HCO(3)- exchangers SLC26A3 and SLC26A6, the cl- channel CFTR, and the regulatory factor SLC9A3R1 in mouse sperm capacitation. Biol Reprod. 2012;86:1–14.
McLachlan RI, Ishikawa T, Osianlis T, Robinson P, Merriner DJ, Healy D, et al. Normal live birth after testicular sperm extraction and intracytoplasmic sperm injection in variant primary ciliary dyskinesia with completely immotile sperm and structurally abnormal sperm tails. Fertil Steril. 2012;97:313–8.
Loges NT, Olbrich H, Fenske L, Mussaffi H, Horvath J, Fliegauf M, et al. DNAI2 mutations cause primary ciliary dyskinesia with defects in the outer dynein arm. Am J Hum Genet. 2008;83:547–58.
Zhao Y, Gao N, Li X, El-ashram S, Wang Z, Zhu L, et al. Identifying candidate genes associated with sperm morphology abnormalities using weighted single-step GWAS in a Duroc boar population. Theriogenology. 2020;141:9–15.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;813:559–75.
Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2008;84:210–23.
Browning BL. Beagle 4.1; 2015.
Wang H, Misztal I, Aguilar I, Legarra A, Fernando RL, Vitezica Z, et al. Genome-wide association mapping including phenotypes from relatives without genotypes in a single-step ( ssGWAS ) for 6-week body weight in broiler chickens. Front Genet. 2014;5:1–10.
Misztal I, Tsuruta S, Strabel T, Auvray B, Druet T, Lee DH. BLUPF90 and related programs (BGF90). Montpellier: Proc. 7th World Congr. Genet. Appl. Livest. Prod; 2002. p. 21–2.
Fangmann A, Sharifi RA, Heinkel J, Danowski K, Schrade H, Erbe M, et al. Empirical comparison between different methods for genomic prediction of number of piglets born alive in moderate sized breeding populations. J Anim Sci. 2017;95:1434–43.
VanRaden PM. Efficient methods to compute genomic predictions. J Dairy Sci. 2008;91:4414–23.
Jensen J, Mantysaari EA, Madsen P, Thompson R. Residual maximum likelihood estimation of (co) variance components in multivariate mixed linear models using average information. J Indian Soc Agric Stat. 1997;49:215–36.
Amaral AJ, Megens HJ, Crooijmans RPMA, Heuven HCM, Groenen MAM. Linkage disequilibrium decay and haplotype block structure in the pig. Genetics. 2008;179:569–79.
Aguilar I, Misztal I, Legarra A, Tsuruta S. Efficient computation of the genomic relationship matrix and other matrices used in single-step evaluation. J Anim Breed Genet. 2011;128:422-8.
Gonzalez-Pena D, Gao G, Baranski M, Moen T, Cleveland BM, Kenney PB, et al. Genome-wide association study for identifying loci that affect fillet yield, carcass, and body weight traits in rainbow trout (Oncorhynchus mykiss). Front Genet. 2016;7:203.
Irano N, de Camargo GMF, Costa RB, Terakado APN, Magalhaes AFB, Silva RM d, et al. Genome-wide association study for indicator traits of sexual precocity in Nellore cattle. PLoS One. 2016;11:e0159502.
Lemos MVA, Chiaia HLJ, Berton MP, Feitosa FLB, Aboujaoud C, Camargo GMF, et al. Genome-wide association between single nucleotide polymorphisms with beef fatty acid profile in Nellore cattle using the single step procedure. BMC Genomics. 2016;17:213.
Kanehisa M, Goto S. KEGG : Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;27:29–34.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25-9.
Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44-57.
Acknowledgements
We would thank Quanshun Mei (phenotypic data collating), Xia Peng (DNA extracting), and Huiwen Zhan (DNA extracting) from Huazhong Agricultural University for their help in this work.
Funding
This study is founded by National Key R&D Program of China (2018YFD0501200), Guangdong Provincial Science and Technology Plan (2014B020202001), and the earmarked fund for Modern Agro-industry Technology Research System (CARS-35).
Author information
Authors and Affiliations
Contributions
NG and YSC conceived this study. NG performed the analyses and wrote the manuscript. YLC and ZST worked on phenotypic and genotypic data collating, respectively. ALL, XYL, and LZ collected the ear tissue and extracted the DNA for genotyping. YXZ, LZ, WJ, and XP performed the semen evaluation and the phenotypic data recording. XHL, CLZ, and YSC guided this study and reviewed the manuscript. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was carried out in accordance with the principles of the Basel Declaration and recommendations of the Institutional Animal Care and Use Committee of Sun Yat-Sen University (SYSU). The protocol was approved by the Institutional Animal Care and Use Committee of SYSU.
Consent for publication
Not applicable.
Competing interests
Authors Yunxiang Zhao, Lin Zhu, Wei Jiang, and Xing Peng were employed by Guangxi Xiubo genetics technology Co., LTD and declare the following conflicts of interest: Xiubo genetics technology Co., LTD. provided all phenotypic data, semen and ear tissue samples and payed for genotyping. The data analysis, results interpretation, and manuscript writing and publication were jointly supported by the funding bodies mentioned in the Funding section. Conglin Zhang was employed by Guangxi Yangxiang Agriculture and Husbandry Co., LTD. Conglin Zhang and all other authors declared no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
Genomic regions explained more than 1% of genetic variance for the number of sperm cells.
Additional file 2: Table S2.
Genomic regions explained more than 1% of genetic variance for sperm motility.
Additional file 3: Table S3.
Genomic regions explained more than 1% of genetic variance for progressive motility.
Additional file 4: Table S4.
Genomic regions explained more than 1% of genetic variance for total morphological abnormalities.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Gao, N., Chen, Y., Liu, X. et al. Weighted single-step GWAS identified candidate genes associated with semen traits in a Duroc boar population. BMC Genomics 20, 797 (2019). https://doi.org/10.1186/s12864-019-6164-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-019-6164-5