Abstract
Background
Fruit ripening is a complex developmental process that depends on a coordinated regulation of numerous genes, including ripening-related transcription factors (TFs), fruit-related microRNAs, DNA methylation and chromatin remodeling. It is known that various TFs, such as MADS-domain, MYB, AP2/ERF and SBP/SPL family proteins play key roles in modulating ripening. However, little attention has been given to members of the large NF-Y TF family in this regard, although genes in this family are known to have important functions in regulating plant growth, development, and abiotic or biotic stress responses.
Results
In this study, the evolutionary relationship between Arabidopsis thaliana and tomato (Solanum lycopersicum) NF-Y genes was examined to predict similarities in function. Furthermore, through gene expression analysis, 13 tomato NF-Y genes were identified as candidate regulators of fruit ripening. Functional studies involving suppression of NF-Y gene expression using virus induced gene silencing (VIGS) indicated that five NF-Y genes, including two members of the NF-YB subgroup (Solyc06g069310, Solyc07g065500) and three members of the NF-YA subgroup (Solyc01g087240, Solyc08g062210, Solyc11g065700), influence ripening. In addition, subcellular localization analyses using NF-Y proteins fused to a green fluorescent protein (GFP) reporter showed that the three NF-YA proteins accumulated in the nucleus, while the two NF-YB proteins were observed in both the nucleus and cytoplasm.
Conclusions
In this study, we identified tomato NF-Y genes by analyzing the tomato genome sequence using bioinformatics approaches, and characterized their chromosomal distribution, gene structures, phylogenetic relationship and expression patterns. We also examined their biological functions in regulating tomato fruit via VIGS and subcellular localization analyses. The results indicated that five NF-Y transcription factors play roles in tomato fruit ripening. This information provides a platform for further investigation of their biological functions.
Similar content being viewed by others
Background
Fleshy fruit ripening is a complex developmental process, that typically involves changes in texture, color and flavor [66]. The regulatory mechanisms underlying these changes, and that coordinate the up- or down-regulation of large numbers of genes, include many ripening-related transcription factors (TFs), fruit-related microRNAs that target some of these TFs, DNA methylation and chromatin remodeling [55]. Members of several families of TFs, such as the MADS-box, MYB, AP2/ERF and SBP/SPL families, participate in the transcriptional regulatory network that modulates ripening [27]. For example, studies of several ripening-related mutants of the fleshy fruit model species tomato (Solanum lycopersicum) [15, 20], including ripening inhibitor (rin) [75], non-ripening (nor) [30] and colorless non-ripening (cnr) [21, 45], resulted in the identification of several ripening-associated TFs. The genes corresponding to these loci were found to encode fruit ripening related TFs, which have subsequently been identified as components of the transcriptional activation cascade that coordinates ripening [27]. Other TF families that participate in fruit ripening, included MYB, AP2/ETHYLENE RESPONSE FACTOR (ERF), HD-zip, basic helix-loop-helix (bHLH) and auxin response factors (ARFs) [27]. For example, SlMYB12 from tomato and MdMYB10 from apple have been shown to influence flavonoid and anthocyanin accumulation during fruit ripening [1, 3, 14], and a member of the AP2/ERF family, AP2a, which acts downstream of RIN, NOR, and CNR, is a negative regulator of tomato ripening [9, 28]. In addition, silencing of LeHB1, which encodes a tomato putative HD-zip protein, led to delayed ripening and reduced expression LeACO1, a gene that encodes an ACC oxidase, which is a key enzyme in the biosynthesis of the ripening associated hormone ethylene [40]. Moreover, members of both bHLH and ARF families have been shown to play diverse roles in fruit development and ripening [32, 72].
While various families of TFs are clearly associated with ripening regulation, there has been no such connection established for the large Nuclear Factor Y (NF-Y) TF family, despite its known functions in regulating plant growth, development, and abiotic or biotic stress responses [4, 36, 57]. NF-Y TFs, which are also known as heme activator proteins (HAPs) or CCAAT binding factors (CBFs), can be categorized as NF-YA (also known as HAP2 or CBF-B), NF-YB (HAP3 or CBF-A) and NF-YC (HAP5 or CBF-C) proteins [39], and they have been shown to bind to CCAAT boxes, which are thought to be present in approximately 30% of eukaryotic promoters [6, 47]. The NF-YB proteins lack a nuclear localization signal (NLS), and depend on an interaction with NF-YC proteins to ensure translocation to the nucleus. Upon arrival in the nucleus, a heterotrimer is formed, comprising the NF-YB and NF-YC heterodimer and NF-YA, which can bind to CCAAT boxes in the promoters or other regions of target genes, [17, 26].
In order to investigate the potential functions of NF-Y TFs in fruit ripening, we evaluated a total of 59 tomato NF-Y genes using a combination of bioinformatic analyses, and high-throughput functional screening, using virus-induced gene silencing (VIGS) [12]; an approach that has been widely used to study tomato fruit development and ripening [18, 53, 60]. We also describe a phylogenetic analysis of NF-Y genes from Arabidopsis thaliana and tomato, and an investigation of their chromosomal distribution, protein motif and exon/intron structure patterns. As a result of these analyses, we propose five candidate NF-Y genes that are likely involved in fruit ripening regulation.
Results
Identification, organization and structure of tomato NF-Y genes
A search of the Plant Transcription Factor Database (PlantTFDB, http://planttfdb.cbi.pku.edu.cn/) revealed a total of fifty-nine predicted tomato NF-Y genes that, based on the encoded subunits, included ten NF-YA, twenty-nine NF-YB and twenty NF-YC genes (Table 1). The physical map positions of the NF-Y genes on the tomato chromosomes were identified (Additional file 1: Figure S1), according to their ascending order of physical position (bp), from the short arm telomere to the long arm telomere. Of the fifty-nine NF-Y genes, fifty-seven could be mapped onto the twelve tomato chromosomes, with the exceptions being Solyc00g107050 and Solyc00g270510. Chromosome 1 contained three NF-YA genes, while chromosomes 4 to 7 and chromosome 9 did not contain any genes from this sub-group. Chromosome 5 contained the largest number of NF-YB genes, seven, almost all of which were located in the upper part of the chromosome. A total of ten NF-Y genes were located on chromosome 5. Chromosome 3 had the largest number of NF-YC genes, six. It appears that the pattern of NF-Y genes across plant genomes is uneven and that the distribution varies among different species. For example, in common bean (Phaseolus vulgaris), PvNF-Y genes were mapped to 10 out of the 11 chromosomes [63] and the nine PvNF-YA genes were uniformly distributed, while five of the PsNF-YB genes were located on chromosome 7. Moreover, members of the PvNF-YC subfamily were found on five chromosomes, with one or two per chromosome [63].
We next analyzed the exon-intron structure of the tomato NF-Y genes (Additional file 2: Figure S2). The NF-YA genes had four to seven exons, and most (eight out of ten had five or six exons, while the majority of the NF-YB genes (twenty out of twenty-nine) had one or two exons. However, the structure of the NF-YC genes was more variable and thirteen out of twenty had one or two exons, while others had three to six exons, and one NF-YC gene, Solyc02g091030, had twenty-two exons. Overall, the gene structure analysis revealed that the members of the NF-YA family had a relatively consistent intron/exon organization (Additional file 2: Figure S2), while the NF-YB and NF-YC sub-groups were more variable among different members, which is similar to the organization reported for common bean NF-Y genes [63].
Multiple alignment of NF-Y protein sequences and analysis of the evolutionary relationship between tomato and A. thaliana families
Multiple sequence alignments were generated of proteins corresponding to members of the tomato and A. thaliana NF-Y subunit family (NF-YA, NF-YB, NF-YC). The A. thaliana NF-Y family was comprised of ten AtNF-YA, thirteen AtNF-YB and fourteen AtNF-YC proteins. Each sub-family was found to have one or more central core regions with extensive homologous motifs (Addtional file 3: Figure S3), that have been reported to be important in subunit interactions and DNA binding in yeast and mammals [29, 48, 70, 78]. The motifs of the tomato NF-Y proteins with putative functions in protein interactions and DNA binding were highlighted using the InterPro protein sequence analysis and classification database (Additional file 4: Fig. S4, http://www.ebi.ac.uk/interpro/). Finally, an un-rooted phylogenetic tree was created using the full length tomato and A. thaliana NF-Y protein sequences in order to gain insight into their evolutionary relationships and to assist with functional predictions (Fig. 1).
Phylogenetic analysis of tomato and A. thaliana NF-Y proteins. Phylogenetic analysis of 96 NF-Y proteins from tomato (59) and A. thaliana (37). Four branches were formed corresponding to genes with different subunits. Pink represents two branches of NF-YC, blue represents NF-YB and yellow represents NF-YA
The tomato and A. thaliana NF-YA proteins were variable in length, ranging from 154-325 and 186-340 amino acids (AAs), respectively (Table 1), and they were observed to share two characteristic conserved domains (Additional file 3: Figure S3). Previous studies with metazoans and yeast have indicated that the first twenty AA conserved domain is required for subunit interactions with NF-YB/YC, and that the other 21 AA domain is necessary for DNA binding and CCAAT sequence recognition specificity [46, 59, 78]. Outside the conserved regions, there was limited AA sequence conservation; however, the sequences were generally rich in Gln (Q) and Ser/Thr (S/T) residues, a feature that has been associated with promoting transcriptional activation [10, 11]. All the tomato NF-YA proteins contained predicted nuclear localization signals that were similar to those of the A. thaliana homologs (red boxes in Fig. 2; [26, 74]).
Expression patterns of NF-Y genes in various tomato organs. RNA-seq expression data corresponding to fifty-nine tomato NF-Y genes were retrieved from The Genome Consortium (TGC) datasets [65] for further analysis. Twelve genes with no detectable transcripts in any tissue/organ were excluded from the heat map. In the heat map, the RPKM (Reads Per Kilobase of exon model per Million mapped reads) values were transformed to log2(value + 1). Thirteen genes are shown labeled with their subunit on the right corresponding to genes targeted for subsequent analysis. The expression in various tomato organs is shown, including bud, flower, leaf, root, F_1cm (fruit with a 1 cm diameter), F_2cm, F_3cm, MG (Mature Green), B (Breaker), B10 (Breaker + 10 days)
The NF-YB proteins from tomato and A. thaliana ranged from 86-357 and 139- 275 AA in length, respectively (Table 1). They were highly similar in a stretch of more than 90 AAs that is considered to be the central conserved domain involved in the interaction with the NF-YC and NF-YA subunits and DNA binding (Additional file 3: Figure S3). Outside this central region, the sequences were variable in both length and AA composition. Evolutionary analysis can be used to predict the function of members from the same clade based on the known function of one or more members of that clade. For example, AtNF-YB6 from A. thaliana, also known as LEC1-LIKE (L1L), is a regulator of embryo development, and L1L RNA accumulates in developing embryos [35]. The tomato L1L paralogs, L1L1 to L1L13, have similar expression patterns in either seed or developing fruit, or both, and may therefore have a similar function [24].
The NF-YC sub-group contained a central domain of approximately 80 AAs that was highly conserved across the different members, and which has been shown to be important for both DNA binding and interactions between NF-Y subunits (Additional file 3: Figure S3) [64]. Most NF-YC proteins were enriched in Q residues (Additional file 3: Figure S3), a feature that has previously been noted in A. thaliana [69], Brachypodium distachyon [7] and other plant species [57]. In mammals and yeast, the Q rich regions of NF-YC proteins have been reported to function in transcriptional activation [10, 11]. The phylogenetic analysis identified two main clades, one containing AtNF-YC10 and four tomato NF-YC proteins, and another containing thirteen AtNF-YC and sixteen tomato NF-YC proteins (Fig. 1). Such gene structure analysis can often provide additional information regarding putative gene function [68] and here it was noted that three pairs of tomato NF-Y genes in the same clade had the same number of exons (Additional file 2: Figure S2).
Expression patterns of NF-Y genes in different tomato organs and during fruit development
Tomato transcript expression (RNA-seq) data sets are publicly available, including expression in 10 tomato organ types: bud, flower, leaf, root, and fruit with 1 cm, 2 cm or 3 cm diameters, and at the Mature Green, Breaker, Breaker + 10 days stages of development and ripening [65]. These datasets were searched using the fifty-nine tomato NF-Y gene sequences and the results were used to construct a hierarchical clustering heat map (Fig. 2) displaying the expression patterns of forty-seven of the fifty-nine NF-Y genes. The other twelve NF-Y genes did not have any corresponding expression data and so were omitted from the analysis. Among remaining forty-seven observed NF-Y genes, more than half had low levels of transcripts, and these were enriched in top part of the heat map (Fig. 2). Specific NF-Y genes such as Solyc12g027650 were more abundantly expressed in buds and flowers, but much lower transcript levels in fruit. In this study, we focused on NF-Y genes with relatively higher expressions in fruit during development and ripening, so thirteen members, comprising four NF-YA, four NF-YB and five NF-YC genes, were selected for further functional analysis (Table 2), which was also labeled with corresponding subunit in the heat map (Fig. 2).
Quantitative RT-PCR analysis of NF-Y gene expression in ripening tomato fruit and their responsiveness to ethylene and 1-MCP
Based on the RNA-seq expression profile analysis, thirteen tomato NF-Y genes were selected as candidates for fruit ripening regulation and their expression was further evaluated by real-time quantitative RT-PCR analysis at different fruit developmental stages: Immature (IM), Mature Green (MG), Breaker (BK), Pink (PK) and Red Ripe (RR). As shown in Fig. 3, the genes collectively displayed a range of expression patterns during ripening, and the changes in expression were in agreement with the RNA-seq profiles, other than extent of the changes for a couple of the genes. The expression patterns of some genes, such as Solyc11g065700 (Fig. 3a), Solyc09g007290 (Fig. 3b), Solyc01g079870 and Solyc01g096710 (Fig. 3c), were consistent with a role in promoting ripening since they showed up-regulated expression at onset of ripening. Others, such as Solyc03g110860 (Fig. 3c) showed reduced expression during ripening, and Solyc01g006930 (Fig. 3a) was down-regulated after the onset of ripening, although its expression increased at the BK stage, which might indicate a role in suppressing ripening. Two NF-Y genes showed a fluctuating expression patterns during development, including Solyc06g069310 (Fig. 3b) and Solyc01g087240 (Fig. 3a), although the pattern were opposite from each other. Other genes, including Solyc08g062210 (Fig. 3a), Solyc07g065500 and Solyc11g068480 (Fig. 3b), Solyc06g016750 and Solyc06g072040 (Fig. 3c) showed minor changes once ripening had initiated.
Expression of tomato NF-Y genes during fruit ripening. Figures are grouped into (a) NF-YA, (b) NF-YB, (c) NF-YC. Relative transcript levels are shown, as determined by quantitative real time RT-PCR, relative to the expression of the tomato ACTIN gene, expressed as 2-△△Ct [43]. The stages of fruit ripening were: Immature (IM), Mature Green (MG), Breaker (BK), Pink (PK), and Red Ripe (RR). Significance values were based on comparisons of expression levels at different ripening stages with expression at the IM stage. Expression levels of the tomato ACS2 gene were used as a positive control. Lowercase letters above the bars indicate values with a significant difference, which were determined by Student’s t-test (* = P < 0.05;** = P < 0.01)
To examine the ethylene responsiveness of these NF-Y genes, their expression levels were monitored in tomato fruits treated with ethylene or the ethylene receptor inhibitor 1-methylcyclopropene (1-MCP). Specifically, the expression of thirteen candidate ripening related NF-Y genes was evaluated in fruit that were harvested at the MG stage and then treated with ethylene or 1-MCP for 24 h, using quantitative RT-PCR analysis. Expression of an ethylene-inducible gene, ACS2 increased in the ethylene treated fruits, and was suppressed in the 1-MCP treated fruits compared with the control (Fig. 4), thereby confirming the induction or inhibition of ethylene signaling by the treatment. The treatment also significantly increased the expression of eleven NF-Y genes (Fig. 4), while the expression of Solyc01g079870 showed no significant change at the MG stage when treated with ethylene or 1-MCP (Fig. 4c), and the expression of Solyc07g065500 declined after treatment with 1-MCP but was not significantly affected by the exogenous ethylene (Fig. 4b), which was in accordance with the flat trend during ripening (Fig. 3b). Solyc03g110860 was the only gene of those tested whose expression was induced by ethylene and decreased to 1-MCP. This pattern was unexpected as we observed that the expression of Solyc03g110860 decreases during tomato ripening (Fig. 3c).
Expression of tomato NF-Y genes in fruit treated with ethylene or 1-MCP. Figures are grouped into (a) NF-YA, (b) NF-YB, (c) NF-YC. Fruit at the MG stage and after 24 h of treatment with ethylene or 1-MCP. Fruits harvested at the MG stage and treated with air for 24 h were used as a control. The change in relative transcript levels of genes in fruits treated with ethylene or 1-MCP are shown, determined by quantitative real time RT-PCR as described above. Asterisks indicate values with a significant difference as determined by Student’s t-test (* = p < 0.05, ** = p < 0.01)
TRV-mediated VIGS of NF-Y genes affects tomato fruit ripening
We next investigated the functions of the thirteen ripening regulator candidates using TRV-mediated virus induced gene silencing (VIGS), an effective tool to down-regulate gene expression [12] that has proven useful for gene functional characterization in tomato fruits [18, 53, 54]. Except for two pairs of genes (Solyc03g110860/Solyc06g072040 and Solyc08g062210/Solyc11g065700), which were highly homologous and therefore shared the same gene fragment in the VIGS vector, gene specific fragments of each gene, ranging from 400 bp to 500 bp, were selected for VIGS plasmid construction. As a result, a total of 11 specific recombinant plasmids and one positive pTRV2 vector carrying a truncated PHYTOENE DESATURASE (tPDS) gene fragment were used for the VIGS experiments.
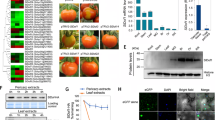
Approximately ten days after infiltration of the fruit stalk with mixed Agrobacterium strain GV3101 containing pTRV1 and pTRV2 or one of the twelve recombinant vectors (similar dual viral vectors construction has been reported by [42]), fruits of the infiltrated plants developed an uneven coloring, which was visible for several days. As expected [31], the PDS gene-silenced fruit (positive control) displayed a photo-bleached phenotype, which led to a partial yellow coloring of the fruits at the red colored stages (Fig. 5a). Silencing of 5 NF-Y genes (Solyc01g087240, Solyc06g069310, Solyc07g065500 and Solyc08g062210/Solyc11g065700) resulted in patchy coloring of the fruit with areas exhibiting different shades of yellow, orange or pink (Fig. 5b to e). In order to ensure the efficiency of the VIGS, the presence of the virus and gene transcript levels were determined (Fig. 5). We observed that mRNA levels of Solyc01g087240, Solyc06g069310 and Solyc08g062210/Solyc11g065700 in the yellow colored areas were reduced by approximately 85 %, 30 %, 35 % and 37 %, respectively, compared with the orange tissues, which suggested that the expression of these genes was upregulated during ripening. In contrast, the mRNA levels of Solyc07g065500 in the red regions were much lower than in the yellow regions (a 74 % reduction), indicating a role in suppressing ripening. Finally, silencing of Solyc01g087240, Solyc06g069310 and Solyc07g065500, resulted in delayed ripening at earlier stages, with a subsequently restored red phenotype at later stages. Only the fruits of plants infiltrated with pTRV2-t2210/t5700, which silenced Solyc08g062210/Solyc11g065700, displayed patchy coloring until the Red Ripe stage (Additional file 5: Figure S5), which may indicate that simultaneous silencing of these two genes strengthens the effect of the VIGS.
TRV-mediated virus induced gene silencing (VIGS) of genes in tomato fruit. The inflorescence peduncles attached to the fruit were injected with Agrobacterium tumefaciens transformed with TRV alone, or with pTRV2 carrying a fragment of a target gene. a pTRV-PDS, (b) pTRV2-Solyc08g062210/Solyc11g065700, (c) pTRV2-Solyc06g069310, (d) pTRV2-Solyc07g065500, (e) pTRV2-Solyc01g087240. Silencing of the target gene led to a decrease in gene expression, and different fruit color phenotypes compared with plants that were transformed with the empty vector control, which displayed normal ripening fruits (a-e). RNA was extracted from the control and the red and yellow areas of the gene-silenced tomato fruits. After reverse transcription, both the control and silenced samples were assayed for the presence of the TRV virus using PCR with primers specific to the TRV2 vector (a, c), and a virus coat protein (CP) gene (b, d). Lane 1 = negative control, Lane 2 = red part of the silenced tomato fruit, lane 3 = yellow part of the silenced tomato fruit, and lane 4 = the positive control. RT-PCR analysis of the target genes in the silenced tomato fruits showed significantly different expression in the yellow areas of the fruits. Asterisks indicate a significant difference as determined by Student’s t-test (* = p < 0.05, ** = p < 0.01)
Subcellular localization of NF-Y genes
From the VIGS experiments it was concluded that five NF-Y TF genes affected tomato fruit ripening. It is known that TFs regulate the transcription of target genes through binding to specific cis-elements in their promoters and that this binding takes place in the nucleus. To assess the subcellular localization of the five TFs, their full-length open reading frames (ORFs) without the stop codon were cloned into a vector in frame with the green fluorescence protein (GFP) reporter gene under the control of the CaMV35S promoter. The resulting constructs and an empty (control) vector were transiently expressed in tobacco BY2 protoplasts. Fluorescence microscopy revealed that TFs with a NF-YA subunit, including Solyc01g087240, Solyc08g062210 and Solyc11g065700 were localized in the nucleus (Fig. 6), which is consistent with the subcellular localization of NF-YA proteins in mammals [26] and A. thaliana [22, 71]. One exception to this localization pattern was the wheat TaNF-YA10-1, which was found to be localized in both the nucleus and the cytoplasm [44]. TFs with a NF-YB subunit, including Solyc07g065500 and Solyc06g069310, were localized in the nucleus and cytoplasm (Fig. 6). It is known that NF-YB lacks a nuclear localization signal, and that only when it interacts with NF-YC, can the heterodimers be transported from the cytoplasm to the nucleus [26]. We infer from this result that heterodimers of tomato NF-YB proteins and endogenous tobacco NF-YC TFs likely formed during the transient expression analysis. Taken together, the subcellular localization analysis of the NF-Y TFs supports their proposed role as regulators of target genes.
Discussion
Based on the current tomato genome sequence and annotations from related databases, including SGN and PlantTFDB, a total of fifty-nine tomato NF-Y genes were identified. In contrast with the well-studied functions of NF-Y genes in regulating plant growth and development, as well as abiotic and biotic stress responses [4, 36, 57], little is known about their role in fruit ripening. To address this deficiency, we used a combination of bioinformatic analysis and high-throughput VIGS based gene functional screening to assess the potential involvement of fifty-nine predicted tomato NF-Y genes in ripening. The bioinformatic analysis included a study of the phylogenetic relationships between A. thaliana and tomato NF-Y proteins and an analysis of the chromosomal distribution, encoded protein motifs and exon/intron structure patterns of the tomato NF-Y genes. This revealed similarities and conservation of NF-Y gene function between A. thaliana and tomato. Subsequent analysis of gene expression patterns, coupled with VIGS-based silencing and subcellular localization studies of candidate genes, supported the involvement of a subset of NF-Y TFs in regulating tomato fruit ripening.
Bioinformatic analysis can be used to help predict the biological functions of gene prediction, and one approach is to use phylogenetic relationships among homologs from different species to infer function. As an example, there have been few functional studies of NF-Y genes in fleshy fruit development but these genes have been studied in more detail in A. thaliana [57], and so we performed a phylogenetic analysis of all the A. thaliana and tomato NF-Y genes. We reasoned that members of the same clade might have similar specific expression patterns, such as that shown by AtNF-YB6 (L1L), which is a critical regulator of embryo development, and is specifically expressed in developing embryos [35]. The tomato homologs, L1L1 to L1L13, were reported to be expressed either in seeds, or in developing fruit, or both [24]. In some cases, similarities in terms of gene structures have also been attributed with biological significance [68]. We observed that genes in group NF-YB with only one exon (e.g. Solyc07g065500, Solyc12g006120, Solyc01g099320, Solyc06g009010 and Solyc09g074760) were in the same clade, while genes in another clade (Solyc03g114400 and Solyc06g069310) had six exons. In addition, similar structures were noted for three NF-YC gene pairs: Solyc03g111460/Solyc03g111470, Solyc00g107050/ Solyc11g016910, and Solyc11g072150/ Solyc01g096710. NF-YA genes had fewer difference among members in gene structures (Additional file 1: Figure S1).
Using this analysis as a foundation, we next examined the expression patterns of the tomato NF-Y genes during ripening to identify those that were specifically expressed, or showed predominant expression, in fruit. Publicly available RNA-seq data, was used to assess the expression of the tomato NF-Y genes, which suggested thirteen candidate fruit ripening related regulators. Real-time quantitative RT-PCR analysis, of these thirteen NF-Y genes revealed a range of expression pattern during fruit ripening and response to treatments with ethylene or 1-MCP. The expression of some of these genes such as Solyc11g065700 (Fig. 3a), Solyc09g007290 (Fig. 3b), Solyc01g079870 and Solyc01g096710 (Fig. 3c), was up-regulated during ripening and induced by ethylene (Fig. 4), whose synthesis in system 2 is key to climacteric fruit including tomato [5]. Expression of Solyc06g069310 was induced by the ethylene treatment (Fig. 4b), which was consistent with its up-regulated expression at the BK stage (Fig. 3b). Other genes, including Solyc11g068480 (Fig. 3b), Solyc06g016750 and Solyc06g072040 (Fig. 3c) were induced by ethylene, but showed relatively minor changes in expressions during ripening, especially at the beginning of the BK stage. We deduced that these might not act as key regulators of ripening, especially when compared to Solyc08g062210 and Solyc07g065500, whose silencing significantly influenced normal ripening (Fig. 5). Expression of some genes, such as Solyc01g087240 and Solyc01g006930 (Fig. 3a) was lower at the BK stage compared to MG but was induced by ethylene treatment (Fig. 4), which suggests that ethylene might not be the only factor regulating their expression at onset of ripening. It was reported that NF-YA genes are the target of the microRNA169 [52, 58], but it is not known whether miR169 regulates their expression at onset of tomato fruit ripening. Interestingly, expression of Solyc03g110860 was not only induced by exogenous ethylene but also reduced by 1-MCP (Fig. 4c); however, its down-regulated expression during ripening indicated a negative association with fruit ripening (Fig. 3c). In climacteric fruit, once ripening is initiated, the burst of system 2 ethylene is accompanied with the activation of many ripening related genes, such as ACS2 and ACS4. In this study, expression of Solyc03g110860 was induced by exogenous but not endogenous ethylene, which may suggest that exogenous ethylene treatments induce stress in tomato fruits. This may be related to the observations that some NF-YC genes play a role in responses to abiotic and biotic stress [23, 80]. It is also not known whether some NF-YC proteins whose functions overlap with that those of Solyc03g110860 competitively recruit NF-YA after forming a complex with NF-YB. Plant NF-Y genes are known to exhibit functional redundancy (Kuminoto et al., 2010) and this might result in varying responses to ethylene when the fruits start to ripen.
The gene expression patterns suggested candidates for further analysis using VIGS, a powerful tool for functional gene studies of tomato fruit development and ripening [18, 53, 60], which has been used to assess the functions of genes encoding ubiquitin-conjugating enzymes in fruit ripening, such as SlUBC32 & SlUBC41 [76]. The VIGS studies, together with confirmed subcellular localization of the candidate proteins in nucleus, indicated that NF-Y TFs likely act to regulate tomato fruit ripening. For example, silencing the expression of either Solyc01g087240, Solyc06g069310 or Solyc07g065500 resulted in altered fruit pigmentation in the earlier stages of ripening, although the fruits were uniformly red at later stages. For these fruits, the restoration of normal ripe coloration suggests that homologous genes may have compensated for the loss of function of the targeted NF-Y genes. Indeed, sequence similarity among the NF-Y genes was apparent and the phylogenetic analysis indicated that Solyc11g065700 is closely related to Solyc01g008490, with which it shares 70 % amino acid identity. Additionally, Solyc07g065500/Solyc12g006120, and Solyc03g114400/Solyc06g069310 share 68 % and 89 % AA identity, respectively (data not shown). As a precedent, complementation of gene functions in the context of gene silencing has been reported for SlUBC32 and SlUBC41 in the ubiquitination process during tomato fruit ripening [76]. Silencing of Solyc07g065500 led to accelerated color development in ripening fruit compared to the control, which suggests a role in suppressing ripening. Indeed, the down-regulation of Solyc07g065500 expression during ripening (Fig. 3b), supports such role at the BK stage. A significant down-regulation tendency was consistent with its proper negative role at BK stage. A similar phenomenon been reported for SlMADS1, the transcript abundance of SlMADS1 decreases significantly during ripening, and SlMADS1-silenced tomato fruits have a reduced ripening time, which suggested that SlMADS1 is a negative regulator of fruit ripening [13].
Taken together, the results of this study suggest that NF-Y TFs play roles in the transcriptional regulation of tomato fruit ripening. The specific functions and modes of action of the different family members that were highlighted, and the interactions among the NF-Y TFs with different subunits will be the subject of further studies.
Conclusion
In this work, fifty-nine tomato NF-Y genes were identified, and the chromosomal distribution, gene structures, phylogenetic relationship and expression patterns were characterized. Among the fifty-nine NF-Y genes, the expressions of thirteen members showed evidence of being related to fruit ripening. Furthermore, we examined their biological functions in ripening regulation using VIGS and subcellular localization studies. We determined that five NF-Y genes, including two members of the NF-YB subgroup (Solyc06g069310, Solyc07g065500) and three members of the NF-YA subgroup (Solyc01g087240, Solyc08g062210, Solyc11g065700), influence ripening. In addition, subcellular localization analyses confirmed the localization of the encoded proteins, fused to the GFP reporter, in the nucleus. We conclude that 5 NF-Y TFs play roles in tomato fruit ripening, and this will provide a platform for further investigation of their biological functions and the evolution of tomato NF-Y gene family.
Methods
NF-Y gene IDs and sequences
Tomato (S. lycopersicum) and A. thaliana NF-Y gene sequences were obtained from the PlantTFDB database v3.0 (http://planttfdb.cbi.edu.cn/) [25]. The annotated S. lycopersicum gene sequences were downloaded from the Sol Genomics Network (SGN, ftp://ftp.solgenomics.net/genomes/Solanum_lycopersicum/annotation/ITAG2.4_release/) database.
NF-Y gene structure and chromosomal location
The tomato NF-Y genes were assigned to chromosomes according to the positions given in the SGN database, as were the exon-intron organizations of individual NF-Y genes. Gene structures were visualized using Fancy gene v1.4 [61].
Alignments and phylogenetic analysis of NF-Y genes
Tomato and A. thaliana NF-Y AA sequences were obtained from the PlantTFDB database v3.0 (http://planttfdb.cbi.edu.cn/) and aligned using the Vector NTI Advance 11.5.1 software (Invitrogen). Alignments of AA sequences of full length NF-Y proteins were made using Clustal X2.1 [37]. An unrooted phylogenetic tree was constructed based on the alignments using MEGA4.0 (http://www.megasoftware.net/mega4/mega.html) [73] and the neighbor-joining (NJ) method. The parameters used in the tree construction were Amino: Poisson correction model plus Uniform rates determined by Pairwise deletion and 1,000 bootstraps. The trees were visualized using the online tool, EvolView [81]. Protein motifs were annotated using InterPro 51.0 [49] and visualized in DOG2.0.1 (http://dog.biocuckoo.org/) [62].
Transcriptome data analysis
The normalized expression levels of tomato genes, based on RNA-seq data, were obtained from The Genome Consortium (TGC) datasets [65]. To visualize the expression patterns of the NF-Y genes in different tomato organs, a heat map was created using R Project (http://www.r-project.org/). To generate a heat map of gene expression, the RPKM (Reads Per Kilobase of exon model per Million mapped reads) values of all genes were transformed to log2 (value + 1) to reveal differences in expression levels between observed organs among observed NF-Y members. A cut-off value of 30 (log2 31 in the heat map) was set to define higher expression, based on a significant distribution difference of published RPKM data [50], RPKM values higher than 30 were considered highly expressed, corresponding a value higher than log2 31 in the heat map. Genes with higher expression in fruit were selected for following experiments.
Plant material and growth conditions
Tomato (cv. Micro-Tom [MT]) seedlings were grown in a greenhouse under long day conditions (16-h light, 8-h dark) at a temperature of 26 °C. For gene expression analysis, organs were collected, frozen in liquid nitrogen and stored at −80 °C until RNA extraction. Three independent samplings were performed.
Ethylene and 1-MCP treatment
Tomato fruit at the Mature Green (MG) stage were placed into an airtight 1-l plastic container with 100 μl/L ethylene and control fruit were placed in the same containers with air. For the 1-MCP treatment MG fruit were placed into the same airtight containers with 10 ppm of 1-MCP that was generated by dissolving 48 mg of 1-MCP-releasing powder in 50 μl of water [19]. The control fruit were placed into the same container with no powder added to the water. Both the treatments were conducted for 24 h in an incubator under 16-h light and 8-h dark at 25 °C conditions. After the treatment, the fruits were sliced, frozen in liquid nitrogen, and subjected to RNA isolation followed by quantitative RT-PCR. The effect of ethylene or 1-MCP was confirmed by quantitative RT-PCR using a pair of primers for ACS2 [79]. Three biological replicates were analyzed with each replicate sample being derived from four tomato fruits.
RNA isolation and real-time quantitative RT-PCR
RNA was isolated from the pericarp of tomato fruits at different developmental stages during ripening: Immature (IM), Mature Green (MG), Breaker (BK), Pink (PK) and Red Ripe (RR) stages, which occurred at on average at 37, 42, 46, 51 and 56 days post-anthesis (DPA), respectively. Total RNA extraction was carried out using DeTRNa reagent (EarthOx, CA, USA) according to the manufacturer’s protocol, and RNA integrity was verified by 1.5 % agar gel electrophoresis. Genomic DNA was removed from the RNA preparations by digestion with DNase I (TaKaRa, China), and RNA quality and quantity was confirmed by spectrophotometry (Thermo Scientific, NanoDrop™ 1000). RNA was reverse-transcribed into cDNA using M-MLV Reverse Transcriptase (Promega, USA) according to the manufacturer's instructions. Real-time quantitative RT-PCR was conducted using TransStart Top Green qPCR SuperMix (Transgen, China) with a real-time PCR System CFX96 (Bio-Rad, CA, USA). The reactions were performed with the following cycling profile: 95 °C for 30 s, 40 cycles at 95 °C for 5 s, and 60 °C for 20s. Melting curve analysis was performed to verify the specificity of the amplification for each primer pair. The tomato ACTIN gene (Solyc03g078400) was used as an internal reference gene, and tomato ripening-related ACS2 (Solyc01g095080) as positive control. Relative gene expression values were calculated using the 2-△△Ct method [43]. Three biological replicates were analyzed with each replicate sample being derived from four tomato fruits. A pairwise Student’s t test was performed to determine whether the qRT-PCR results were statistically different between two samples (*P < 0.05;**P < 0.01). Primers designed for Real-time quantitative RT-PCR are listed in Additional file 6: Table S1.
Plasmid construction for VIGS
The pTRV1 and pTRV2 VIGS vectors have been previously described [42]. In order to improve the cloning efficiency, we used a Cloning and Assembly Kit (Transgen, China). All the primers designed for PCR-amplification had a 15-bp overlap with the linearized pTRV2 vector and contained a BamHI restriction site. Since Solyc03g110860 and Solyc06g072040 are highly homologous, they shared the same gene fragment in the VIGS vector, as did Solyc08g062210 and Solyc11g065700. The primers are listed in Additional file 7: Table S2.
TRV mediated VIGS in tomato fruit
Agrobacterium tumefaciens strain GV3101 containing pTRV1 or pTRV2 and its derivatives were used for the VIGS experiments. GV3101 containing the TRV-VIGS vectors were grown at 28 °C in LB medium containing 10 mM MES buffer (pH5.6) and 20 mM acetosyringone with appropriate antibiotics (gentamicin and rifampicin for GV3101, kanamycin for pTRV1 or pTRV2). After overnight culturing (28 °C, 200 rpm), A. tumefaciens cells were harvested and resuspended in infiltration buffer (10 mM MgCl2, 10 mM MES [pH 5.6] and 150 mM acetosyringone) to a final OD600 of 2.0 (for both pTRV1 or pTRV2 and its derivatives). A. tumefaciens containing pTRV1 and pTRV2 or the recombinant vectors were mixed in a 1:1 ratio, and left for 3-5 h at room temperature before infiltration, as previously described [18]. The tomato inflorescence pedicels attached to the fruit were injected with cultures of A. tumefaciens harboring the vectors, using a 1-ml syringe. For biological replicates, at least 12 individual Micro-Tom tomato plants were injected for each gene silenced.
VIGS phenotype analysis, virus assay and quantitative RT-PCR
To detect the accumulation of the virus in the fruit, total RNA was extracted (as above) from different regions of tomato fruit showing different external coloration, and reverse-transcribed into cDNA using the M-MLV Reverse Transcriptase (Promega, USA) with a TRV-RNA2-specific primer, 5’-GGGCGTAATAACGCTTACGTAGGC-3’. The RNA2 cDNA of TRV was amplified with the RNA2-specific primers (GenBank accession number AF406991), 5’-CGGTCTAGAGGCACTCAACTTTATAAACC-3’ and 5’-CGGGGATCCCTTCAGTTTTCTGTCAAACC-3’. The RNA2 cDNA of the viral coat protein (CP) was amplified with the primers, 5’-CTGACTTGATGGACGATTCTT-3’ and 5’-TGTTCGCCTTGGTAGTAGTA-3’. To detect the silencing efficiency of specific genes, the isolated total RNA was reverse-transcribed into cDNA using the M-MLV Reverse Transcriptase (Promega, USA) with an oligo (dT)18 primer. Real-time quantitative RT-PCR was performed to analyze gene expression patterns, as above. A pairwise Student’s t test was performed to determine whether the qRT-PCR results were statistically different between two samples (* = P < 0.05;** = P < 0.01). The primers used are listed in Additional file 8: Table S3. Each analysis was analyzed separately for three different fruits.
Subcellular localization
The coding sequence (CDS) of the five NF-Y genes without the stop codon was amplified by PCR (primers are listed in Additional file 9: Table S4, CDS are listed in Additional file 10: Table S5) and sub-cloned into the pBI221-GFP vector, in frame with the green fluorescent protein (GFP) sequence, resulting in a set of 35S::GFP-NF-Y vectors. These fusion constructs and the control GFP vector were transformed into tobacco (Nicotiana tabacum) BY-2 suspension culture cell protoplasts using the polyethylene glycol (PEG) method [2, 67]. GFP fluorescence was observed with a fluorescence microscope (Zeiss7 10-3 channel). All transient expression assays were repeated at least three times.
Availability of supporting data
The RNA-seq data, are downloaded from The Genome Consortium (TGC) datasets, and can be obtained from http://www.nature.com/nature/journal/v485/n7400/extref/nature11119-s3.zip.
Other data supporting the results of this article are included within the article and its additional files.
Abbreviations
- 1-MCP:
-
1-methylcyclopropene
- AA:
-
amino acid
- ARF:
-
auxin respons factor
- BK:
-
Breaker
- CDS:
-
coding sequence
- ERF:
-
ethylene response factor
- GFP:
-
green fluorescence protein
- IM:
-
immature
- L1L:
-
LEC1-LIKE
- MG:
-
mature green
- NF-Y:
-
nuclear factor Y
- NJ:
-
neighbor-joining
- ORFs:
-
open reading frames
- PEG:
-
polyethylene glycol
- PK:
-
pink
- RPKM:
-
reads per kilobase of exon model per million mapped reads
- RR:
-
red ripe
- SGN:
-
Sol Genomics Network
- TFs:
-
transcription factors
- TGC:
-
the genome consortium
- tPDS:
-
truncated PHYTOENE DESATURASE
- VIGS:
-
virus induced gene silencing
References
Adato A, Mandel T, Mintz-Oron S, Venger I, Levy D, Yativ M, et al. Fruit-surface flavonoid accumulation in tomato is controlled by a SlMYB12-regulated transcriptional network. PLoS Genet. 2009;5:e1000777.
Ba LJ, Shan W, Kuang JF, Feng BH, Xiao YY, Lu WJ, et al. The banana MaLBD (lateral organ boundaries domain) transcription factors regulate EXPANSIN expression and are involved in fruit ripening. Plant Mol Biol Rep. 2014;32:1103–13.
Ballester AR, Molthoff J, de Vos R, Hekkert B, Orzaez D, Fernandez-Moreno JP, et al. Biochemical and molecular analysis of pink tomatoes: deregulated expression of the gene encoding transcription factor SlMYB12 leads to pink tomato fruit color. Plant Physiol. 2010;152:71–84.
Ballif J, Endo S, Kotani M, MacAdam J, Wu Y. Over-expression of HAP3b enhances primary root elongation in Arabidopsis. Plant Physiol Biochem. 2011;49:579–83.
Barry C, Llop-Tous M, Grierson D. The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol. 2000;123:979–86.
Bucher P, Trifonov EN. CCAAT box revisited: bidirectionality, location and context. J Biomol Struct Dyn. 1988;5:1231–6.
Cao S, Kumimoto RW, Siriwardana CL, Risinger JR, Holt 3rd BF. Identification and characterization of NF-Y transcription factor families in the monocot model plant Brachypodium distachyon. PLoS ONE. 2011;6:e21805.
Chen NZ, Zhang XQ, Wei PC, Chen QJ, Ren F, Chen J, et al. AtHAP3b plays a crucial role in the regulation of flowering time in Arabidopsis during osmotic stress. J BiochemMol Biol. 2007;40:1083–9.
Chung MY, Vrebalov J, Alba R, Lee J, McQuinn R, Chung JD, et al. A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J. 2010;64:936–47.
Coustry F, Maity SN, Sinha S, de Crombrugghe B. The transcriptional activity of the CCAAT-binding factor CBF is mediated by two distinct activation domains, one in the CBF-B subunit and the other in the CBF-C subunit. J Biol Chem. 1996;271:14485–91.
de Silvio A, Imbriano C, Mantovani R. Dissection of the NF-Y transcriptional activation potential. Nucleic Acids Res. 1999;27:2578–84.
Dinesh-Kumar SP, Anandalakshmi R, Marathe R, Schiff M, Liu Y. Virus-induced gene silencing. Methods Mol Biol. 2003;236:287–94.
Dong TT, Hu ZL, Deng L, Wang Y, Zhu MK, Zhang JL, et al. A tomato MADS-box transcription factor, SlMADS1, acts as a negative regulator of fruit ripening. Plant Physiol. 2013;163:1026–36.
Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007;49:414–27.
Fernandez AI, Viron N, Alhagdow M, Karimi M, Jones M, Amsellem Z, et al. Flexible tools for gene expression and silencing in tomato. Plant Physioloy. 2009;151:1729–40.
Fornari M, Calvenzani V, Masiero S, Tonelli C, Petroni K. The Arabidopsis NF-YA3 and NF-YA8 gene are functionally reduntant and are required in early embryogenesis. PLoS One. 2013;8:e82043.
Frontini M, Imbriano C, Manni I, Mantovani R. Cell cycle regulation of NF-YC nuclear localization. Cell Cycle. 2004;3:217–22.
Fu DQ, Zhu BZ, Zhu HL, Jiang WB, Luo YB. Virus-induced gene silencing in tomato fruit. Plant J. 2005;43:299–308.
Fujisawa M, Nakano T, Shima Y, Ito Y. A large-scale identification of direct targets of the tomato MADS Box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell. 2013;25:371–86.
Giovannoni JJ. Genetic regulation of fruit development and ripening. Plant Cell. 2004;16:Suppl S170–180.
Giovannoni JJ. Fruit ripening mutants yield insights into ripening control. Curr Opin Plant Biol. 2007;10:283–9.
Hackenberg D, Wu Y, Voigt A, Adams R, Schramm P, Grimm B. Studies on differential nuclear translocation mechanism and assembly of the three subunits of the Arabidopsis thaliana transcription factor NF-Y. Mol Plant. 2012;5:876–88.
Hackenberg D, Keetman U, Grimm B. Homologous NF-YC2 subunit from Arabidopsis and tobacco is activated by photooxidative stress and induces flowering. Int J Mol Sci. 2012;13:3458–77.
Hilioti Z, Ganopoulos I, Bossis I, Tsaftaris A. LEC1-LIKE paralog transcription factor: how to survive extinction and fit in NF-Y protein complex. Gene. 2014;543:220–33.
Jin J, Zhang H, Kong L, Gao G, Luo J. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014;42:D1182–7.
Kahle J, Baake M, Doenecke D, Albig W. Subunits of the heterotrimeric transcription factor NF-Y are imported into the nucleus by distinct pathways involving importin beta and importin 13. Mol Cell Biol. 2005;25:5339–54.
Karlova R, Chapman N, David K, Angenent GC, Seymour GB, de Maagd RA. Transcriptional control of fleshy fruit development and ripening. J Exp Bot. 2014;65:4527–41.
Karlova R, Rosin FM, Busscher-Lange J, Parapunova V, Do PT, Fernie AR, et al. Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell. 2011;23:923–41.
Kim IS, Sinha S, de Crombrugghe B, Maity SN. Determination of functional domains in the C subunit of the CCAAT-binding factor (CBF) necessary for formation of a CBF-DNA complex: CBF-B interacts simultaneously with both the CBF-A and CBF-C subunits to form a heterotrimeric CBF molecule. Mol Cell Biol. 1996;16:4003–13.
Klee HJ, Giovannoni JJ. Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet. 2011;45:41–59.
Kumagai MH, Donson J, Della-Cioppa G, Harvey D, Hanley K, Grill LK. Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc Natl Acad Sci U S A. 1995;92:1679–83.
Kumar R, Tyagi AK, Sharma AK. Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Mol Genet Genomics. 2011;285:245–60.
Kumimoto RW, Adam L, Hymus GJ, Repetti PP, Reuber TL, Marion CM, et al. The nuclear factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta. 2008;228:709–23.
Kumimoto RW, Zhang Y, Siefers N, Holt III BF. NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J. 2010;63:379–91.
Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, et al. LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell. 2003;15:5–18.
Laloum T, De Mita S, Gamas P, Baudin M, Niebel A. CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 2013;18:157–66.
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8.
Leyva-Gonzalez MA, Ibarra-Laclette E, Cruz-Ramirez A, Herrera-Estrella L. Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS One. 2012;7:e48138.
Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, et al. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell. 2008;20:2238–51.
Lin Z, Hong Y, Yin M, Li C, Zhang K, Grierson D. A tomato HD-zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J. 2008;55:301–10.
Liu JX, Howell SH. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell. 2010;22:782–96.
Liu Y, Schiff M, Dinesh-Kumar SP. Virus-induced gene silencing in tomato. Plant J. 2002;31:777–86.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8.
Ma X, Zhu X, Li C, Song Y, Zhang W, Xia G, et al. Overexpression of wheat NF-YA10 gene regulates the salinity stress response in Arabidopsis thaliana. Plant Physiol Biochem. 2015;86:34–43.
Manning K, Tor M, Poole M, Hong Y, Thompson AJ, King GJ, et al. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet. 2006;38:948–52.
Mantovani R, Li XY, Pessara U, Hooft Van Huisjduijnen R, Benoist C, Mathis D. Dominant negative analogs of NF-YA. J Biol Chem. 1994;269:20340–6.
Mantovani R. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 1998;26:1135–43.
McNabb DS, Tseng KA, Guarente L. The Saccharomyces cerevisiae Hap5p homolog from fission yeast reveals two conserved domains that are essential for assembly of heterotetrameric CCAAT-binding factor. Mol Cell Biol. 1997;17:7008–18.
Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43:D213–21.
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008;5:621–8.
Mu JY, Tan HL, Hong SL, Liang Y, Zuo JR. Arabidopsis transcription factor genes NF-YA1, 5, 6, and 9 play redundant roles in male gametogenesis, embryogenesis, and seed development. Mol Plant. 2013;6:188–201.
Ni ZY, Hu Z, Jiang QY. GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol Biol. 2013;82:113–29.
Orzaez D, Medina A, Torre S, Fernandez-Moreno JP, Rambla JL, Fernandez-del-Carmen A, et al. A visual reporter systemfor virus-induced gene silencing in tomato fruit based on anthocyanin accumulation. Plant Physiol. 2009;150:1122–34.
Orzaez D, Mirabel S, Wieland WH, Granell A. Agroinjection of tomato fruits: a tool for rapid functional analysis of transgenes directly in fruit. Plant Physiol. 2006;140:3–11.
Osorio S, Scossa F, Fernie AR. Molecular regulation of fruit ripening. Front Plant Sci. 2013;4:198.
Peng WT, Lee YW, Nester EW. The phenolic recognition profiles of the Agrobacterium tumefaciens VirA protein are broadened by a high level of the sugar binding protein ChvE. J Bacteriol. 1998;180:5632–8.
Petroni K, Kumimoto RW, Gnesutta N, Calvenzani V, Fornari M, Tonelli C, et al. The promiscuous life of plant nuclear factor transcription factors. Plant Cell. 2012;24:4777–92.
Potkar R, Recla J, Busov V. Ptr-miR169 is a posttranscriptional repressor of PtrHAP2 during vegetative bud dormancy period of aspen (Populus tremuloides) trees. Biochem Bioph Res Co. 2013;431:512–8.
Quach TN, Nguyen HT, Valliyodan B, Joshi T, Xu D, Nguyen HT. Genome-wide expression analysis of soybean NF-Y genes reveals potential function in development and drought response. Mol Genet Genomics. 2015;290:1095–115.
Quadrana L, Rodriguez MC, López M, Bermúdez L, Nunes-Nesi A, Fernie AR, et al. Coupling virus-induced gene silencing to exogenous green fluorescence protein expression provides a highly efficient system for functional genomics in Arabidopsis and across all stages of tomato fruit development. Plant Physiol. 2011;156:1278–91.
Rambaldi D, Ciccarelli FD. FancyGene: dynamic visualization of gene structures and protein domain architectures on genomic loci. Bioinformatics. 2009;25:2281–2.
Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. DOG 1.0: illustrator of protein domain structures. Cell Res. 2009;19:271–3.
Rípodas C, Castaingts M, Clúa J, Blanco F, Zanetti ME. Annotation, phylogeny and expression analysis of the nuclear factor Y families in common bean (Phaseolus vulgaris). Front Plant Sci. 2015;14:761.
Romier C, Cocchiarella F, Mantovani R, Moras D. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J Biol Chem. 2003;278:1336–45.
Sato S, Tabata S, Hirakawa H, Asamizu E, Shirasawa K, Isobe S, et al. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–41.
Seymour G, Poole M, Manning K, King GJ. Genetics and epigenetics of fruit development and ripening. Curr Opin Plant Biol. 2008;11:58–63.
Shan W, Kuang JF, Chen L, Xie H, Peng HH, Xiao YY, et al. Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening. J Exp Bot. 2012;63:5171–87.
Shiu SH, Bleecker AB. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003;132:530–43.
Siefers N, Dang KK, Kumimoto RW, Bynum 4th WE, Tayrose G, Holt 3rd BF. Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 2009;149:625–41.
Sinha S, Kim IS, Sohn KY, de Crombrugghe B, Maity SN. Three classes of mutations in the A subunit of the CCAAT-binding factor CBF delineate functional domains involved in the three-step assembly of the CBF-DNA complex. Mol Cell Biol. 1996;16:328–37.
Siriwardana CL, Kumimoto RW, Jones DS, Holt 3rd BF. Gene family analysis of the Arabidopsis NF-YA transcription factors reveals opposing abscisic acid responses during seed germination. Plant Mol Biol Report. 2014;32:971–86.
Sun H, Fan HJ, Ling HQ. Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genomics. 2015;22:16–9.
Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9.
Thon M, Al Abdallah Q, Hortschansky P, Scharf DH, Eisendle M, Haas H, et al. The CCAAT-binding complex coordinates the oxidative stress response in eukaryotes. Nucleic Acids Res. 2010;38:1098–113.
Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, et al. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science. 2002;296:343–6.
Wang Y, Wang W, Cai J, Zhang Y, Qin G, Tian S. Tomato nuclear proteome reveals the involvement of specific E2 ubiquitin-conjugating enzymes in fruit ripening. Genome Biol. 2014;15:548.
Warpeha KM, Upadhyay S, Yeh J, Adamiak J, Hawkins SI, Lapik YR, et al. The GCR1, GPA1, PRN1, NF-Y signal chainmediates both blue light and abscisic acid responses in Arabidopsis. Plant Physiol. 2007;143:1590–600.
Xing Y, Zhang S, Olesen JT, Rich A, Guarente L. Subunit interaction in the CCAAT-binding heteromeric complex is mediated by a very short alpha-helix in HAP2. Proc Natl Acad Sci U S A. 1994;91:3009–13.
Yokotani N, Nakano R, Imanishi S, Nagata M, Inaba A, Kubo Y. Ripening-associated ethylene biosynthesis in tomato fruit is autocatalytically and developmentally regulated. J Exp Bot. 2009;60:3433–42.
Zanetti ME, Blanco FA, Beker MP, Battaglia M, Aguilar OM.A C subunit of the plant nuclear factor NF-Y required for rhizobial infection and nodule development affects partner selection in the common bean-Rhizobium etli symbiosis. Plant Cell. 2010;22:4142–57.
Zhang H, Gao S, Lercher MJ, Hu S, Chen WH. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 2012;40:W569–72.
Acknowledgements
We thank Professor Jianye Chen and Dr. Jianfei Kuang (South China Agricultural University) for the generous gift of tobacco BY2 suspension cells and the pBI221-GFP vector. We thank Yifan Wang for help with the subcellular localization experiment. This work was supported by the National Natural Science Foundation of China (NSFC 31271959 and NSFC 31571894). We thank PlantScribe (www.plantscribe.com) for careful editing of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conceived and designed the experiments: BZ. Performed the experiments: SL KL. Analyzed the data: DC ZJ. Contributed reagents/materials/analysis tools: YL. Edited the manuscript: YL HZ DF. Wrote the paper: SL. All authors read and approved the final manuscript.
Additional files
Additional file 1: Figure S1.
Chromosome distribution of NF-Y genes in the tomato genome. The gene IDs below denotes each specific NF-Y gene. The position of each gene on the bar denotes the relative location on the corresponding chromosome. Arrows following each gene indicate the orientation of the gene. (JPG 7258 kb)
Additional file 2: Figure S2.
Exon-intron structure of NF-Y genes in tomato. The exon and intron distribution of each gene is shown. Rectangles represent the location and length of exons and broken lines indicate the location and length of introns. Arrows at the end of the last exon indicate the direction of the gene on the chromosome. (JPG 7098 kb)
Additional file 3: Figure S3.
Alignment of tomato and A. thaliana NF-YA domains. Numbers in parentheses correspond to the start and end sites of the NF-YA domain in the given protein. The amino acids that are predicted to be important for DNA binding and subunit interaction are based on Quach et al. [59]. The three boxes in red are the basic residual clusters required for nuclear targeting ([56]. (JPG 8734 kb)
Additional file 4: Figure S4.
Motif composition of tomato NF-Y proteins. A schematic representation of motif composition of tomato NF-Y proteins is shown based on InterPro protein sequence analysis and classification, including NF-YA (a), NF-YB (b) and NF-YC (c) subunits. (JPG 9995 kb)
Additional file 5: Figure S5.
TRV-mediated VIGS of Solyc08g062210/Solyc11g065700 in tomato fruit. External fruit color changes induced by VIGS infiltration were maintained until the Red Ripe stage. (JPG 7768 kb)
Additional file 6: Table S1.
Primers for real time qPCR analysis of expression during fruit ripening. (PDF 97 kb)
Additional file 7: Table S2.
Primers for VIGS plasmid construction. (PDF 142 kb)
Additional file 8: Table S3.
Primers for real time qPCR analysis of the VIGS assay. (PDF 84 kb)
Additional file 9: Table S4.
Primers for plasmid construction used in subcellular localization assays. (PDF 79 kb)
Additional file 10: Table S5.
CDS of NF-Y genes in subcellular localization. The CDS of each NF-Y gene was cloned as a fusion with EGFP with a deletion of termination codon. (PDF 118 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Li, S., Li, K., Ju, Z. et al. Genome-wide analysis of tomato NF-Y factors and their role in fruit ripening. BMC Genomics 17, 36 (2016). https://doi.org/10.1186/s12864-015-2334-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-015-2334-2