Abstract
Background
High-throughput proteomics was used to determine the role of the fish liver in defense responses to bacterial infection. This was done using a rainbow trout (Oncorhynchus mykiss) model following infection with Aeromonas salmonicida, the causative agent of furunculosis. The vertebrate liver has multifaceted functions in innate immunity, metabolism, and growth; we hypothesize this tissue serves a dual role in supporting host defense in parallel to metabolic adjustments that promote effective immune function. While past studies have reported mRNA responses to A. salmonicida in salmonids, the impact of bacterial infection on the liver proteome remains uncharacterized in fish.
Results
Rainbow trout were injected with A. salmonicida or PBS (control) and liver extracted 48 h later for analysis on a hybrid quadrupole-Orbitrap mass spectrometer. A label-free method was used for protein abundance profiling, which revealed a strong innate immune response along with evidence to support parallel rewiring of metabolic and growth systems. 3076 proteins were initially identified against all proteins (n = 71,293 RefSeq proteins) annotated in a single high-quality rainbow trout reference genome, of which 2433 were maintained for analysis post-quality filtering. Among the 2433 proteins, 109 showed significant differential abundance following A. salmonicida challenge, including many upregulated complement system and acute phase response proteins, in addition to molecules with putative functions that may support metabolic re-adjustments. We also identified novel expansions in the complement system due to gene and whole genome duplication events in salmonid evolutionary history, including eight C3 proteins showing differential changes in abundance.
Conclusions
This study provides the first high-throughput proteomic examination of the fish liver in response to bacterial challenge, revealing novel markers for the host defense response, and evidence of metabolic remodeling in conjunction with activation of innate immunity.
Similar content being viewed by others
Background
Vertebrate immune function requires coordination of a complex set of regulatory processes and signaling pathways. The immediate innate response to pathogenic insult is underpinned by cellular and humoral components that are fairly well characterized in teleost fishes [1, 2], with many known contributing genetic components [3]. For example, the conserved cytokines IL-1β, IL-8 and tumor necrosis factor α (TNFα) each activate nuclear factor kappa-light-chain enhancer of activated B cells (NF-κB) signaling pathways to regulate early inflammatory responses to bacterial infection [4], which are followed by the acute phase response (APR), defined by production of plasma proteins such as complement system components, cerebellin-like proteins, lectins, haptoglobin and ferritin [5, 6].
The innate immune response of teleost fishes occurs primarily in lymphoid organs such as head kidney and spleen, and a variety of mucosal-associated sites (e.g. gills, gut, skin and nostrils) [7, 8], which produce cells and humoral parameters responsible for clearing a pathogen [1]. Monocytes, macrophages, and neutrophils degrade pathogenic particles through phagocytosis, while nonspecific cytotoxic cells help eliminate pathogens by apoptosis [9]. Humoral innate immune components include a wide range of receptors and molecules that are soluble in plasma and other body fluids, consisting of cytokines, APR proteins, antimicrobial peptides and protease inhibitors, among others [1]. Despite its traditional perception as a metabolic, nutrient storage, and detoxification center, the vertebrate liver is also an important immune organ and produces cytokines, chemokines, complement components and APR proteins in response to pathogenic challenge (reviewed in [10]). Resident immune cells contributing to liver immune function include macrophages (Kupffer cells), neutrophils, B lymphocytes, T lymphocytes and NK cells [11]. The liver’s dual role in immune function and metabolism makes it an interesting candidate for linking host defense with metabolic readjustments upon pathogen challenge.
Significant energy reserves are required to maintain immune function and to activate defense responses, for example the production of high titres of pro-inflammatory cytokines and APR proteins [5, 12, 13]. Energetic reserves allocated towards an immune response cannot be invested in other physiological systems, including anabolic processes linked to growth and reproduction, which may be essential to survival and fitness. Given that such systems also demand significant energetic resources, trade-offs between immune and other functions should have evolved under selection to maximize fitness and survival depending on environmental conditions (e.g. the presence or absence of a pathogen) and life-history strategy [14]. In turn, such trade-offs presumably require ‘cross-talk’ between different physiological systems to facilitate re-mobilization of energy towards an effective immune response upon infection [15]. In teleosts, there is growing evidence for such interrelationships between immune function and key pathways regulating growth [16,17,18], as well as stress (see [19,20,21]), thyroid function [22, 23] and other endocrine factors (reviewed by [24]).
Here we hypothesize that the liver plays a dual role in supporting the host defense response during infection, while mediating parallel metabolic adjustments that promote rewiring of energetic resources towards immune function. As a study system we used rainbow trout (Oncorhynchus mykiss) exposed to Aeromonas salmonicida, the causative agent of furunculosis, a disease with profound negative effects on aquacultured fish including salmonids. Past studies of salmonids have characterized responses to bacterial pathogens using transcriptomic analyses of different tissues, including liver [25,26,27], spleen [27, 28], head kidney [25, 27] and gill [25]. Additional research has documented the effects of bacterial pathogens in various fish immune tissues using proteomics [28,29,30], though these studies are yet to include liver. We have employed high-throughput proteomics to discover a set of proteins showing differential abundance in rainbow trout liver following A. salmonicida infection, revealing many known and novel markers for host defense, along with candidate proteins that may support cross-talk between immune function and metabolism.
Results
Experimental design and validation of immune response
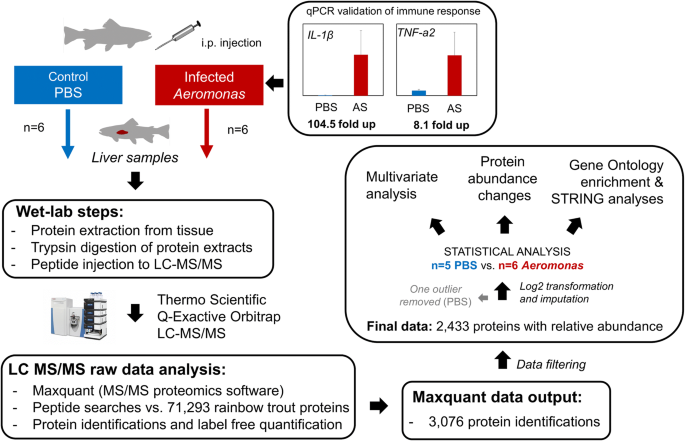
The experimental design of our study is summarized in Fig. 1. We performed a high-throughput proteomic analysis of rainbow trout liver comparing controls (48 h after PBS-injection) to bacterial-challenged (48 h post-infection of Aeromonas salmonicida; ‘AS’) samples (Fig. 1). While we did not confirm liver pathology in the AS-challenged trout, past studies in other fish species have provided evidence for liver pathology post-infection with another Aeromonas species (A. hydrophila) at similar timescales (e.g. for catfish: [31]; for Golden Mahseer: [32]). In this study, to validate that a systematic immune response occurred to AS challenge, we used quantitative PCR gene expression profiling to demonstrate a strong transcriptional up-regulation of the pro-inflammatory cytokines IL-1β and TNF-α2 in head kidney (i.e. primary immune tissue) sampled from the same animals (see Fig. 1). Specifically, IL-1β and TNF-α2 increased by approx. 105- and 8.1-fold, respectively, in the AS samples compared to controls (respective one-way ANOVA tests: P < 0.0001 and P < 0.001). The strong response of these immune markers in head kidney is consistent with a systematic immune response to AS infection in the animals used for proteomics. In addition, we observed a markedly enlarged spleen in the AS treated animals only (not shown), which is a clinical sign of bacterial infection (e.g. [33]), and further indicates a successful infection challenge.
Our choice to sample animals 48 h post-AS challenge is supported by past work done by authors on this paper, which reported the temporal dynamics of liver and head kidney responses to AS challenge in rainbow trout using a similar infection model, including expression responses for several immune genes [25]. That study revealed a robust induction of immune genes at 6 h, 12 h, 24 h and 48 h, with some variation across time-points [25]. By selecting a single sampling time point, we acknowledge that our study fails to capture the full temporal dynamic of bacterial infection, but nonetheless, taken with our gene expression profiling data at 48 h (Fig. 1) we can be confident that a mature immune response was underway, providing a strong basis for proteomic exploration.
Global liver proteome response to bacterial challenge
Our high-throughput proteomics analysis of rainbow trout liver led to the initial identification of 3076 proteins (Additional File 1: Table S1), 2433 of which were maintained for statistical analysis after quality control steps (Methods; data in Additional file 1: Table S2). Nonmetric multidimensional scaling was used to view all samples in the same multivariate space, revealing a clear separation of individuals from the AS and control groups (Fig. 2). Separation of AS and control samples at the proteomic level was confirmed using a PERMANOVA (9999 permutations, pseudo-F1,9 = 3.07, P = 0.002). Overall, these multivariate analyses demonstrate a major proteomic remodeling of rainbow trout liver following AS challenge.
Nonmetric multidimensional scaling showing separation of AS vs. PBS (control) injected rainbow trout. Each label represents an individual fish and their entire quantified liver proteome. The proximity of labels indicates the relatedness of proteomic profiles among individuals. Ellipses represent 95% confidence intervals
Proteins responsive to bacterial challenge
Using a general linear model separately for all proteins in our dataset, 69 and 109 proteins showed differential abundance between the AS and control groups at two significance cut-offs correcting for multiple comparisons (FDR-adjusted P < 0.05 and P < 0.1, respectively) (Full data in Additional file 1: Table S3). Given the large number of proteins in our dataset, the FDR adjustment is likely very stringent, so we tolerated the higher type-I error rate (i.e. FDR-adjusted P < 0.1) to allow inclusion of a greater number of proteins in downstream analyses (Table 1), particularly when considering that known immune proteins were included in the list falling between P = 0.05 and 0.1. Hierarchical clustering revealed a clear separation of AS and PBS samples, with 70 upregulated and 39 downregulated proteins (Fig. 3). The upregulated proteins includes many with putative roles in innate immunity (e.g. complement proteins, leukocyte cell-derived chemotaxin-2-like, C-type lectin, alpha2-macroglobulin, nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 [NF-κB]), innate immune accessory proteins (e.g. sequestosome, haptoglobin, eosinophil peroxidase, ferritin heavy subunit, furin-1), translation and transcription factors (e.g. eukaryotic translation initiation factor 1, transcription factor BTF3 homolog 4), transport proteins (e.g. Golgi phosphoprotein 3, phosphatidylinositol transfer protein beta isoform-like), hydrolases (e.g. tyrosine-protein phosphatase non-receptor type 1-like, phosphotriesterase related), proteins involved in the stress response (e.g. 78 kDa glucose-regulated protein, SIL1 nucleotide exchange factor, dnaJ homolog subfamily B member 11-like, endoplasmin-like), and the metalloreductase STEAP4 (STAMP2) (Table 1, Additional file 1: Table S3).
Notably, 11 of the proteins upregulated in AS were from the complement system, representing ~ 10% of all differentially abundant proteins and > 40% of all complement system proteins identified in our dataset (Fig. 4). Identified complement proteins were mapped onto a characterized pathway to highlight the potential impacts of increased abundance on bactericidal activity (Fig. 4a). A phylogenetic analysis was performed with eight unique complement C3 proteins identified in our analysis, which revealed novel teleost and salmonid specific paralogues showing distinct levels of upregulation (Fig. 4c). Interestingly, three significantly upregulated C3 proteins in rainbow trout liver are encoded by genes on chromosome Om02 that were evidently expanded by tandem duplication after divergence between Oncorhynchus and Salmo (Fig. 4c). In addition, our analysis distinguished distinct complement C3 proteins encoded by rainbow trout gene duplicates retained on different chromosomes, which according to our phylogenetic analysis, are orthologous to Atlantic salmon (Salmo salar) genes located within genomic regions retained from the salmonid-specific whole genome duplication event [34] (Fig. 4c).
Response of the complement system in liver of AS-challenged rainbow trout. a Complement system pathway annotated with protein components quantified in our study. The numbers in parentheses highlight how many unique proteins were identified in our dataset for each complement component. The number of significantly upregulated complement proteins for each system component is shown by orange arrows. b Clustering of all identified complement system proteins; titles in bold show significantly differential abundance for AS (n = 6) vs. PBS (n = 5). c Maximum likelihood phylogenetic analysis of C3 proteins from: Human Homo sapiens (“Hs”), zebrafish Danio rerio (“Dr”), tilapia Oreochromis niloticus (“On”), northern pike Esox lucius (“El”, a sister lineage to salmonids that did not undergo the salmonid-specific whole genome duplication [53]), Atlantic salmon Salmo salar (“Ss”), and rainbow trout Oncorhynchus mykiss (“Om”). The tree is annotated to show potential teleost genome duplication events in the teleost (“3R”) and salmonid ancestor (“4R”). Branch support values are shown as circles on each node. Vertebrate C5 proteins provided a validated outgroup to vertebrate C3 proteins [55]. Abundance fold-change values for proteins identified in this study are displayed using bubble plots, with closed circles depicting significantly upregulated proteins
The more limited set of 39 proteins downregulated by AS included proteins regulating translation (e.g. eukaryotic translation initiation 2B), C-reactive protein, transferrin receptor, beta-glucuronidase-like, lysosome membrane proteins, and apolipoprotein B-100 (Table 1).
Protein-protein interaction analysis
A putative protein-protein interaction (PPI) network was analyzed and a visual representation created using STRING (Fig. 5). This analysis revealed a very significant enrichment (P = 1.35e-11) of PPIs among the 109 proteins showing significantly differential abundance between control and AS. The majority of the PPIs (edges) were centered on four main node (protein) clusters, representing the complement (e.g. C3, C4A, C4B, C5), and molecular chaperone systems (e.g. HSP90B1, HSPA5, calreticulin [CALR], DNAJB11), as well as a cluster of APR proteins (e.g. albumin [ALB], haptoglobin [HP], alpha-2-macroglobulin [A2M] and ceruloplasmin [CP]) connected to the molecular chaperone cluster through a smaller cluster of proteins comprised of apolipoprotein B (APOB), transferrin receptor (TFRC) and scavenger receptor class B (SCARB2). Abbreviations and protein annotations from STRING are available in Table S7.
STRING protein-protein interaction network. 109 proteins showing significantly differential abundance between controls and AS were included in the analysis, but only 99 are included in this visual network. Proteins are represented as nodes while interactions appear as edges. The quantity of edges relates to the strength of the interaction relationship
Gene ontology analyses
We tested whether proteins showing differential abundance between control and AS groups showed an enrichment of particular functions using Gene Ontology (GO) analyses. 103 GO Biological Process (BP) terms showed significant enrichment (Additional file 1: Table S4), many linked to the immune system; such as complement activation, alternative & classical pathway, positive regulation of apoptotic cell clearance, and acute inflammatory response (Additional file 1: Table S5). Four complement C3 proteins contributed to the most significant immune-related GOBP enrichments (Additional file 1: Table S5). Other enriched terms included cholesterol transport, negative regulation of metabolic process, and response to steroid hormone. Owing to the complexity of these systems, both upregulated and downregulated proteins contributed to these signals. For example, cholesterol transport is predominantly explained by downregulated proteins (6 down vs. 2 up), while negative regulation of metabolic process (19 up vs. 13 down) and response to steroid hormone (10 up vs. 3 down) are mainly accounted for by upregulated proteins (Additional file 1: Table S5). Several GOBP terms associated with metabolism, including positive regulation of ERK1 and ERK2 cascade, regulation of triglyceride biosynthetic process, and positive regulation of lipid storage were largely accounted for by the same four complement C3 factors explaining the immune-related GOBP terms (Additional file 1: Table S5).
GO slim analysis revealed 6 significantly enriched terms (Table 2), including immune system process, homeostatic process, response to stress, and vesicle-mediated transport (Table 2). A large proportion of proteins are recurrent across terms, contributing ~ 56% of the total explanatory proteins for all 6 significant GO slim terms. Additionally, the GO slim analysis was predominantly driven by upregulated proteins, with ~ 69% of unique contributing proteins showing increased abundance across the 6 significant terms, which predominantly matches with individual GO slim terms (Additional file 1: Table S6).
Discussion
High-throughput proteomics is gaining rapid traction in teleost physiology [35,36,37,38,39,40], underpinned by the rapid progression in generation of new genomics resources, for example reference genomes for multiple salmonid lineages with high-quality proteome predictions (e.g. [41]). Our current study highlights the power of such approaches, revealing many proteins with altered abundances in rainbow trout liver following bacterial infection, including molecules supporting innate immune defense (notably, the APR) and candidate proteins that potentially facilitate energetic re-allocation towards immune function. These findings have implications for our understanding of salmonid health in aquaculture, where bacterial infections such as A. salmonicida cause major issues; the proteins we identified may serve as valuable markers both for infection and vaccination responses, and also as candidate immunostimulants that could be explored with the aim of boosting vaccination responses or disease resistance.
Previous proteomics studies of fish responses to bacterial pathogens have not considered liver, instead focusing on spleen, kidney, and intestinal mucosa [28,29,30]. A recent high-throughput analysis of Atlantic salmon reported remodeling of the liver proteome in response to elevated temperature [42], identifying a number of proteins in common with our study, however the major difference in treatment compared to our immune-focused study limits a useful biological interpretation of parallel changes in protein abundance. Nonetheless, it is interesting that leukocyte cell-derived chemotaxin 2, which was highly upregulated in our study following AS challenge, was strongly downregulated in salmon liver following thermal stress. Our findings are consistent with previous mRNA expression profiling of teleost tissues exposed to bacterial pathogens [25,26,27, 43,44,45,46]. For example, many APR proteins that increased in our study were reported as upregulated in transcriptomic studies of liver responses to A. salmonicida, including complement proteins, haptoglobin, ferritin and cerebellin-like protein [25, 27, 43]. Additionally, several non-immune proteins, including 78 kDa glucose-regulated protein precursor (GRP 78), dnaJ homolog subfamily B member 11-like, serine protease, and endoplasmin all showed congruent responses across studies [25, 27, 43]. However, there was not always a direct relationship between liver transcript responses and protein abundance changes across studies. For example, glutathione S-transferase kappa 1 showed upregulation here, but was previously reported to decrease at the mRNA level [25]. It is important to note differences that confound comparisons with past work, including starvation [43] and a distinct experimental infection route [27], while Martin et al. [25] used an attenuated A. salmonicida strain known to promote the development of immunological memory [47].
A notable study finding was the strong upregulation of a genetically expanded salmonid complement system following AS challenge. The complement system is activated through the classical, alternative and lectin pathways [48, 49], leading to bactericidal actions through pathogen opsonization, phagocytic activity, and eventual lysis [50,51,52]. Our analysis distinguished eight C3 proteins (four that were significantly upregulated) encoded by distinct genes, which is a significant expansion on past studies that have reported three distinct C3 proteins in rainbow trout [53, 54]. Our phylogenetic analysis indicated origins for the novel C3 proteins via a history of gene duplication events involving both small-scale and whole genome duplication (WGD) mechanisms, consistent with a past study focused on zebrafish [55] that revealed an ancient teleost-specific C3 member that was identified in our analysis. WGD events are well established in salmonid evolution, including an ancestral autotetraploidization that occurred 88–103 Mya (e.g. [56]). We also observed a > 15-fold upregulation of complement component 1q (C1q), with moderate increases in protein abundance for additional complement components beyond C3, including C4, C5 and factor B. C1q plays a key role in the classical pathway, linking the innate and adaptive systems by binding immunoglobulin molecules to pathogens [57]. Until recently, complement proteins were thought to serve solely immune functions, but emerging evidence suggests metabolic functions, mainly though insulin-like roles and by facilitating triglyceride metabolism [58, 59]. Such roles may contribute to metabolic changes during bacterial infection in rainbow trout, but further work is needed to test this idea. C-reactive protein (CRP) is another innate immune factor that activates complement pathways [60, 61], assists in clearing apoptotic cells [62], and increases liver Kupffer cell phagocytotic activity [63]. Interestingly, CRP showed downregulation in this study, but this nonetheless agrees with past studies of fish liver exposed to A. salmonicida [13, 64, 65]. However, increased CRP was detected in serum of rainbow trout exposed to anti-ectoparasitic chemicals [66] and in common carp (Cyprinus carpio) challenged with A. hydrophila, but not with Escherichia coli lipopolysaccharide [67]. These results suggest either a tissue-specific immune role for CRP or yet to be identified liver functions unrelated to immunity. Additionally, the STRING analysis implicated the complement system as being a tightly connected PPI network. But we caution against over-interpretation of STRING results using salmonids, due to genetic expansions in many protein families caused by ancestral WGD events.
We identified other immune proteins, upregulated by AS treatment, which extend far beyond the complement system. For example, leukocyte cell-derived chemotaxin-2-like, which was > 7-fold increased, contributes to adaptive immunity through its chemotactic properties for neutrophils [68]. C-type lectin domain family 4 member E-like(macrophage-inducible C-type lectin), which increased > 2-fold, is an innate immune receptor that induces expression of proinflammatory cytokines [69, 70]. The gene encoding this protein has shown contradictory expression patterns in past studies of Atlantic salmon liver, being either upregulated [71] or unchanged [46] during AS infection. It is notable that NF-κB (p100/p52 subunit) was upregulated by ~ 2-fold following AS treatment. This transcription factor plays a key role in coordinating pathways that drive immune responses [72] and is rapidly activated by pathogens, stress signals, and pro-inflammatory cytokines, leading to the production of cytokines, chemokines, antimicrobial peptides, stress-response proteins, and anti-apoptotic proteins [72].
Several APR proteins showed altered abundance following AS treatment. The metalloreductase six-transmembrane epithelial antigen of prostate 4 (STEAP4), also known as six-transmembrane protein of prostate 2 (STAMP2), was > 2-fold upregulated, and provides a key role in cellular iron and copper homeostasis, crucial to innate immune function [73]. STAMP2 has been suggested to play an important role in metabolic homeostasis by linking inflammation and nutrient signaling [74, 75]. Ferritin heavy subunit, which binds and sequesters blood plasma iron, was increased > 5-fold in AS, and was previously shown to be upregulated by pro-inflammatory cytokines and bacterial infection [76, 77]. Two distinct proteins identified as haptoglobin, best known for its role in binding free plasma hemoglobin, were robustly increased by AS, agreeing with previous work [25, 27, 43] and the characterized immune functions of this APR [78]. Transferrin receptor 1, which was downregulated in our study, serves as the cellular entry point for transferrin-bound iron [79], reducing the availability of iron for bacterial pathogens [5]. Taken with the lack of transferrin regulation observed in our study, which contradicts past work reporting up-regulation of transferrin by AS [27], it is possible that the transferrin system responds to bacterial infection with a complex temporal dynamic that was not captured by our study.
Protein tyrosine phosphatases (PTPs) have important roles in cellular signaling. One specific PTP of interest for cross-talk between growth and immunity is tyrosine-protein phosphatase non-receptor type 1-like (PTP1B), which was upregulated by AS. PTP1B inhibits glucose uptake in the mammalian liver by dephosphorylating the insulin receptor [80,81,82], potentially making energetic reserves available to other physiological systems (e.g. immune response). PTP1B can also regulate transcription through the attenuation of leptin and JAK2 signaling, which subsequently activate STAT3 [80, 82,83,84]. PTP1B can also increase protein synthesis by activating Src, an integral part of the PI3K/Akt pathway [85,86,87,88], but no significant difference was measured for Src in this study (Additional file 1: Table S3). Contrary to such anabolic effects, there is also evidence for decreased protein synthesis through increased PTP1B induced phosphorylation of PERK and eukaryotic initiation-factor 2α (eIF2α) in mouse pancreatic cells [89]. Further research also suggests a role for PTP1B in immune function, including by negatively regulating cytokine signaling through the dephosphorylation of JAK2 [90, 91] and by TNFα, which promotes PTP1B expression in the liver, partly through NF-κB [92]. The involvement of PTP1B in both growth and immune function makes it a potential candidate for mediating cross-talk between both systems.
An interesting protein upregulated by AS in the context of potential metabolic re-adjustments is the molecular chaperone 78 kDa glucose-regulated protein (GRP78/HSPA5/BiP), which controls activation of the unfolded protein response (UPR) in the endoplasmic reticulum and is upregulated in response to cellular stress [93, 94]. This protein has also been assigned immune functions, including in a past proteomic analysis of an Atlantic salmon head kidney cell line stimulated with the cytokine interferon-γ [95]. Two other proteins that increased following AS treatment, dnaJ homolog subfamily B member 11-like and SIL1 nucleotide exchange factor, serve as co-chaperones to GRP78 during an UPR [96, 97]. The proteinase inhibitor alpha-2-macroglobulin, ~ 2-fold upregulated by AS infection, interacts with GRP78 at the cell surface to activate the PI3K/Akt, ERK1, and MAPK pathways leading to cellular proliferation [98]. Also involved in this pathway is Akt, which serves to activate NF-κB, leading to anti-apoptotic signaling and cellular survival [98]. These interactions of GRP78 indicate a possible role in the rewiring of energetic resources away from growth and protein synthesis and, via NF-κB, into accelerated immune function.
A further subset of proteins was identified that may assist remodeling of liver metabolism in response to infection. We observed downregulation of the eukaryotic translation initiation factor 2B, subunit 5 epsilon (EIF2B5), which likely reduces translation generally, but concomitantly may increase the translation of stress response mRNAs [99, 100]. Phosphatidylinositol transfer protein beta isoform-like, also decreased in AS, is an important component of the polyphosphoinositide synthesis machinery, which is required for epidermal growth factor signaling [101, 102]. Golgi phosphoprotein 3 was increased in AS and is essential for a properly functioning Golgi and morphology [103], but also enhances signaling of the mTORC1 and mTORC2 complexes through increased phosphorylation of their respective substrates, S6K and Akt-S473 [104], leading to increased translation. ATP-binding cassette sub-family A member 1-like (ABCA1), increased > 2.5-fold in AS, facilitates the transfer of cholesterol and lipophilic molecules across cellular membranes, but also has a complicated relationship with cytokines, with some (interferon-γ and interleukin-1β) inhibiting expression, and others (interleukin-10 and transforming growth factor-β1) promoting ABCA1 expression [105]. Another upregulated protein was probable ATP-dependent RNA helicase DDX5 (p68 DEAD box RNA helicase) which has an important role in transcription initiation, elongation, and post-transcriptional processes [106]. Lastly, apolipoprotein B is the primary protein for transporting and distributing lipids throughout the body, especially cholesterol used for plasma membrane and steroid hormone biosynthesis [107] and is generally decreased during the APR [108], matching the observed downregulation in liver of AS-challenged rainbow trout and potentially facilitating metabolic changes. Previous work has also indicated important immune functions for teleost apolipoprotein A [109], which is consistent with the upregulation of one apolipoprotein A protein in response to AS in our study.
Conclusions
This study has revealed a range of proteins induced by bacterial infection in rainbow trout liver, along with proteins that may contribute to accompanying metabolic readjustments. While this study focused on total protein levels, future proteomics work should be aimed at better understanding the signaling changes, many at the level of reversible phosphorylation modifications, driving immune responses and cross-talk between immunity and metabolism during infection.
Methods
Fish husbandry and injection protocols
Rainbow trout (n = 25, approx. Weight: 100 g) were kept in two separate 250 L freshwater tanks at the University of Aberdeen’s aquarium facilities. Water temperature was maintained at 14 °C, and fish were fed a commercial pellet diet at 2% body weight per day. The fish were maintained under these conditions for seven weeks prior to experimental infection challenge. Ten fish (five per tank) were randomly selected to receive either a bacterial or PBS injection (approx. Weight 200 g). The pathogenic Hooke strain of the Gram-negative bacterium A. salmonicida (AS) [110] was used for the challenge. Animals were anaesthetized then injected intraperitoneally (i.p.) with 2 × 105 colony forming units (cfu)/mL AS in PBS (0.5 mL/fish). The same volume (0.5 mL) of PBS was injected i.p. as a control. Sampling occurred 48 h post-infection. The fish were killed using a Schedule 1 method following prior anaesthetization using 2-phenoxyethanol (0.1% v/v) and whole liver and head kidney was immediately sampled, flash frozen in liquid nitrogen and stored at -70 °C until analysis. Samples were taken from ten fish per group (Aeromonas vs. PBS), from which n = 6 biological replicates per group were randomly selected for proteomics (liver), and gene expression profiling to validate a systematic immune response to AS challenge.
Gene expression validation of immune responses
To validate a systematic immune response to AS challenge, we performed qPCR analyses on first-strand cDNA synthesized from total RNA of head kidney samples matched to the same fish used in proteomics (n = 6 control, n = 6 AS). RNA extraction and cDNA synthesis were done as detailed elsewhere [111]. qPCR was performed on a Roche LightCycler® 480 using 2× SYBR® Green I (Invitrogen™) qPCR Master Mix, made with a Immolase DNA Polymerase kit (Bioline), using 10 μL reaction mixtures in 384-well plates (Roche), containing 4 μL diluted cDNA in each reaction and 500 nmol of forward and reverse primers (primer details for IL-1β, TNF-a2 and EF-1α published in Hu et al. [112]). Raw data were analyzed using LightCycler® 480 Software 1.5.1 (Roche). The copy number of each gene was quantified using internal references, by serial dilution of equimolar amounts of PCR product from each gene. Relative gene expression values were separately calculated by normalizing copy number values for IL-1β and TNF-a2 against EF-1α (i.e. reference gene) values. To test for differences between the AS and control samples, a one-way ANOVA was completed in Minitab 18 (Minitab, Inc). As the model residuals either showed non-normality or unequal variances for both target genes, a Box-Cox transformation was performed, leading to data that conformed to the assumptions of normality and equal variances.
Sample preparation for proteomics
Sample preparation, liquid chromatography–mass spectrometry (LC-MS), data analysis, and statistical analysis were performed as reported previously [40]. Briefly, liver tissue was thawed on ice, weighed and lysis buffer (0.5 M pH 6.8 Tris-HCl, 0.2 M EDTA, 8 M Urea, 0.5 M DTT, 10% v/v Glycerol, 10% v/v NP40, pH 3–10 ampholytes) added for a final ratio of ~ 2 mg/μL. The tissue was ground within the buffer using a micropestle, followed by sonication (Fischer Scientific, Sonic Dismembrator) on ice. The resulting suspension was centrifuged at 13,000 g for 5 min, before the supernatant was separated and stored at -80 °C until further analysis. The supernatant was thawed on ice, diluted 50% with molecular grade water, followed by protein precipitation using a ReadyPrep 2-D clean up kit (Bio-Rad Laboratories) according to the manufacturer’s instructions. The resulting pellet was dissolved in 100 μL of 3–10 pH Reswell buffer (Urea, Thiourea, CHAPS, DTT, MilliQ water, and IPG buffer). 5 μL of 3X dissociation buffer (0.5 M pH 6.8 Tris-HCl, 25% SDS, 2-mercaptoethanol, glycerol) was combined with 10 μL of the Reswell solution and incubated for 5 min at 100 °C. A small, 3 μL aliquot of this solution was run a short distance into a 10% acrylamide 1-D gel, then stained with colloidal Coomassie Blue G250 (Fisher Scientific). The protein band was excised for an in-gel tryptic (Promega, sequencing grade) digestion (Digilab ProGest robot). The resulting peptide solutions were dried via centrifugal evaporation (Savant SpeedVac Plus) then dissolved in 20 μL 0.1% formic acid and centrifuged for 5 min at 14,000 g prior to LC-MS.
LC-MS
An UltiMate 3000 RSLCnano (Dionex/Thermo Scientific) coupled to a Q Exactive Plus quadrupole-equipped Orbitrap MS/MS system was used to analyze samples, where 4 μL of the tryptic peptide solution was injected per sample. A loading solvent of water/acetonitrile/formic acid (98:2:0.1) with a flow rate of 10 μL/min was used to concentrate peptides on a μ-precolumn (C18 PepMap; 300 μm i.d. × 5 mm). The μ-precolumn was switched to the analytical flow path after 5 min. Peptides were separated at a flow rate of 0.3 μL/min along a C18 PepMap RSLC column (2 μm i.d. × 50 cm) fitted to an EASY-Spray nano ESI source. Two solvents were used to separate peptides: Solvent A constituted water/formic acid (1000:1) and Solvent B water/acetonitrile/formic acid (200:800:1). An increasing proportion of solvent B was used along a gradient for the separation of peptides: 3–10% from 5 to 25 min: 10–45% from 25 to 185 min; 45–90% from 185 to 190 min; 90% from 190 to 205 min, 90–3% from 205 to 210 min, followed by re-equilibration (3% solvent B, 30 min). A “Top 10” data-dependent acquisition (DDA) method was used, beginning at 5 min into the LC method and lasting for 200 min. The electrospray voltage was 1.9 kV, capillary temperature 270 °C and S-lens RF level 60. The MS scans were performed between 375 and 1750 m/z at resolution 70,000 (m/z 200) with an automatic gain control of 3E + 6 and maximum injection time of 50 ms. The 10 most intense ions of charge state 2–5 were sequentially selected (isolation window 1.6 m/z), followed by fragmentation in the higher-energy collisional dissociation (HCD) cell at a normalized collision energy of 26%. MS2 scans were conducted at resolution 17,500, with an automatic gain control of 5E + 4 and maximum injection time of 100 ms. Additional data-dependent settings included; peptide match preferred, exclude isotopes turned on, and a 40 s dynamic exclusion.
Data analysis
Raw data files from the Q-Exactive were analyzed using MaxQuant (v1.5.3.30; [113]) with a label-free quantification (LFQ) method [114]. MaxQuant default and recommended settings were predominantly used (after: [115]) excepted that unmodified counterpart peptides were not discarded. Trypsin digestion was selected, with a maximum of two missed cleavages. Variable modifications allowed were oxidation of methionine and protein acetylation at the N-terminus, while carbamidomethylation of cysteine was a fixed modification. Peptide and protein identifications were subject to a 1% false-discovery rate (FDR), with a first search mass tolerance for precursor ions set to 20 ppm and a 4.5 ppm setting for the main search. The ‘Match between runs’ option was used to identify missing spectra across samples. Peptides were matched against a high-quality rainbow trout genome annotation including 71,293 RefSeq proteins (NCBI accession; GCA_002163495). Contaminants, reverse identifications, and identifications only by site were filtered from the MaxQuant ‘proteingroups.txt’ file and only proteins that had LFQ values in five samples were kept for statistical analysis. LFQ values were log2 transformed and missing values imputed using missForest, a random forest based non-parametric method [116]. MaxQuant output (Additional file 1: Table S1) and imputed data (Additional file 1: Table S2) can be accessed within the Supplementary Tables (Additional file 1).
Statistical analysis
R version 3.3.2 (“Sincere Pumpkin Patch”) interfacing with R-studio v1.0.136 (Rstudio, Boston, MA) was used for statistical modeling and graphics production. A linear model was used to determine differences between AS and control treatments, done in the ‘limma’ package with smoothing of the standard errors using an empirical Bayes approach [117] and an applied false-discovery rate of 0.1 (rationale in Results section). Heatmaps were produced by comparing Z-scores of normalized LFQ values using the ‘gplots’ and ‘seriation’ packages [118, 119]. Hierarchical clustering was achieved by minimizing Hamiltonian path length through optimal leaf ordering [120]. Multivariate analyses were performed on the same filtered, log2 transformed, and imputed LFQ values, using ‘vegan’ [121]. Non-metric multidimensional scaling (nMDS) was used to visualize the data over a PCA to better preserve the distance between data points. Simultaneous changes across the liver proteomes of AS and control samples was determined using a permutational ANOVA (PERMANOVA, 9999 permutations) [122]. The multivariate homogeneity of group dispersion (variance) was assessed and revealed no dispersion effect [123]. One control (PBS) individual grouped with the AS-challenged fish in the nMDS analysis and was taken as an outlier that was removed from the study.
Gene ontology (GO) analyses
GO enrichment analysis was conducted for significantly differentially abundant proteins identified from the AS vs control comparison. All rainbow trout proteins identified from MaxQuant were used in BLASTp [124] searches against Atlantic salmon RefSeq proteins predicted from the reference ICSASG_v2 genome [34] (NCBI accession: GCA_000233375.4), which are assigned with GO terms [125]. This allowed us to provide all identified rainbow trout proteins with GO terms for use in enrichment analyses (Additional file 1: Table S1). GO biological process (BP) enrichment was determined using the ‘topGO’ package with the ‘weight01’ algorithm and Fisher’s test statistic [126]. GO slim was conducted using the ‘GOstats’ and ‘GSEABase’ packages [127, 128].
STRING PPI analysis
PPIs were determined using the STRING database (http://string-db.org/) that determines both physical and functional associations between proteins [129]. The subset of 109 proteins that showed significant differential abundance due to AS treatment were entered into the STRING database. The Homo sapiens orthologues were determined within the STRING database from each O. mykiss amino acid sequence. Default settings were used, with the interaction score set to “high confidence (0.700)”. Each node represents a protein while the edges indicate the strength of the relationship between proteins (i.e. more edges give higher confidence).
Phylogenetic analyses of C3 proteins
Phylogenetic analysis of rainbow trout C3 proteins identified in our dataset was performed using sequences gathered from a standardized set of vertebrate taxa. We began with the Human C3 protein and used BLASTp [124] against the NCBI non-redundant protein database to extract putative C3 orthologues from a range of teleost taxa. Previous studies identified three rainbow trout C3 proteins [53, 54] and eight zebrafish (Danio rerio) C3 proteins [55] that provided key reference points for the analysis. As done previously [55], vertebrate C5 proteins were gathered using the same approach as an outgroup for phylogenetic analyses. The collected protein sequences (n = 36) were aligned using Mafft V7 [130] with default settings before alignment quality filtering was completed using the Guidance2 algorithm [131], leading to a high-confidence 1243 amino acid sequence alignment (provided as Additional File 2: Supplementary Dataset 1). The maximum likelihood approach IQ-tree [132] and server [133] was used to determine the best-fitting amino acid substitution model (WAG+F + I + G4) and build a consensus tree employing the same model, along with 1000 ultrafast bootstrap pseudoreplicates [134] to gain branch support values. The consensus tree was visualized and rendered using FigTree V1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
Abbreviations
- ABCA1:
-
ATP-binding cassette sub-family A member 1-like
- APR:
-
Acute phase response
- AS:
-
Aeromonas salmonicida-injected
- BP:
-
Biological Process
- C1q:
-
Complement component 1q
- C3–5:
-
Complement component 3, 4, or 5
- cfu:
-
Colony forming units
- CRP:
-
C-reactive protein
- DDA:
-
Data-dependent acquisition
- DDX5:
-
p68 DEAD box RNA helicase
- EIF2B5:
-
Eukaryotic translation initiation factor 2B, subunit 5 epsilon
- eIF2α:
-
Eukaryotic initiation-factor 2α
- Erk:
-
Extracellular signal-regulated kinase
- FDR:
-
False-discovery rate
- GO:
-
Gene Ontology
- GRP78:
-
78 kDa glucose-regulated protein
- HCD:
-
Higher-energy collisional dissociation
- i.p.:
-
Interperitoneally
- IL:
-
Interleukin
- JAK2:
-
Janus kinase 2
- LFQ:
-
Label-free quantification
- mTORC1 & 2:
-
Mammalian target of rapamycin complex 1 & 2
- NF-κB:
-
Nuclear factor kappa-light-chain enhancer of activated B cells
- PERK:
-
Protein kinase R (PKR)-like endoplasmic reticulum kinase
- PERMANOVA:
-
Permutational ANOVA
- PI3K:
-
Phosphatidylinositol-4,5-bisphosphate 3-kinase
- PPI:
-
Protein-protein interaction
- PTP:
-
Protein tyrosine phosphatase
- PTP1B:
-
Tyrosine-protein phosphatase non-receptor type 1-like
- S6K:
-
Ribosomal protein S6 kinase
- STAMP2:
-
Six-transmembrane protein of prostate 2
- STAT3:
-
Signal transducer and activator of transcription 3
- STEAP4:
-
Six-transmembrane epithelial antigen of prostate 4
- TNFα:
-
Tumor necrosis factor α
- UPR:
-
Unfolded protein response
- WGD:
-
Whole genome duplication
References
Magnadóttir B. Innate immunity of fish (overview). Fish Shellfish Immunol. 2006;20:137–51.
Secombes CJ. What’s new in fish cytokine research? Fish Shellfish Immunol. 2016;53:1–3.
Bird S, Zou J, Secombes CJ. Advances in fish cytokine biology give clues to the evolution of a complex network. Curr Pharm Des. 2006;12:3051–69.
Zou J, Secombes CJ. The function of fish cytokines. Biology. 2016;5:23.
Bayne CJ, Gerwick L. The acute phase response and innate immunity of fish. Dev Comp Immunol. 2001;25:725–43.
Bayne CJ, Gerwick L, Fujiki K, Nakao M, Yano T. Immune-relevant (including acute phase) genes identified in the livers of rainbow trout, Oncorhynchus mykiss, by means of suppression subtractive hybridization. Dev Comp Immunol. 2001;25:205–17.
Gomez D, Sunyer JO, Salinas I. The mucosal immune system of fish: the evolution of tolerating commensals while fighting pathogens. Fish Shellfish Immunol. 2013;35:1729–39.
Xu Z, Parra D, Gómez D, Salinas I, Zhang Y-A, Jørgensen L von G, et al. Teleost skin, an ancient mucosal surface that elicits gut-like immune responses. Proc Natl Acad Sci. 2013;110:13097–102.
Uribe C, Folch H, Enriquez R, Moran G. Innate and adaptive immunity in teleost fish: a review. Vet Med. 2011;56:486–503.
Robinson MW, Harmon C, O’Farrelly C. Liver immunology and its role in inflammation and homeostasis. Cell Mol Immunol. 2016;13:267–76.
Freitas-Lopes MA, Mafra K, David BA, Carvalho-Gontijo R, Menezes GB. Differential location and distribution of hepatic immune cells. Cell. 2017;6. https://doi.org/10.3390/cells6040048.
Fraser KPP, Rogers AD. Protein metabolism in marine animals: the underlying mechanism of growth. Adv Mar Biol. 2007;52:267–362.
Jensen LE, Hiney MP, Shields DC, Uhlar CM, Lindsay AJ, Whitehead AS. Acute phase proteins in salmonids: evolutionary analyses and acute phase response. J Immunol. 1997;158:384–92.
Rauw WM. Immune response from a resource allocation perspective. Front Genet. 2012;3:267. https://doi.org/10.3389/fgene.2012.00267.
Houston AI, McNamara JM, Barta Z, Klasing KC. The effect of energy reserves and food availability on optimal immune defence. Proc R Soc Lond B Biol Sci. 2007;274:2835–42.
Alzaid A, Castro R, Wang T, Secombes CJ, Boudinot P, Macqueen DJ, et al. Cross-talk between growth and immunity: coupling of the insulin-like growth factor axis to conserved cytokine pathways in rainbow trout. Endocrinology. 2016;157:2015–24.
Alzaid A, Kim J-H, Devlin RH, Martin SAM, Macqueen DJ. Growth hormone transgenesis in coho salmon disrupts muscle immune function impacting cross-talk with growth systems. J Exp Biol. 2018;221. https://doi.org/10.1242/jeb.173146.
Martin SAM, Król E. Nutrigenomics and immune function in fish: new insights from omics technologies. Dev Comp Immunol. 2017;75(Suppl C):86–98.
Holland JW, Pottinger TG, Secombes CJ. Recombinant interleukin-1 beta activates the hypothalamic-pituitary-interrenal axis in rainbow trout, Oncorhynchus mykiss. J Endocrinol. 2002;175:261–7.
Tort L. Stress and immune modulation in fish. Dev Comp Immunol. 2011;35:1366–75.
Weyts FAA, Cohen N, Flik G, Verburg-van Kemenade BML. Interactions between the immune system and the hypothalamo-pituitary-interrenal axis in fish. Fish Shellfish Immunol. 1999;9:1–20.
Lam SH, Sin YM, Gong Z, Lam TJ. Effects of thyroid hormone on the development of immune system in zebrafish. Gen Comp Endocrinol. 2005;142:325–35.
Quesada-García A, Valdehita A, Kropf C, Casanova-Nakayama A, Segner H, Navas JM. Thyroid signaling in immune organs and cells of the teleost fish rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 2014;38:166–74.
Harris J, Bird DJ. Modulation of the fish immune system by hormones. Vet Immunol Immunopathol. 2000;77:163–76.
Martin SAM, Blaney SC, Houlihan DF, Secombes CJ. Transcriptome response following administration of a live bacterial vaccine in Atlantic salmon (Salmo salar). Mol Immunol. 2006;43:1900–11.
Skugor S, Jørgensen SM, Gjerde B, Krasnov A. Hepatic gene expression profiling reveals protective responses in Atlantic salmon vaccinated against furunculosis. BMC Genomics. 2009;10:503.
Vanya Ewart K, Belanger JC, Williams J, Karakach T, Penny S, Tsoi SCM, et al. Identification of genes differentially expressed in Atlantic salmon (Salmo salar) in response to infection by Aeromonas salmonicida using cDNA microarray technology. Dev Comp Immunol. 2005;29:333–47.
Long M, Zhao J, Li T, Tafalla C, Zhang Q, Wang X, et al. Transcriptomic and proteomic analyses of splenic immune mechanisms of rainbow trout (Oncorhynchus mykiss) infected by Aeromonas salmonicida subsp. salmonicida. J Proteome. 2015;122:41–54.
Liu P-F, Du Y, Meng L, Li X, Liu Y. Proteomic analysis in kidneys of Atlantic salmon infected with Aeromonas salmonicida by iTRAQ. Dev Comp Immunol. 2017;72:140–53.
Di G, Li H, Zhang C, Zhao Y, Zhou C, Naeem S, et al. Label-free proteomic analysis of intestinal mucosa proteins in common carp (Cyprinus carpio) infected with Aeromonas hydrophila. Fish Shellfish Immunol. 2017;66:11–25.
Abdelhamed H, Ibrahim I, Nho SW, Banes MM, Wills RW, Karsi A, et al. Evaluation of three recombinant outer membrane proteins, OmpA1, Tdr, and TbpA, as potential vaccine antigens against virulent Aeromonas hydrophila infection in channel catfish (Ictalurus punctatus). Fish Shellfish Immunol. 2017;66:480–6.
Kumar R, Pande V, Singh L, Sharma L, Saxena N, et al. Pathological findings of experimental Aeromonas hydrophila infection in Golden Mahseer (Tor putitora). Fish Aquac J. 2016;7:1–6.
Hadidi S, Glenney GW, Welch TJ, Silverstein JT, Wiens GD. Spleen size predicts resistance of rainbow trout to Flavobacterium psychrophilum challenge. J Immunol. 2008;180:4156–65.
Lien S, Koop BF, Sandve SR, Miller JR, Kent MP, Nome T, et al. The Atlantic salmon genome provides insights into rediploidization. Nature. 2016;533:200–5.
Chiozzi RZ, Capriotti AL, Cavaliere C, Barbera GL, Montone CM, Piovesana S, et al. Label-free shotgun proteomics approach to characterize muscle tissue from farmed and wild European Sea bass (Dicentrarchus labrax). Food Anal Methods. 2018;11:292–301.
Dong M, Tang SW, Shen W, Au DWT, Lam YW. Characterization of the interactions between fish serum proteins and pathogenic bacteria by using label-free quantitative proteomics. J Immunol. 2016;196(1 Suppl):63.5.
Li C, Tan XF, Lim TK, Lin Q, Gong Z. Comprehensive and quantitative proteomic analyses of zebrafish plasma reveals conserved protein profiles between genders and between zebrafish and human. Sci Rep. 2016;6:24329.
Nuez-Ortín WG, Carter CG, Wilson R, Cooke IR, Amoroso G, Cobcroft JM, et al. Triploid Atlantic salmon shows similar performance, fatty acid composition and proteome response to diploids during early freshwater rearing. Comp Biochem Physiol Part D Genomics Proteomics. 2017;22:67–77.
Piovesana S, Capriotti AL, Caruso G, Cavaliere C, La Barbera G, Zenezini Chiozzi R, et al. Labeling and label free shotgun proteomics approaches to characterize muscle tissue from farmed and wild gilthead sea bream (Sparus aurata). J Chromatogr A. 2016;1428:193–201.
Causey DR, Kim J-H, Stead DA, SAM M, Devlin RH, Macqueen DJ. Proteomic comparison of selective breeding and growth hormone transgenesis in fish: Unique pathways to enhanced growth. J Proteomics. 2018. https://doi.org/10.1016/j.jprot.2018.08.013.
Macqueen DJ, Primmer CR, Houston RD, Nowak BF, Bernatchez L, Bergseth S, et al. Functional annotation of all salmonid genomes (FAASG): an international initiative supporting future salmonid research, conservation and aquaculture. BMC Genomics. 2017;18:484.
Nuez-Ortín WG, Carter CG, Nichols PD, Cooke IR, Wilson R. Liver proteome response of pre-harvest Atlantic salmon following exposure to elevated temperature. BMC Genomics. 2018;19:133.
Martin SA, Douglas A, Houlihan DF, Secombes CJ. Starvation alters the liver transcriptome of the innate immune response in Atlantic salmon (Salmo salar). BMC Genomics. 2010;11:418.
Raida MK, Buchmann K. Innate immune response in rainbow trout (Oncorhynchus mykiss) against primary and secondary infections with Yersinia ruckeri O1. Dev Comp Immunol. 2009;33:35–45.
Rise ML, Jones SRM, Brown GD, von Schalburg KR, Davidson WS, Koop BF. Microarray analyses identify molecular biomarkers of Atlantic salmon macrophage and hematopoietic kidney response to Piscirickettsia salmonis infection. Physiol Genomics. 2004;20:21–35.
Tsoi SCM, Ewart KV, Penny S, Melville K, Liebscher RS, Brown LL, et al. Identification of immune-relevant genes from Atlantic Salmon using suppression subtractive hybridization. Mar Biotechnol. 2004;6:199–214.
Marsden MJ, Devoy A, Vaughan LM, Foster TJ, Secombes CJ. Use of a genetically attenuated strain of Aeromonas salmonicida to vaccinate salmonid fish. Aquac Int. 1996;4:55–66.
Merle NS, Noe R, Halbwachs-Mecarelli L, Fremeaux-Bacchi V, Roumenina LT. Complement system part II: role in immunity. Front Immunol. 2015;6:257. https://doi.org/10.3389/fimmu.2015.00257.
Zhu L, Nie L, Zhu G, Xiang L, Shao J. Advances in research of fish immune-relevant genes: a comparative overview of innate and adaptive immunity in teleosts. Dev Comp Immunol. 2013;39:39–62.
Ellis AE. Immunity to bacteria in fish. Fish Shellfish Immunol. 1999;9:291–308.
Ellis AE. Innate host defense mechanisms of fish against viruses and bacteria. Dev Comp Immunol. 2001;25:827–39.
Holland MCH, Lambris JD. The complement system in teleosts. Fish Shellfish Immunol. 2002;12:399–420.
Sunyer JO, Zarkadis IK, Sahu A, Lambris JD. Multiple forms of complement C3 in trout that differ in binding to complement activators. Proc Natl Acad Sci. 1996;93:8546–51.
Zarkadis IK, Sarrias MR, Sfyroera G, Sunyer JO, Lambris JD. Cloning and structure of three rainbow trout C3 molecules: a plausible explanation for their functional diversity. Dev Comp Immunol. 2001;25:11–24.
Forn-Cuní G, Reis ES, Dios S, Posada D, Lambris JD, Figueras A, et al. The evolution and appearance of C3 duplications in fish originate an exclusive teleost c3 gene form with anti-inflammatory activity. PLoS One. 2014;9:e99673.
Macqueen DJ, Johnston IA. A well-constrained estimate for the timing of the salmonid whole genome duplication reveals major decoupling from species diversification. Proc R Soc Lond B Biol Sci. 2014;281:20132881.
Boshra H, Gelman AE, Sunyer JO. Structural and functional characterization of complement C4 and C1s-like molecules in teleost fish: insights into the evolution of classical and alternative pathways. J Immunol. 2004;173:349–59.
Barbu A, Hamad OA, Lind L, Ekdahl KN, Nilsson B. The role of complement factor C3 in lipid metabolism. Mol Immunol. 2015;67:101–7.
King BC, Blom AM. Non-traditional roles of complement in type 2 diabetes: metabolism, insulin secretion and homeostasis. Mol Immunol. 2017;84:34–42.
Cook MT, Hayball PJ, Birdseye L, Bagley C, Nowak BF, Hayball JD. Isolation and partial characterization of a pentraxin-like protein with complement-fixing activity from snapper (Pagrus auratus, Sparidae) serum. Dev Comp Immunol. 2003;27:579–88.
Jiang HX, Siegel JN, Gewurz H. Binding and complement activation by C-reactive protein via the collagen-like region of C1q and inhibition of these reactions by monoclonal antibodies to C-reactive protein and C1q. J Immunol. 1991;146:2324–30.
Nauta AJ, Daha MR, van KC, Roos A. Recognition and clearance of apoptotic cells: a role for complement and pentraxins. Trends Immunol. 2003;24:148–54.
Inatsu A, Manabu K, Nakashima H, Shimizu J, Saitoh D, Seiichi T, et al. Novel mechanism of C-reactive protein for enhancing mouse liver innate immunity. Hepatology. 2009;49:2044–54.
Lee PT, Bird S, Zou J, Martin SAM. Phylogeny and expression analysis of C-reactive protein (CRP) and serum amyloid-P (SAP) like genes reveal two distinct groups in fish. Fish Shellfish Immunol. 2017;65:42–51.
Lund V, Olafsen JA. A comparative study of pentraxin-like proteins in different fish species. Dev Comp Immunol. 1998;22:185–94.
Kodama H, Matsuoka Y, Tanaka Y, Liu Y, Iwasaki T, Watarai S. Changes of C-reactive protein levels in rainbow trout (Oncorhynchus mykiss) sera after exposure to anti-ectoparasitic chemicals used in aquaculture. Fish Shellfish Immunol. 2004;16:589–97.
MacCarthy EM, Burns I, Irnazarow I, Polwart A, Greenhough TJ, Shrive AK, et al. Serum CRP-like protein profile in common carp Cyprinus carpio challenged with Aeromonas hydrophila and Escherichia coli lipopolysaccharide. Dev Comp Immunol. 2008;32:1281–9.
Chenoweth DE, Rowe JG, Hugli TE. A modified method for chemotaxis under agarose. J Immunol Methods. 1979;25:337–53.
Holmskov U, Thiel S, Jensenius JC. Collectins and Ficolins: humoral lectins of the innate immune defense. Annu Rev Immunol. 2003;21:547–78.
Patin EC, Orr SJ, Schaible UE. Macrophage inducible C-type lectin as a multifunctional player in immunity. Front Immunol. 2017;8:861. https://doi.org/10.3389/fimmu.2017.00861.
Soanes KH, Figuereido K, Richards RC, Mattatall NR, Ewart KV. Sequence and expression of C-type lectin receptors in Atlantic salmon (Salmo salar). Immunogenetics. 2004;56:572–84.
Li Q, Verma IM. NF-κB regulation in the immune system. Nat Rev Immunol. 2002;2:725–34.
Scarl RT, Lawrence CM, Gordon HM, Nunemaker CS. STEAP4: its emerging role in metabolism and homeostasis of cellular iron and copper. J Endocrinol. 2017;234:R123–34.
Hotamisligil GS, Erbay E. Nutrient sensing and inflammation in metabolic diseases. Nat Rev Immunol. 2008;8:923–34.
Wellen KE, Fucho R, Gregor MF, Furuhashi M, Morgan C, Lindstad T, et al. Coordinated regulation of nutrient and inflammatory responses by STAMP2 is essential for metabolic homeostasis. Cell. 2007;129:537–48.
Lee J-H, Pooley NJ, Mohd-Adnan A, Martin SAM. Cloning and characterisation of multiple ferritin isoforms in the Atlantic Salmon (Salmo salar). PLoS One. 2014;9:e103729. https://doi.org/10.1371/journal.pone.0103729.
Torti FM, Torti SV. Regulation of ferritin genes and protein. Blood. 2002;99:3505–16.
Huntoon KM, Wang Y, Eppolito CA, Barbour KW, Berger FG, Shrikant PA, et al. The acute phase protein haptoglobin regulates host immunity. J Leukoc Biol. 2008;84:170–81.
Hentze MW, Muckenthaler MU, Galy B, Camaschella C. Two to tango: regulation of mammalian Iron metabolism. Cell. 2010;142:24–38.
Delibegovic M, Zimmer D, Kauffman C, Rak K, Hong E-G, Cho Y-R, et al. Liver-specific deletion of protein-tyrosine phosphatase 1B (PTP1B) improves metabolic syndrome and attenuates diet-induced endoplasmic reticulum stress. Diabetes. 2009;58:590–9.
Tiganis T, Bennett AM. Protein tyrosine phosphatase function: the substrate perspective. Biochem J. 2007;402:1–15.
Zabolotny JM, Bence-Hanulec KK, Stricker-Krongrad A, Haj F, Wang Y, Minokoshi Y, et al. PTP1B regulates leptin signal transduction in vivo. Dev Cell. 2002;2:489–95.
Bates SH, Stearns WH, Dundon TA, Schubert M, Tso AWK, Wang Y, et al. STAT3 signalling is required for leptin regulation of energy balance but not reproduction. Nature. 2003;421:856–9.
Cheng A, Uetani N, Simoncic PD, Chaubey VP, Lee-Loy A, McGlade CJ, et al. Attenuation of leptin action and regulation of obesity by protein tyrosine phosphatase 1B. Dev Cell. 2002;2:497–503.
Bjorge JD, Pang A, Fujita DJ. Identification of protein-tyrosine phosphatase 1B as the major tyrosine phosphatase activity capable of dephosphorylating and activating c-Src in several human breast Cancer cell lines. J Biol Chem. 2000;275:41439–46.
Cheng A, Bal GS, Kennedy BP, Tremblay ML. Attenuation of adhesion-dependent signaling and cell spreading in transformed fibroblasts lacking protein tyrosine phosphatase-1B. J Biol Chem. 2001;276:25848–55.
Liang F, Lee S-Y, Liang J, Lawrence DS, Zhang Z-Y. The role of protein-tyrosine phosphatase 1B in integrin signaling. J Biol Chem. 2005;280:24857–63.
Wu C, You J, Fu J, Wang X, Zhang Y. Phosphatidylinositol 3-kinase/Akt mediates integrin signaling to control RNA polymerase I transcriptional activity. Mol Cell Biol. 2016;36:1555–68.
Bettaieb A, Liu S, Xi Y, Nagata N, Matsuo K, Matsuo I, et al. Differential regulation of endoplasmic reticulum stress by protein tyrosine phosphatase 1B and T cell protein tyrosine phosphatase. J Biol Chem. 2011;286:9225–35.
Heinonen KM, Dubé N, Bourdeau A, Lapp WS, Tremblay ML. Protein tyrosine phosphatase 1B negatively regulates macrophage development through CSF-1 signaling. Proc Natl Acad Sci. 2006;103:2776–81.
Myers MP, Andersen JN, Cheng A, Tremblay ML, Horvath CM, Parisien J-P, et al. TYK2 and JAK2 are substrates of protein-tyrosine phosphatase 1B. J Biol Chem. 2001;276:47771–4.
Zabolotny JM, Kim Y-B, Welsh LA, Kershaw EE, Neel BG, Kahn BB. Protein-tyrosine phosphatase 1B expression is induced by inflammation in vivo. J Biol Chem. 2008;283:14230–41.
Ni M, Zhang Y, Lee AS. Beyond the endoplasmic reticulum: atypical GRP78 in cell viability, signalling and therapeutic targeting. Biochem J. 2011;434:181–8.
Lee AS. Mammalian stress response: induction of the glucose-regulated protein family. Curr Opin Cell Biol. 1992;4:267–73.
Martin SAM, Mohanty BP, Cash P, Houlihan DF, Secombes CJ. Proteome analysis of the Atlantic salmon (Salmo salar) cell line SHK-1 following recombinant IFN-γ stimulation. Proteomics. 2007;7:2275–86.
Chung KT, Shen Y, Hendershot LM. BAP, a mammalian BiP-associated protein, is a nucleotide exchange factor that regulates the ATPase activity of BiP. J Biol Chem. 2002;277:47557–63.
Shen Y, Hendershot LM. ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a CoFactor for BiP’s interactions with unfolded substrates. Mol Biol Cell. 2004;16:40–50.
Misra UK, Deedwania R, Pizzo SV. Activation and cross-talk between Akt, NF-κB, and unfolded protein response signaling in 1-LN prostate Cancer cells consequent to ligation of cell surface-associated GRP78. J Biol Chem. 2006;281:13694–707.
Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–35.
Pavitt GD. eIF2B, a mediator of general and gene-specific translational control. Biochem Soc Trans. 2005;33:1487–92.
Thomas GMH, Cunningham E, Fensome A, Ball A, Totty NF, Truong O, et al. An essential role for phosphatidylinositol transfer protein in phospholipase C-mediated inositol lipid signaling. Cell. 1993;74:919–28.
Kauffmann-Zeh A, Thomas GM, Ball A, Prosser S, Cunningham E, Cockcroft S, et al. Requirement for phosphatidylinositol transfer protein in epidermal growth factor signaling. Science. 1995;268:1188–90.
Dippold HC, Ng MM, Farber-Katz SE, Lee S-K, Kerr ML, Peterman MC, et al. GOLPH3 bridges Phosphatidylinositol-4- phosphate and Actomyosin to stretch and shape the Golgi to promote budding. Cell. 2009;139:337–51.
Scott KL, Kabbarah O, Liang MC, Ivanova E, Anagnostou V, Wu J, et al. GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature. 2009;459:1085–90.
Yin K, Liao D, Tang C. ATP-binding membrane cassette transporter A1 (ABCA1): a possible link between inflammation and reverse cholesterol transport. Mol Med. 2010;16:438–49.
Fuller-Pace FV, Ali S. The DEAD box RNA helicases p68 (Ddx5) and p72 (Ddx17): novel transcriptional co-regulators. Biochem Soc Trans. 2008;36:609–12.
Mahley RW, Innerarity TL, Rall SC, Weisgraber KH. Plasma lipoproteins: apolipoprotein structure and function. J Lipid Res. 1984;25:1277–94.
Hoffman JS, Benditt EP. Changes in high density lipoprotein content following endotoxin administration in the mouse. Formation of serum amyloid protein-rich subfractions. J Biol Chem. 1982;257:10510–7.
Concha MI, Smith VJ, Castro K, Bastías A, Romero A, Amthauer RJ. Apolipoproteins A-I and A-II are potentially important effectors of innate immunity in the teleost fish Cyprinus carpio. Eur J Biochem. 2004;271:2984–90.
Scott CJW, Austin B, Austin DA, Morris PC. Non-adjuvanted flagellin elicits a non-specific protective immune response in rainbow trout (Oncorhynchus mykiss, Walbaum) towards bacterial infections. Vaccine. 2013;31:3262–7.
Wang T, Secombes CJ. Rainbow trout suppressor of cytokine signalling (SOCS)-1, 2 and 3: molecular identification, expression and modulation. Mol Immunol. 2008;45:1449–57.
Hu Y, Maisey K, Subramani PA, Liu F, Flores-Kossack C, Imarai M, et al. Characterisation of rainbow trout peripheral blood leucocytes prepared by hypotonic lysis of erythrocytes, and analysis of their phagocytic activity, proliferation and response to PAMPs and proinflammatory cytokines. Dev Comp Immunol. 2018;88:104–13.
Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72.
Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–26.
Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11:2301–19.
Stekhoven DJ, Bühlmann P. MissForest—non-parametric missing value imputation for mixed-type data. Bioinformatics. 2012;28:112–8.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47.
Hahsler M, Buchta C, Hornik K, Murtagh F, Brusco M, Stahl S, et al. Seriation: infrastructure for ordering objects using seriation. 2016. https://cran.r-project.org/web/packages/seriation/index.html.
Warnes GR, Bolker B, Bonebakker L, Gentleman R, Liaw WHA, Lumley T, et al. Gplots: various R programming tools for plotting data. 2016. https://cran.r-project.org/web/packages/gplots/index.html.
Bar-Joseph Z, Gifford DK, Jaakkola TS. Fast optimal leaf ordering for hierarchical clustering. Bioinformatics. 2001;17(suppl_1):S22–9.
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, et al. Vegan: community ecology package. 2016. https://cran.r-project.org/web/packages/vegan/index.html.
Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46.
Anderson MJ. Distance-based tests for homogeneity of multivariate dispersions. Biometrics. 2006;62:245–53.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
Robertson FM, Gundappa MK, Grammes F, Hvidsten TR, Redmond AK, Lien S, et al. Lineage-specific rediploidization is a mechanism to explain time-lags between genome duplication and evolutionary diversification. Genome Biol. 2017;18:111.
Alexa A, Rahnenfuhrer J. topGO: enrichment analysis for gene ontology. 2010. http://bioconductor.org/packages/release/bioc/html/topGO.html.
Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–8.
Morgan M, Falcon S, Gentleman R. GSEABase: Gene set enrichment data structures and methods. 2015. http://bioconductor.org/packages/release/bioc/html/GSEABase.html.
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–52.
Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80.
Sela I, Ashkenazy H, Katoh K, Pupko T. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 2015;43:W7–14.
Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74.
Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44:W232–5.
Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–22.
Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–56.
Acknowledgements
We thank Dr. Tiehui Wang (University of Aberdeen) for help with the bacterial challenge, Dr. Abdullah Alzaid (formerly University of Aberdeen) for collecting liver samples, and Evelyn Argo / Craig Pattinson (Aberdeen Proteomics Facility) for performing sample preparation.
Funding
Proteomics research in DJM’s lab was supported by the Royal Society (grant ref.: RG130823) and the Biotechnology and Biological Sciences Research Council (grant ref.: BB/M026345/1). DRC’s PhD studentship is supported through The Developmental Trust and Elphinstone Scholarship Programme of the University of Aberdeen.
Availability of data and materials
All mass spectrometry proteomics data generated in the study was deposited to the ProteomeXchange Consortium via the PRIDE [135] partner repository with the dataset identifier PXD010186. All other data generated or analyzed during the study are included in this published article and its supplementary information files.
Author information
Authors and Affiliations
Contributions
Conceived study and designed: DJM, SAMM, CJS, MAP; performed animal experiments and sampling: MAP; performed LC-MS: DAS; analyzed proteomic data: DRC; performed statistics: DRC; drafted Figures and Tables: DRC; drafted manuscript: DRC, DJM. All authors contributed to editing and finalization of the submitted manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
All animal work was carried out in compliance with the Animals 145 (Scientific Procedures) Act 1986 by a UK Home Office license holder, approved by the ethics committee of the University of Aberdeen.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional File 1:
Table S1. Results from BLASTp of Oncorhynchus mykiss protein sequences against Salmo salar and MaxQuant output after filtering contaminants, reverse sequences, proteins identified by site, and proteins with only 1 peptide for identification (3076 proteins). Table S2. Imputed protein abundance data after implementing all filtering criteria (2433 proteins). Table S3. Full statistical results from the linear model for each of the 2433 proteins included in the analysis. Table S4. Full results for GO Biological Process (GOBP) enrichment analysis. Table S5. Significantly different proteins that contribute to each significant GOBP term. Table S6. Significantly different proteins that contribute to each significant GO slim term. Table S7. Abbreviations and proteins annotations from STRING. (XLSX 3388 kb)
Additional File 2:
Supplementary Dataset 1. Sequence alignment used for phylogenetic analysis of C3 complement sequences. (FASTA 47 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Causey, D.R., Pohl, M.A.N., Stead, D.A. et al. High-throughput proteomic profiling of the fish liver following bacterial infection. BMC Genomics 19, 719 (2018). https://doi.org/10.1186/s12864-018-5092-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-018-5092-0