Abstract
Background
Colorectal cancer (CRC) is one of the most common malignant tumors worldwide. CRC molecular pathogenesis is heterogeneous and may be followed by mutations in oncogenes and tumor suppressor genes, chromosomal and microsatellite instability, alternative splicing alterations, hypermethylation of CpG islands, oxidative stress, impairment of different signaling pathways and energy metabolism. In the present work, we have studied the alterations of alternative splicing patterns of genes related to energy metabolism in CRC.
Results
Using CrossHub software, we analyzed The Cancer Genome Atlas (TCGA) RNA-Seq datasets derived from colon tumor and matched normal tissues. The expression of 1014 alternative mRNA isoforms involved in cell energy metabolism was examined. We found 7 genes with differentially expressed alternative transcripts whereas overall expression of these genes was not significantly altered in CRC. A set of 8 differentially expressed transcripts of interest has been validated by qPCR. These eight isoforms encoded by OGDH, COL6A3, ICAM1, PHPT1, PPP2R5D, SLC29A1, and TRIB3 genes were up-regulated in colorectal tumors, and this is in concordance with the bioinformatics data. The alternative transcript NM_057167 of COL6A3 was also strongly up-regulated in breast, lung, prostate, and kidney tumors. Alternative transcript of SLC29A1 (NM_001078177) was up-regulated only in CRC samples, but not in the other tested tumor types.
Conclusions
We identified tumor-specific expression of alternative spliced transcripts of seven genes involved in energy metabolism in CRC. Our results bring new knowledge on alternative splicing in colorectal cancer and suggest a set of mRNA isoforms that could be used for cancer diagnosis and development of treatment methods.
Similar content being viewed by others
Background
Colorectal cancer (CRC) is the third most common cancer in the world [1]. It accounts for more than 1,360,000 new cancer incidences and about 9% of all cancer deaths worldwide [2], [http://gco.iarc.fr/]. The risk of CRC increases with age, and CRC incidence rates are higher among males, than females [3]. CRC starts in either the colon or the rectum and is represented by several histological types. More than 90% of CRC are adenocarcinomas [4]. Generally CRC patients are characterized by a lack of clinical symptoms in early stages, and this leads to poor prognosis and high mortality rate [5]. Approximately 20% of patients with CRC have already developed metastatic disease at the time of diagnosis [6, 7]. Colorectal cancer metastases are found in the liver, lung, skin, and brain [8]. The median survival of patients with advanced metastatic disease is less than 24 months [9]. Although the medical management of CRC has improved, there are limited therapeutic options for advanced CRC. Identification of new therapeutic targets and biomarkers is imperative for the development of CRC therapies and diagnosis.
Energy metabolism in cancer cells is characterized by increased glucose uptake and aerobic glycolysis [10]. Even in the presence of oxygen, most cancer cells produce lactate instead of oxidation of glycolytic pyruvate in the mitochondrial tricarboxylic acid (TCA) cycle [11, 12]. This phenomenon was originally observed by Otto Warburg and termed the “Warburg effect” [13]. Increased aerobic glycolysis is associated with the alteration of gene expression, protein modifications and mutations [14]. Warburg effect is the tumor adaptation mechanism to oxidative stress and hypoxia [15]. Each type of cancer is characterized by a distinct metabolic signature due to its unique transformation process [16, 17]. Elucidation of the metabolic specificities of several cancers and metabolic differences between cancer and normal cells has provided important biomarker findings [18, 19].
The aim of the study was to identify tumor-associated expression of alternatively spliced transcripts related to energy metabolism in CRC. Obtained data suggest several ones that could be involved in the development of CRC through altered energy metabolism. Identified tumor-specific mRNA isoforms may be used for the development of cancer diagnosis and treatment methods.
Methods
Bioinformatics analysis
We analyzed TCGA RNA-Seq datasets (read counts) for colon cancer using CrossHub software [20]. Here is a brief description of TCGA RNA-Seq Version 2 pipeline. Illumina reads were aligned to hg19 UCSC reference genome using MapSplice [21]. The alignment results were translated to transcriptome coordinates prior to transcript level quantification using the UNC Bioinformatics Utilities (https://github.com/mozack/ubu). RSEM was used to estimate gene and transcript abundances [22]. The further analysis of read count data was performed using CrossHub [20]. Read counts were normalized with TMM (trimmed mean of M-values) method and then two expression test were performed: for two pools of samples and for paired samples only (paired samples comprise only about 10% of all TCGA samples). We excluded lowly expressed isoforms; only genes with at least 70 reads in each of 50% samples (either normal or tumor) have passed expression level threshold. Then comparison of trimmed (4% from each tail) mean of expression values between normal and tumor tissues using t-test was performed. Behjamini-Hochberg p-value adjustment was performed in order to calculate FDR. We selected only isoforms with concordant results in paired and pooled tests.
The selection of genes participating cell energy metabolism was performed using Gene Ontology database and the following keywords: glucose, glycolytic, glycolysis, cell respiration, respiratory, TCA cycle, oxidative phosphorylation, Krebs. Finally, a set of 277 genes with 1014 alternatively spliced transcripts was selected for the further analysis. Unfortunately, TCGA read count data were derived for the previous genome assembly (hg19) using UCSC genome annotation. When possible, UCSC transcript identifiers were converted to RefSeq accession numbers. Using CrossHub, we analyzed associations with disease stage, follow-up status, TNM indexes. We paid special attention to the alternatively spliced transcripts that are strongly overexpressed in colon tumors against the background of low changes in overall gene expression level or its down-regulation.
Tissue specimens
A total of 40 colorectal, 30 breast, 30 non-small cell lung, 30 prostate, and 30 kidney cancer specimens and matched morphologically normal tissues were obtained after surgical resection prior to radiation or chemotherapy. The samples were frozen and stored in liquid nitrogen. The morphological classification of the tumor was performed according to the American Joint Committee on Cancer (AJCC) staging system [23]. Only samples with 70% or more tumor cells were studied. Written informed consent was obtained from all patients. The study was approved by The Ethics committee of Herzen Moscow Cancer Research Institute, Ministry of Health of the Russian Federation. The study was done in accordance with the principles outlined in the Declaration of Helsinki (1964). The sample information for colorectal tumors is presented in Table 1.
Isolation of RNA and cDNA synthesis
Total RNA was isolated from frozen tissues using RNeasy Mini kit (Qiagen, Germany) according to manufacturer's instructions. RNA quality was measured using the RIN method (RNA Integrity Number) on Agilent RNA Bioanalyzer 2100 (Agilent Technologies, USA). The RNA quantification was carried out on a NanoDrop 1000 (NanoDrop Technologies Inc., USA). cDNA synthesis was done using M-MLV Reverse Transcriptase (Thermo Fisher Scientific, USA) and random hexamers.
Quantitative PCR
Gene and transcript expression levels were estimated by quantitative PCR (qPCR) analysis. All probes contained the dye FAM at 5′-end and RTQ1 at 3′-end. Specific primer pairs and probes were synthesized for the alternative splicing isoforms of target genes (Table 2). Primers and probes for reference genes were designed as previously described [24, 25]. PCR was carried out in triplicates on AB 7500 Real-Time PCR System (Thermo Fisher Scientific, USA) following the manufacturer's instructions. PCR program was as follows: 10 min at 95 °C and then 50 two-step cycles 15 s at 95 °C and 60 s at 60 °C. The total reaction volume was 20 μL in triplicate. PCR products were analyzed in 2% agarose gels, purified and submitted for Sanger sequencing on ABI Prism 3100 Genetic Analyzer (Thermo Fisher Scientific, USA).
Analysis of qPCR data
The Relative Quantitation software (Thermo Fisher Scientific, USA) and ATG (Analysis of Transcription of Genes) tool were used to analyze the obtained qPCR data taking into account the efficiency of the PCR amplification [26, 27]. The expression levels of target genes were normalized to GAPDH and ACTB reference genes and finally relative (T/N) expression level of target genes was calculated using ΔΔC t method. The relative inner variability between mRNA levels of reference genes do not exceed two times, and, therefore, 2-fold or more expression alterations of the target genes/isoforms were considered significant. Inter-group and intra-group comparisons were performed using nonparametric Wilcoxon/Mann-Whitney U–test and Kruskal-Wallis test. Differences with p < 0.05 were considered statistically significant. The statistical procedures were performed with BioStat software (AnalystSoft Inc., USA).
Results
Analysis of TCGA data with CrossHub
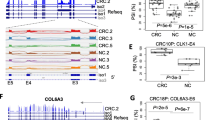
Differential expression profiles were derived for 277 genes (1014 alternatively spliced transcripts) which participate in cell energy metabolism. 285 tumor and 41 matched normal colon tissue samples were used in the analysis. Among differentially expressed transcripts, we paid a special attention to the following eight mRNA isoforms, which were overexpressed in colon tumors: OGDH (uc011kby/AK296400), COL6A3 (uc002vwo/NM_057167), ICAM1 (uc010xle/AK301412), PHPT1 (uc004cjq/NM_014172), PPP2R5D (uc010jyd/NM_180977), SLC29A1 (uc003owz/NM_001078177), TRIB3 (uc002wdm/NM_021158 and uc002wdn/AK297546). Most of them are minor mRNA isoforms that are selectively up-regulated against the background of intact or under-expressed other isoforms and intact expression of entire gene (sum across all transcripts). The list of LogFC, p-values, FDR, CPM, RNA-Seq expression profiles and associations with clinical characteristics is provided in the Additional file 1. Differential expression of these transcripts was further validated with qPCR. Most of the selected isoforms have unique splice event which do not occur in the other isoforms (e.g. exon boundaries). This allows accurate detection and quantification of these transcripts with spliced reads spanning this unique exon junction.
OGDH (oxoglutarate dehydrogenase) mRNA isoform uc011kby lacks three exons and this unique feature allows quantification of this transcripts with RNA-Seq data. Compared to the reference isoform (UniProt ID Q02218-1), the protein encoded with uc011kby transcript lacks three regions: 75–123 a.a., 139–172 a.a., and 211–263 a.a. Full-length isoform of OGDH is normally regulated by Ca2+, adenine nucleotides, and NADH. Mutations in the second region (D154A for Q02218-1) results in 6-fold decrease of calcium sensitivity [28]. Transcript uc003owz is very minor isoform of SLC29A1 mRNA (CPM = 0.3 and 2.1 for normal and tumor accordingly). The first exon of this transcript almost completely covers CpG island in the promotor region of SLC29A1. Frequent induction of this minor isoform suggests altered mechanisms of SLC29A1 expression regulation in colon tumors.
Two other transcripts of interest, uc010xle and uc010jyd are minor isoforms of ICAM1 (Intercellular Adhesion Molecule 1) and PPP2R5D (Protein Phosphatase 2 Regulatory Subunit B' Delta) genes, respectively. Compared to the major isoforms, they do not include some exons. As the result, protein encoded with ICAM1 uc010xle has deletion at 22–244 a.a. (UniProt P05362-1) which spans two Ig-like C2-type domains. The signal peptide sequence is almost completely kept (1–28 a.a.). This isoform is strongly overexpressed in tumor (CPM is 0.6 and 5.2 for normal and tumor) whereas major isoform is overexpressed only 1.5-times (CPM = 36 and 53). Protein encoded with PPP2R5D transcript uc010jyd has deletion at 10–107 a.a. (UniProt Q14738-1).
Up-regulation of eight alternative mRNA transcripts in CRC
Quantitative expression estimation was performed for eight alternative mRNA isoforms of seven genes involved in energy metabolism in CRC (Fig. 1). Bioinformatics results showed good concordance with qPCR results. All the transcripts showed up-regulation in more than 50% of CRC cases (Table 3). The highest frequency and extent of the mRNA level increase were observed for alternative transcripts of COL6A3 (uc002vwo/ NM_057167, 90% and 10.6-fold average increase), TRIB3 (uc002wdm/NM_021158, 90% and 10.5-fold average increase; uc002wdn/AK297546, 67.5% and 3.6-fold average increase), and SLC29A1 (uc003owz/NM_001078177, 77.5% and 4.6-fold average increase) genes.
Simultaneous up-regulation of alternative mRNA transcripts in colorectal, breast, lung, prostate, and kidney cancers
To evaluate the possibility of tumor-specific expression of alternative transcripts, we analyzed their expression in breast, lung, prostate, and kidney cancers. We revealed the significantly increased expression of alternative mRNA isoform uc002vwo/NM_057167 of COL6A3 in all these tumors (Fig. 2). The mRNA level of the transcript was up-regulated in 96.7% (29 of 30, p < 0.05) of breast, 63.3% (19 of 30, p < 0.05) of lung, 76.7% (23 of 30, p < 0.05) of prostate, and 50% (15 of 30, p < 0.05) of kidney cancer samples.
We also observed the increased expression of two alternative transcripts (TRIB3: uc002wdm/NM_021158 and ICAM1: uc010xle/AK301412) in all the cancers with one exception (Fig. 3). We found that the uc002wdm/NM_021158 (TRIB3) mRNA level was increased in 73.3% (22 of 30, p < 0.05), 50% (15 of 30, p < 0.05), and 93.3% (28 of 30, p < 0.05) cases of breast, prostate, and kidney cancers, respectively. The stable expression of uc002wdm/NM_021158 (TRIB3) was detected in most cases of lung cancer. The up-regulation of uc010xle/AK301412 (ICAM1) level was detected in breast (50%, 30 of 15, p < 0.05), prostate (50%, 30 of 15, p < 0.05), and kidney (70%, 21 of 30, p < 0.05) cancers. The alternative transcript uc010xle/AK301412 (ICAM1) was differentially expressed in lung cancer; the mRNA level of one was increased in 36.7% (11 of 30, p < 0.05) and decreased in 40% (12 of 30, p < 0.05). The expression of uc003owz/NM_001078177 SLC29A1 transcript was not significantly changed in more than 50% cases of all tumors studied, but was up-regulated in 77.5% (31 of 40, p < 0.05) colorectal cancer samples (Fig. 4).
Discussion
Alternative splicing plays a critical role in multiple cellular processes and development programs [29]. In recent years, alternative splicing has been recognized to contribute to many human disorders, including cancer [30]. Alternative splicing is one more mechanism that allows expressing transcripts involved in the regulation of apoptosis, signaling pathways and cellular metabolism [31]. Changes in splicing patterns occur widely in cancer cells and was shown to be associated with the resistance to therapeutic treatments [32]. Alterations in the trans-acting splicing regulatory elements are the most frequent in cancer [33, 34]. Mutations in various splicing regulatory factors such as U2AF1, ZRSR2, SRSF2, SF3B1, and RBM10 have been described in multiple tumor types [35–37]. Overexpression of a positive splicing factor, serine/arginine-rich (SR), was found in colorectal, thyroid, small intestine, kidney, and lung cancers [38]. On the other hand, an alternative splicing repressor, heterogeneous nuclear ribonucleoprotein (hnRNP), was primary down-regulated in these tumor types. Mutations in cis-acting splicing elements were shown in both oncogenes (KIT, CDH17, and BRCA1/2) and tumor suppressors (LKB1 and KLF6), which have causal role in cancer initiation and progression [34, 39, 40]. Cancer-associated alterations of splicing patterns have been also reported for other cancer-related genes [39]. For instance, it has been shown that splicing pattern of Ron and Rac1 genes were altered in tumors and overexpression of their tumor-associated isoforms was sufficient to culture cell transformation [41, 42]. The alternatively spliced isoforms of MDM2/HDM2 gene, that is a regulator of p53 protein, have been identified in many cancers. Moreover, some of their protein products were shown to have transforming properties [43, 44].
Alternative splicing is known contributor to cancer pathogenesis. For instance, activation of splicing factor hnRNP by EGFRvIII mutation promotes glycolytic gene expression in glioblastoma [45]. Alternative splicing of pyruvate kinase M (PKM) pre-mRNA generates the PKM2 isoform in all cancer cells [46]. PKM2 is a critical enzyme for aerobic glycolysis that mediates Warburg effect and facilitates tumor growth [47]. PKM2 is highly expressed in embryonic and tumor cells, whereas PKM1 is primarily expressed in normal tissues [47, 48]. Splicing repressors hnRNPA1 and hnRNPA2 have been found to regulate PKM alternative splicing in cancer cells [49]. Down-regulation of these factors in cancer cells resulted in an increase in the PKM1/PKM2 protein ratio and decrease in lactate production [31]. These data indicate that alternative splicing is involved in the switch from oxidative phosphorylation to aerobic glycolysis in cancer. Mitochondrial damage modulates alternative splicing in neuronal cells leading to changes in the abundance of certain isoforms [50]. Thus, mitochondrial dysfunction, as a notable feature of cancer, may be also the mechanism underlying the changes in alternative splicing patterns.
Our study revealed tumor-associated changes in alternative splicing patterns of seven genes involved in energy metabolism, including OGDH, COL6A3, ICAM1, PHPT1, PPP2R5D, SLC29A1, and TRIB3. OGDH gene encodes a subunit of the multi-enzyme 2-oxoglutarate dehydrogenase complex (OGDHC) that is the first and rate-limiting component of one [51]. OGDHC plays a major role in TCA cycle and involved in the regulation of the glutamine and glutamate metabolism [52]. OGDHC is often implied to be inactive in cancer [53]. Oncogenic mutations reduce the enzyme activity of NADP+-dependent isocitrate dehydrogenases isoforms 1 and 2 (IDH1/2) resulting in increased 2-hydroxyglutarate levels and decreased concentrations of the OGDHC substrate 2-oxoglutarate [54, 55]. In a previous study, we showed that OGDHL, encoding one more component of the OGDHC, is down-regulated by promoter hypermethylation in CRC [56]. The promoter hypermethylation in OGDHL gene was also observed in breast, cervix, lung, oesophagus, and pancreas cancers [57, 58]. It has been shown that re-expression of OGDHL induced apoptosis through a PI3K/AKT pathway in cervical cancer cells [51]. The alterations in OGDHC expression were shown to be functional in various cancer cells [53]. Thus, we assumed that up-regulation of OGDH alternative mRNA transcript may indicate the presence of active OGDH complex in colorectal cancer that is required to control energy and glutamine metabolism.
Overexpression of SLC29A1 alternative transcript was found in colorectal cancer and was not significantly changed in breast, lung, prostate, and kidney cancers. Transporter SLC29A1 has been reported relating to multidrug resistance (MDR). Significant up-regulation of SLC29A1 in colorectal, astroglial, and breast cancer cells contributed to cisplatin resistance and increased cell viability [59]. On the other hand, knockdown of SLC29A1 reduced sensitivity of leukemia and lung cancer to drugs since it plays a role in cellular uptake [60, 61]. Increased SLC29A1 mRNA level was suggested as a critical factor of pancreatic and biliary tract cancer cells sensitivity to chemotherapy [62, 63]. The expression of SLC29A1 alternative transcripts in cancer has not been previously analyzed. We first observed the tumor-specific up-regulation of the uc003owz/NM_001078177 SLC29A1 transcript in CRC. The functional role of a protein encoded by the transcript in CRC is not obvious. The further investigation will address how the transcript is associated with the resistance and sensitivity of colorectal cancer cells to therapy by various agents and may be useful for prediction of its efficacy.
Using qPCR method, we showed the tumor-specific overexpression of uc002vwo/NM_057167 COL6A3 alternative transcript in colorectal, breast, lung, prostate, and kidney cancers. Collagen VI, a protein of the extracellular matrix (ECM), is significant in the progression of cancer and resistance to chemotherapy [64, 65]. COL6A3 encodes one of the three α chains of type VI collagen which is involved in the regulation of metabolic health by ECM [66, 67]. Recent studies have demonstrated that COL6A3 was up-regulated in gastric, pancreatic, and ovarian cancers [68–71]. Exon array analysis revealed the expression of COL6A3 alternative long isoform in colon, bladder, pancreatic, and prostate cancers [72, 73].
Intercellular adhesion molecule 1 ICAM1 (CD54) is known to play a major role in immune response, inflammation, regulation of energy balance, and angiogenesis [74–77]. Increased levels of ICAM1 were reported in several human malignances and cancer cell lines [78]. In melanoma and gastric cancer, ICAM1 expression was associated with an increase in metastases [79, 80]. This can be explained by ICAM1-mediated activation of leukocytes and induction of cell migration [81]. On the other hand, immunohistochemistry studies reported better prognosis for patients with ICAM1-positive tumors (including lymphoma, ovarian, colorectal, head and neck cancers) [82–84]. Cancer cell can expresses and release soluble ICAM1, that is regulated by TNF-α and INF-γ [85]. It is an essential mechanism used by tumors to escape immune recognition [86, 87]. For example, elevated serum levels of ICAM1 in colorectal cancer patients were correlated with tumor stage and tendency to metastasis formation [84, 88, 89]. Thus, ICAM1 seems to have different roles in tumorigenesis. Tumor-specific expression of ICAM1 alternative splice variants was not previously reported. We found up-regulation of uc010xle/AK301412 ICAM1 alternative splice isoform in colorectal, breast, prostate, and kidney cancers that may be important prognostic factor.
PHPT1 and PPP2R5D genes encode proteins belonging to phosphatase activity and glucose metabolism [90–92]. PHPT1 has been found to be overexpressed in lung cancer and playing a role in cancer progression, migration and invasion [93, 94]. PPP2R5D gene, encoding a regulatory B subunit of protein phosphatase 2A (PP2A), was reported to be involved in Myc activation and degradation [95]. We first showed up-regulation of PHPT1 and PPP2R5D alternative transcripts in colorectal cancer.
The protein encoded by TRIB3 gene is tribbles pseudokinase-3 that has been proposed as inhibitor of AKT and interaction partner of transcription factors (including ATF-4, CHOP9, and several MAPKs) that regulate cell growth, differentiation and metabolism [96–100]. Schwarzer and co-authors showed that TRIB3 emerges as a transcriptional target of PI3K/Akt signaling pathway and is involved in regulation of glucose metabolism [101]. Recent studies reported that increase in TRIB3 expression promoted cancer cell death through apoptosis [102–105]. Genetic inhibition of TRIB3 resulted in activation of mTORC2/AKT/FOXO pathway and was associated with more aggressive phenotype in several animal models of cancer [106]. However, TRIB3 was up-regulated in CRC samples, gastrointestinal and colorectal cancer cell lines [107]. These data are consistent with the overexpression of both TRIB3 alternative splice variants in CRC observed in this study. Thus, tumor-associated changes in alternative splicing lead to overexpression of certain TRIB3 splice isoforms which can be involved in development of colorectal cancer.
Conclusion
In the present study, using our previously developed bioinformatics tools and TCGA data, we evaluated alternative splicing profiles of genes associated with energy metabolism in CRC samples and then validated the results by qPCR. Differential expression of the transcripts of seven genes (OGDH, COL6A3, ICAM1, PHPT1, PPP2R5D, SLC29A1, and TRIB3) was confirmed. Alternative transcript uc003owz/ NM_001078177 of SLC29A1 was characterized with tumor-specific overexpression in CRC that can be associated with drug resistance and sensitivity. Changes in alternative splicing patterns of OGDH gene may play an important role in the regulation of energy and glutamine metabolism in CRC. Overexpression of COL6A3 alternative transcript in all examined tumor types indicates its significant contribution to disease development and pathogenesis. Increase in expression of PHPT1, PPP2R5D, and two TRIB3 transcripts indicates that tumor-associated changes in alternative splicing can affect glucose metabolism in colorectal cancer. Several alternative transcripts may be suggested as potential cancer biomarkers, although further studies must be performed to confirm these results.
References
Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg. 2009;22(4):191–7.
Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64(2):104–17.
Murphy G, Devesa SS, Cross AJ, Inskip PD, McGlynn KA, Cook MB. Sex disparities in colorectal cancer incidence by anatomic subsite, race and age. Int J Cancer. 2011;128(7):1668–75.
Fleming M, Ravula S, Tatishchev SF, Wang HL. Colorectal carcinoma: pathologic aspects. J Gastrointest Oncol. 2012;3(3):153–73.
Wang X, Kuang YY, Hu XT. Advances in epigenetic biomarker research in colorectal cancer. World J Gastroenterol. 2014;20(15):4276–87.
Cook AD, Single R, McCahill LE. Surgical resection of primary tumors in patients who present with stage IV colorectal cancer: an analysis of surveillance, epidemiology, and end results data, 1988 to 2000. Ann Surg Oncol. 2005;12(8):637–45.
Qiu M, Hu J, Yang D, Cosgrove DP, Xu R. Pattern of distant metastases in colorectal cancer: a SEER based study. Oncotarget. 2015;6(36):38658–66.
Patel NN, Shah PR, Wilson E, Haray PN. An unexpected supraclavicular swelling. World J Surg Oncol. 2007;5:90.
Davies JM, Goldberg RM. Treatment of metastatic colorectal cancer. Semin Oncol. 2011;38(4):552–60.
Ganapathy-Kanniappan S, Geschwind JF. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer. 2013;12:152.
Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–33.
Kudryavtseva AV, Krasnov GS, Dmitriev AA, Alekseev BY, Kardymon OL, Sadritdinova AF, Fedorova MS, Pokrovsky AV, Melnikova NV, Kaprin AD, et al. Mitochondrial dysfunction and oxidative stress in aging and cancer. Oncotarget. 2016;7(29):44879–905.
Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–14.
Qiu Y, Cai G, Zhou B, Li D, Zhao A, Xie G, Li H, Cai S, Xie D, Huang C, et al. A distinct metabolic signature of human colorectal cancer with prognostic potential. Clin Cancer Res. 2014;20(8):2136–46.
Wellen KE, Thompson CB. Cellular metabolic stress: considering how cells respond to nutrient excess. Mol Cell. 2010;40(2):323–32.
Hirayama A, Kami K, Sugimoto M, Sugawara M, Toki N, Onozuka H, Kinoshita T, Saito N, Ochiai A, Tomita M, et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009;69(11):4918–25.
Beyoglu D, Imbeaud S, Maurhofer O, Bioulac-Sage P, Zucman-Rossi J, Dufour JF, Idle JR. Tissue metabolomics of hepatocellular carcinoma: tumor energy metabolism and the role of transcriptomic classification. Hepatology. 2013;58(1):229–38.
Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457(7231):910–4.
Kudryavtseva AV, Lipatova AV, Zaretsky AR, Moskalev AA, Fedorova MS, Rasskazova AS, Shibukhova GA, Snezhkina AV, Kaprin AD, Alekseev BY, et al. Important molecular genetic markers of colorectal cancer. Oncotarget. 2016;7(33):53959–83.
Krasnov GS, Dmitriev AA, Melnikova NV, Zaretsky AR, Nasedkina TV, Zasedatelev AS, Senchenko VN, Kudryavtseva AV. CrossHub: a tool for multi-way analysis of The Cancer Genome Atlas (TCGA) in the context of gene expression regulation mechanisms. Nucleic Acids Res. 2016;44(7), e62.
Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38(18), e178.
Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics. 2010;26(4):493–500.
Hari DM, Leung AM, Lee JH, Sim MS, Vuong B, Chiu CG, Bilchik AJ. AJCC Cancer Staging Manual 7th edition criteria for colon cancer: do the complex modifications improve prognostic assessment? J Am Coll Surg. 2013;217(2):181–90.
Krasnov GS, Dmitriev AA, Sadtritdinova AF, Fedorova MS, Snezhkina AV, Melnikova NV, Poteryakhina AV, Nyushko KM, Belyakov MM, Kaprin AD, et al. Evaluation of gene expression of hexokinases in colorectal cancer with the use of bioinformatics methods. Biofizika. 2015;60(6):1050–6.
Krasnov GS, Oparina NY, Dmitriev AA, Kudryavtseva AV, Anedchenko EA, Kondrat'eva TT, Zabarovsky ER, Senchenko VN. RPN1, a new reference gene for quantitative data normalization in lung and kidney cancer. Mol Biol. 2011;45(2):211–20.
Senchenko VN, Anedchenko EA, Kondratieva TT, Krasnov GS, Dmitriev AA, Zabarovska VI, Pavlova TV, Kashuba VI, Lerman MI, Zabarovsky ER. Simultaneous down-regulation of tumor suppressor genes RBSP3/CTDSPL, NPRL2/G21 and RASSF1A in primary non-small cell lung cancer. BMC Cancer. 2010;10:75.
Senchenko VN, Krasnov GS, Dmitriev AA, Kudryavtseva AV, Anedchenko EA, Braga EA, Pronina IV, Kondratieva TT, Ivanov SV, Zabarovsky ER, et al. Differential expression of CHL1 gene during development of major human cancers. PLoS One. 2011;6(3), e15612.
Armstrong CT, Anderson JL, Denton RM. Studies on the regulation of the human E1 subunit of the 2-oxoglutarate dehydrogenase complex, including the identification of a novel calcium-binding site. Biochem J. 2014;459(2):369–81.
Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10(11):741–54.
Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med. 2012;18(8):472–82.
Clower CV, Chatterjee D, Wang Z, Cantley LC, Vander Heiden MG, Krainer AR. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci U S A. 2010;107(5):1894–9.
David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24(21):2343–64.
Bonomi S, Gallo S, Catillo M, Pignataro D, Biamonti G, Ghigna C. Oncogenic alternative splicing switches: role in cancer progression and prospects for therapy. Int J Biochem Cell Biol. 2013;2013:962038.
Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8(10):749–61.
Brooks AN, Choi PS, de Waal L, Sharifnia T, Imielinski M, Saksena G, Pedamallu CS, Sivachenko A, Rosenberg M, Chmielecki J, et al. A pan-cancer analysis of transcriptome changes associated with somatic mutations in U2AF1 reveals commonly altered splicing events. PLoS One. 2014;9(1), e87361.
Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64–9.
Maguire SL, Leonidou A, Wai P, Marchio C, Ng CK, Sapino A, Salomon AV, Reis-Filho JS, Weigelt B, Natrajan RC. SF3B1 mutations constitute a novel therapeutic target in breast cancer. J Pathol. 2015;235(4):571–80.
Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14(3):185–93.
Srebrow A, Kornblihtt AR. The connection between splicing and cancer. J Cell Sci. 2006;119(Pt 13):2635–41.
Mucaki EJ, Ainsworth P, Rogan PK. Comprehensive prediction of mRNA splicing effects of BRCA1 and BRCA2 variants. Hum Mutat. 2011;32(7):735–42.
Singh A, Karnoub AE, Palmby TR, Lengyel E, Sondek J, Der CJ. Rac1b, a tumor associated, constitutively active Rac1 splice variant, promotes cellular transformation. Oncogene. 2004;23(58):9369–80.
Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, Green MR, Riva S, Biamonti G. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 2005;20(6):881–90.
Bartel F, Taubert H, Harris LC. Alternative and aberrant splicing of MDM2 mRNA in human cancer. Cancer Cell. 2002;2(1):9–15.
Lukas J, Gao DQ, Keshmeshian M, Wen WH, Tsao-Wei D, Rosenberg S, Press MF. Alternative and aberrant messenger RNA splicing of the mdm2 oncogene in invasive breast cancer. Cancer Res. 2001;61(7):3212–9.
Babic I, Anderson ES, Tanaka K, Guo D, Masui K, Li B, Zhu S, Gu Y, Villa GR, Akhavan D, et al. EGFR mutation-induced alternative splicing of Max contributes to growth of glycolytic tumors in brain cancer. Cell Metab. 2013;17(6):1000–8.
Chen M, Zhang J, Manley JL. Turning on a fuel switch of cancer: hnRNP proteins regulate alternative splicing of pyruvate kinase mRNA. Cancer Res. 2010;70(22):8977–80.
Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452(7184):230–3.
Eigenbrodt E, Basenau D, Holthusen S, Mazurek S, Fischer G. Quantification of tumor type M2 pyruvate kinase (Tu M2-PK) in human carcinomas. Anticancer Res. 1997;17(4B):3153–6.
David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463(7279):364–8.
Maracchioni A, Totaro A, Angelini DF, Di Penta A, Bernardi G, Carri MT, Achsel T. Mitochondrial damage modulates alternative splicing in neuronal cells: implications for neurodegeneration. J Neurochem. 2007;100(1):142–53.
Sen T, Sen N, Noordhuis MG, Ravi R, Wu TC, Ha PK, Sidransky D, Hoque MO. OGDHL is a modifier of AKT-dependent signaling and NF-kappaB function. PLoS One. 2012;7(11), e48770.
Bunik VI, Fernie AR. Metabolic control exerted by the 2-oxoglutarate dehydrogenase reaction: a cross-kingdom comparison of the crossroad between energy production and nitrogen assimilation. Biochem J. 2009;422(3):405–21.
Bunik V, Mkrtchyan G, Grabarska A, Oppermann H, Daloso D, Araujo WL, Juszczak M, Rzeski W, Bettendorff L, Fernie AR, et al. Inhibition of mitochondrial 2-oxoglutarate dehydrogenase impairs viability of cancer cells in a cell-specific metabolism-dependent manner. Oncotarget. 2016;7(18):26400–21.
Reitman ZJ, Parsons DW, Yan H. IDH1 and IDH2: not your typical oncogenes. Cancer Cell. 2010;17(3):215–6.
Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2010;465(7300):966.
Fedorova MS, Kudryavtseva AV, Lakunina VA, Snezhkina AV, Volchenko NN, Slavnova EN, Danilova TV, Sadritdinova AF, Melnikova NV, Belova AA, et al. Downregulation of OGDHL expression is associated with promoter hypermethylation in colorectal cancer. Mol Biol. 2015;49(4):608–17.
Ostrow KL, Park HL, Hoque MO, Kim MS, Liu J, Argani P, Westra W, Van Criekinge W, Sidransky D. Pharmacologic unmasking of epigenetically silenced genes in breast cancer. Clin Cancer Res. 2009;15(4):1184–91.
Hoque MO, Kim MS, Ostrow KL, Liu J, Wisman GB, Park HL, Poeta ML, Jeronimo C, Henrique R, Lendvai A, et al. Genome-wide promoter analysis uncovers portions of the cancer methylome. Cancer Res. 2008;68(8):2661–70.
Gotovdorj T, Lee E, Lim Y, Cha EJ, Kwon D, Hong E, Kim Y, Oh MY. 2,3,7,8-Tetrachlorodibenzo-p-dioxin induced cell-specific drug transporters with acquired cisplatin resistance in cisplatin sensitive cancer cells. J Korean Med Sci. 2014;29(9):1188–98.
Huang Y, Anderle P, Bussey KJ, Barbacioru C, Shankavaram U, Dai Z, Reinhold WC, Papp A, Weinstein JN, Sadee W. Membrane transporters and channels: role of the transportome in cancer chemosensitivity and chemoresistance. Cancer Res. 2004;64(12):4294–301.
Li L, Schaid DJ, Fridley BL, Kalari KR, Jenkins GD, Abo RP, Batzler A, Moon I, Pelleymounter L, Eckloff BW, et al. Gemcitabine metabolic pathway genetic polymorphisms and response in patients with non-small cell lung cancer. Pharmacogenet Genomics. 2012;22(2):105–16.
Mori R, Ishikawa T, Ichikawa Y, Taniguchi K, Matsuyama R, Ueda M, Fujii Y, Endo I, Togo S, Danenberg PV, et al. Human equilibrative nucleoside transporter 1 is associated with the chemosensitivity of gemcitabine in human pancreatic adenocarcinoma and biliary tract carcinoma cells. Oncol Rep. 2007;17(5):1201–5.
Nakano Y, Tanno S, Koizumi K, Nishikawa T, Nakamura K, Minoguchi M, Izawa T, Mizukami Y, Okumura T, Kohgo Y. Gemcitabine chemoresistance and molecular markers associated with gemcitabine transport and metabolism in human pancreatic cancer cells. Br J Cancer. 2007;96(3):457–63.
Sherman-Baust CA, Weeraratna AT, Rangel LB, Pizer ES, Cho KR, Schwartz DR, Shock T, Morin PJ. Remodeling of the extracellular matrix through overexpression of collagen VI contributes to cisplatin resistance in ovarian cancer cells. Cancer Cell. 2003;3(4):377–86.
Iyengar P, Espina V, Williams TW, Lin Y, Berry D, Jelicks LA, Lee H, Temple K, Graves R, Pollard J, et al. Adipocyte-derived collagen VI affects early mammary tumor progression in vivo, demonstrating a critical interaction in the tumor/stroma microenvironment. J Clin Invest. 2005;115(5):1163–76.
Zanussi S, Doliana R, Segat D, Bonaldo P, Colombatti A. The human type VI collagen gene. mRNA and protein variants of the alpha 3 chain generated by alternative splicing of an additional 5-end exon. J Biol Chem. 1992;267(33):24082–9.
McCulloch LJ, Rawling TJ, Sjoholm K, Franck N, Dankel SN, Price EJ, Knight B, Liversedge NH, Mellgren G, Nystrom F, et al. COL6A3 is regulated by leptin in human adipose tissue and reduced in obesity. Endocrinology. 2015;156(1):134–46.
Sun H. Identification of key genes associated with gastric cancer based on DNA microarray data. Oncol Lett. 2016;11(1):525–30.
Xie X, Liu X, Zhang Q, Yu J. Overexpression of collagen VI alpha3 in gastric cancer. Oncol Lett. 2014;7(5):1537–43.
Ismail RS, Baldwin RL, Fang J, Browning D, Karlan BY, Gasson JC, Chang DD. Differential gene expression between normal and tumor-derived ovarian epithelial cells. Cancer Res. 2000;60(23):6744–9.
Arafat H, Lazar M, Salem K, Chipitsyna G, Gong Q, Pan TC, Zhang RZ, Yeo CJ, Chu ML. Tumor-specific expression and alternative splicing of the COL6A3 gene in pancreatic cancer. Surgery. 2011;150(2):306–15.
Thorsen K, Sorensen KD, Brems-Eskildsen AS, Modin C, Gaustadnes M, Hein AM, Kruhoffer M, Laurberg S, Borre M, Wang K, et al. Alternative splicing in colon, bladder, and prostate cancer identified by exon array analysis. Mol Cell Proteomics. 2008;7(7):1214–24.
Gardina PJ, Clark TA, Shimada B, Staples MK, Yang Q, Veitch J, Schweitzer A, Awad T, Sugnet C, Dee S, et al. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;7:325.
Mackay CR, Imhof BA. Cell adhesion in the immune system. Immunol Today. 1993;14(3):99–102.
Yasuda M, Shimizu S, Ohhinata K, Naito S, Tokuyama S, Mori Y, Kiuchi Y, Yamamoto T. Differential roles of ICAM-1 and E-selectin in polymorphonuclear leukocyte-induced angiogenesis. Am J Physiol Cell Physiol. 2002;282(4):C917–25.
Babic AM, Wang HW, Lai MJ, Daniels TG, Felbinger TW, Burger PC, Stricker-Krongrad A, Wagner DD. ICAM-1 and beta2 integrin deficiency impairs fat oxidation and insulin metabolism during fasting. Mol Med. 2004;10(7-12):72–9.
Gho YS, Kleinman HK, Sosne G. Angiogenic activity of human soluble intercellular adhesion molecule-1. Cancer Res. 1999;59(20):5128–32.
Rokhlin OW, Cohen MB. Expression of cellular adhesion molecules on human prostate tumor cell lines. Prostate. 1995;26(4):205–12.
Natali P, Nicotra MR, Cavaliere R, Bigotti A, Romano G, Temponi M, Ferrone S. Differential expression of intercellular adhesion molecule 1 in primary and metastatic melanoma lesions. Cancer Res. 1990;50(4):1271–8.
Koyama S, Ebihara T, Fukao K. Expression of intercellular adhesion molecule 1 (ICAM-1) during the development of invasion and/or metastasis of gastric carcinoma. J Cancer Res Clin Oncol. 1992;118(8):609–14.
de Groote ML, Kazemier HG, Huisman C, van der Gun BT, Faas MM, Rots MG. Upregulation of endogenous ICAM-1 reduces ovarian cancer cell growth in the absence of immune cells. Int J Cancer. 2014;134(2):280–90.
Terol MJ, Lopez-Guillermo A, Bosch F, Villamor N, Cid MC, Rozman C, Campo E, Montserrat E. Expression of the adhesion molecule ICAM-1 in non-Hodgkin's lymphoma: relationship with tumor dissemination and prognostic importance. J Clin Oncol. 1998;16(1):35–40.
Arnold JM, Cummings M, Purdie D, Chenevix-Trench G. Reduced expression of intercellular adhesion molecule-1 in ovarian adenocarcinomas. Br J Cancer. 2001;85(9):1351–8.
Maeda K, Kang SM, Sawada T, Nishiguchi Y, Yashiro M, Ogawa Y, Ohira M, Ishikawa T, Hirakawa YSCK. Expression of intercellular adhesion molecule-1 and prognosis in colorectal cancer. Oncol Rep. 2002;9(3):511–4.
Witkowska AM, Borawska MH. Soluble intercellular adhesion molecule-1 (sICAM-1): an overview. Eur Cytokine Netw. 2004;15(2):91–8.
Gearing AJ, Newman W. Circulating adhesion molecules in disease. Immunol Today. 1993;14(10):506–12.
Becker JC, Dummer R, Hartmann AA, Burg G, Schmidt RE. Shedding of ICAM-1 from human melanoma cell lines induced by IFN-gamma and tumor necrosis factor-alpha. Functional consequences on cell-mediated cytotoxicity. J Immunol. 1991;147(12):4398–401.
Mantur M, Snarska J, Koper O, Dzieciol J, Plonski A, Lemancewicz D. Serum sICAM, sVCAM and sE-selectin levels in colorectal cancer patients. Folia histochemica et cytobiologica / Polish Academy of Sciences, Polish Histochemical and Cytochemical Society. 2009;47(4):621–5.
Dymicka-Piekarska V, Guzinska-Ustymowicz K, Kuklinski A, Kemona H. Prognostic significance of adhesion molecules (sICAM-1, sVCAM-1) and VEGF in colorectal cancer patients. Thromb Res. 2012;129(4):e47–50.
Ek P, Pettersson G, Ek B, Gong F, Li JP, Zetterqvist O. Identification and characterization of a mammalian 14-kDa phosphohistidine phosphatase. Eur J Biochem / FEBS. 2002;269(20):5016–23.
McCright B, Brothman AR, Virshup DM. Assignment of human protein phosphatase 2A regulatory subunit genes b56alpha, b56beta, b56gamma, b56delta, and b56epsilon (PPP2R5A-PPP2R5E), highly expressed in muscle and brain, to chromosome regions 1q41, 11q12, 3p21, 6p21.1, and 7p11.2 -->> p12. Genomics. 1996;36(1):168–70.
Kamath V, Kyathanahalli CN, Jayaram B, Syed I, Olson LK, Ludwig K, Klumpp S, Krieglstein J, Kowluru A. Regulation of glucose- and mitochondrial fuel-induced insulin secretion by a cytosolic protein histidine phosphatase in pancreatic beta-cells. Am J Physiol Endocrinol Metab. 2010;299(2):E276–86.
Tian T, Hao J, Xu A, Hao J, Luo C, Liu C, Huang L, Xiao X, He D. Determination of metastasis-associated proteins in non-small cell lung cancer by comparative proteomic analysis. Cancer Sci. 2007;98(8):1265–74.
Xu A, Hao J, Zhang Z, Tian T, Jiang S, Hao J, Liu C, Huang L, Xiao X, He D. 14-kDa phosphohistidine phosphatase and its role in human lung cancer cell migration and invasion. Lung Cancer. 2010;67(1):48–56.
Liu L, Eisenman RN. Regulation of c-Myc protein abundance by a protein phosphatase 2A-glycogen synthase kinase 3beta-negative feedback pathway. Genes Cancer. 2012;3(1):23–36.
Zhang J, Wen HJ, Guo ZM, Zeng MS, Li MZ, Jiang YE, He XG, Sun CZ. TRB3 overexpression due to endoplasmic reticulum stress inhibits AKT kinase activation of tongue squamous cell carcinoma. Oral Oncol. 2011;47(10):934–9.
Du K, Herzig S, Kulkarni RN, Montminy M. TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science. 2003;300(5625):1574–7.
Ohoka N, Yoshii S, Hattori T, Onozaki K, Hayashi H. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J. 2005;24(6):1243–55.
Kiss-Toth E, Bagstaff SM, Sung HY, Jozsa V, Dempsey C, Caunt JC, Oxley KM, Wyllie DH, Polgar T, Harte M, et al. Human tribbles, a protein family controlling mitogen-activated protein kinase cascades. J Biol Chem. 2004;279(41):42703–8.
Boudeau J, Miranda-Saavedra D, Barton GJ, Alessi DR. Emerging roles of pseudokinases. Trends Cell Biol. 2006;16(9):443–52.
Schwarzer R, Dames S, Tondera D, Klippel A, Kaufmann J. TRB3 is a PI 3-kinase dependent indicator for nutrient starvation. Cell Signal. 2006;18(6):899–909.
Carracedo A, Gironella M, Lorente M, Garcia S, Guzman M, Velasco G, Iovanna JL. Cannabinoids induce apoptosis of pancreatic tumor cells via endoplasmic reticulum stress-related genes. Cancer Res. 2006;66(13):6748–55.
Carracedo A, Lorente M, Egia A, Blazquez C, Garcia S, Giroux V, Malicet C, Villuendas R, Gironella M, Gonzalez-Feria L, et al. The stress-regulated protein p8 mediates cannabinoid-induced apoptosis of tumor cells. Cancer Cell. 2006;9(4):301–12.
Salazar M, Carracedo A, Salanueva IJ, Hernandez-Tiedra S, Lorente M, Egia A, Vazquez P, Blazquez C, Torres S, Garcia S, et al. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J Clin Invest. 2009;119(5):1359–72.
Nicoletti-Carvalho JE, Nogueira TC, Gorjao R, Bromati CR, Yamanaka TS, Boschero AC, Velloso LA, Curi R, Anhe GF, Bordin S. UPR-mediated TRIB3 expression correlates with reduced AKT phosphorylation and inability of interleukin 6 to overcome palmitate-induced apoptosis in RINm5F cells. J Endocrinol. 2010;206(2):183–93.
Salazar M, Lorente M, Garcia-Taboada E, Perez Gomez E, Davila D, Zuniga-Garcia P, Maria Flores J, Rodriguez A, Hegedus Z, Mosen-Ansorena D, et al. Loss of Tribbles pseudokinase-3 promotes Akt-driven tumorigenesis via FOXO inactivation. Cell Death Differ. 2015;22(1):131–44.
Miyoshi N, Ishii H, Mimori K, Takatsuno Y, Kim H, Hirose H, Sekimoto M, Doki Y, Mori M. Abnormal expression of TRIB3 in colorectal cancer: a novel marker for prognosis. Br J Cancer. 2009;101(10):1664–70.
Acknowledgments
Authors thank National Medical Research Center of Radiology, State Hospital №57, and A.V. Vishnevsky Institute of Surgery for supplying tumor specimens and providing patient data; Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Insilico Medicine, Inc., and Moscow Institute of Physics and Technology for the opportunity to use computational resources.
Part of this work (isolation of RNA, cDNA synthesis and quantitative PCR) was performed using the equipment of EIMB RAS “Genome” center (http://www.eimb.ru/rus/ckp/ccu_genome_c.php).
Declarations
This article has been published as part of BMC Genetics Vol 17 Suppl 14, 2016: Selected articles from BGRS\SB-2016: genomics. The full contents of the supplement are available online at http://bmcgenomics.biomedcentral.com/articles/supplements/volume-17-supplement-14.
Funding
This work and publication costs were financially supported by the Russian Science Foundation (RSCF), grant # 14-15-01083.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Authors’ contributions
SAV, KGS, DAA, KAV wrote the manuscript; ZAR, MAA, KIY, AAI, KoDV, KAV conceived and designed the work; SAV, NKM, AAI, FMS, VNN, SDV, PAY, KaDV, KAD, ABY performed the experiments; SAV, FMS, SAF, MNV analyzed the experiments; KGS, LAV, DAA, ZA performed bioinformatics analysis. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The study was approved by The Ethics committee of Herzen Moscow Cancer Research Institute, Ministry of Health of the Russian Federation. The study was done in accordance with the principles outlined in the Declaration of Helsinki (1964).
Author information
Authors and Affiliations
Corresponding author
Additional file
Additional file 1: Table S1.
Differential expression profiles of genes and transcripts in colon adenocarcinomas compared to matched normal tissues (TCGA data analyzed by CrossHub). (XLSX 43 MB)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Snezhkina, A.V., Krasnov, G.S., Zaretsky, A.R. et al. Differential expression of alternatively spliced transcripts related to energy metabolism in colorectal cancer. BMC Genomics 17 (Suppl 14), 1011 (2016). https://doi.org/10.1186/s12864-016-3351-5
Published:
DOI: https://doi.org/10.1186/s12864-016-3351-5