Abstract
Background
Evolution of reproductive isolation is an important process, generating biodiversity and driving speciation. To better understand this process, it is necessary to investigate factors underlying reproductive isolation through various approaches but also in various taxa. Previous studies, mainly focusing on diploid animals, supported the prevalent view that reproductive barriers evolve gradually as a by-product of genetic changes accumulated by natural selection by showing a positive relationship between the degree of reproductive isolation and genetic distance. Haplodiploid animals are expected to generate additional insight into speciation, but few studies investigated the prevalent view in haplodiploid animals. In this study, we investigate whether the relationship also holds in a haplodiploid spider mite, Amphitetranychus viennensis (Zacher).
Results
We sampled seven populations of the mite in the Palaearctic region, measured their genetic distance (mtDNA) and carried out cross experiments with all combinations. We analyzed how lack of fertilization rate (as measure of prezygotic isolation) as well as hybrid inviability and hybrid sterility (as measures of postzygotic isolation) varies with genetic distance. We found that the degree of reproductive isolation varies among cross combinations, and that all three measures of reproductive isolation have a positive relationship with genetic distance. Based on the mtDNA marker, lack of fertilization rate, hybrid female inviability and hybrid female sterility were estimated to be nearly complete (99.0–99.9% barrier) at genetic distances of 0.475–0.657, 0.150–0.209 and 0.145–0.210, respectively. Besides, we found asymmetries in reproductive isolation.
Conclusions
The prevalent view on the evolution of reproductive barriers is supported in the haplodiploid spider mite we studied here. According to the estimated minimum genetic distance for total reproductive isolation in parent population crosses in this study and previous work, a genetic distance of 0.15–0.21 in mtDNA (COI) appears required for speciation in spider mites. Variations and asymmetries in the degree of reproductive isolation highlight the importance of reinforcement of prezygotic reproductive isolation through incompatibility and the importance of cytonuclear interactions for reproductive isolation in haplodiploid spider mites.
Similar content being viewed by others
Background
Reproductive isolation facilitates divergence of closely related groups by restricting gene flow. Therefore, evolution of reproductive isolation is an important process for understanding speciation [1, 2]. Mechanisms of reproductive isolation and its evolutionary factors have been investigated by genetic, theoretical, ecological, molecular and comparative approaches [1, 2]. These various approaches are important for understanding evolution of reproductive isolation, since various factors and mechanisms contribute to it. However, it is also important to focus on various taxa, because that allows us to generalize insights from major model organisms, to reveal the details in mechanisms by taking advantageous traits of taxa other than major model organisms, and to figure out factors creating differences in its evolution among taxa.
Major model organisms in speciation studies are often diploid animals such as Drosophila [2]. Haplodiploid animals, however, are also widespread in the animal kingdom; for instance, approximately 15% of arthropods is haplodiploid [3]. They are different from diploids in genetic and sex determination system: females develop from diploid eggs (2n) and males develop from haploid eggs (n). The differences affect various ecological and evolutionary aspects such as mating system, sexual selection, resistance to inbreeding and rapid adaptation (e.g. against pesticides) [3]. Therefore, the differences may also affect speciation process and evolution of reproductive isolation.
Besides, haplodiploids have several advantages for studies on reproductive isolation. For example, the occurrence of a postmating, prezygotic barrier is typically called ‘cryptic’ reproductive isolation [4, 5]. In haplodiploids, however, postmating, prezygotic barrier can be easily detected in the offspring sex ratio, because females and males develop from fertilized and unfertilized eggs, respectively [6,7,8,9]. Hence, if fewer eggs are fertilized this results in a more male-biased sex ratio. It is also easy to investigate incompatible allelic interactions in haplodiploids, because males are haploid, therefore, it is not necessary to take effects of dominant allelic interactions in males into account [10]. Therefore, haplodiploids are expected to generate additional insight into speciation [11], and indeed, several studies have focused on the role of reproductive isolation in speciation using haplodiploids [9, 10, 12,13,14,15]. However, more studies on haplodiploids and evolutionary factors of reproductive isolation are needed before generalizations and comparisons with diploid species can be made.
Spider mites are tiny arthropod herbivores with a haplodiploid, arrhenotokous genetic system (i.e., males develop from unfertilized haploid eggs). Some of them are agricultural pests, therefore, their biology and ecology have been investigated quite well [16]. Various stages of reproductive isolation (e.g. premating, prezygotic and postzygotic) have been found, not only among closely related species [17,18,19] but also among populations or strains within a species [9, 10, 20,21,22,23]. Endosymbiont infections such as Wolbachia, Cardinium and Spiroplasma are also often associated with reproductive isolation in spider mites [24,25,26,27].
Many papers have reported presence of reproductive isolation in spider mites, however, few studies have addressed the evolutionary mechanisms. For example, it is suggested that reproductive isolation evolves gradually by accumulation of genetic changes caused by natural selection and genetic drift [1, 2]. This view is supported by comparative analyses showing a positive relationship between intensity of reproductive isolation and genetic distance among groups in several taxa [28,29,30,31,32,33,34,35,36,37,38]. However, in haplodiploids only one paper, which focused on the spider mite Stigmaeopsis miscanthi (Saito) species group, tested this idea [9]. The study found a positive relationship between intensity of reproductive isolation and genetic distance as in other taxa. However, to verify that the reproductive isolation patterns found in S. miscanthi species group are general in spider mites, studies using other spider mites are necessary. Besides, the study using S. miscanthi species group did not include hybrid sterility in the analyses because few hybrids were obtained in the study. To investigate the effect of genetic distance on hybrid sterility, it would be required to focus on younger groups which are still able to produce hybrids.
Here, we study evolution of reproductive isolation in the Hawthorn spider mite Amphitetranychus viennensis (Zacher), a herbivorous arthropod that feeds on rosaceous trees and occurs widespread in the Palaearctic region [39]. It is thought that A. viennensis originates from Eastern Asia because of the origin of its host plants [40]. Since it infests and thrives on cultivated rosaceous trees such as apples, peaches and cherries, it is under scrutiny as an orchard pest in several countries. A previous study [21] investigated the genetic and reproductive relationships between A. viennensis populations collected from France and Japan, and found genetic divergence in COI (mtDNA) and ITS (nDNA) and also incomplete reproductive isolation between them, suggesting that French and Japanese populations of A. viennensis are quite young, diverging taxa. Besides, they partly produced hybrids, indicating that A. viennensis allows us to analyze the relationship between the degree of reproductive isolation and genetic distance, including hybrid sterility. Therefore, in this study, we collected the mite from seven locations in the Palaearctic region including France and Japan, measured genetic distances and carried out cross experiments among them. We also checked endosymbiont infection status of each population, since reproductive incompatibility in spider mites may be caused by endosymbiont infection. We analyzed the relationships of pre- and post-zygotic reproductive isolation as well as hybrid sterility with genetic distance.
Results
Phylogeny and genetic distance
We collected A. viennensis from seven locations in the Eurasian continent: France (F), Turkey (T), Iran (I), Inner Mongolia (CIM, China), Eastern China (CN), Korea (K) and also from Japan (J) (Table 1). A maximum likelihood (ML) tree of A. viennensis populations based on the COI sequences (Fig. 1) showed that A. viennensis consists of two clades: one comprises the populations from France (F) and Turkey (T) and the other comprises the other five populations. In the latter clade, the populations from Korea (K), Eastern China (CN) and Inner Mongolia (CIM) seemed to be derived from populations of Iran (I) and Japan (J), although bootstrap values supporting the branches were not sufficiently high (Fig. 1). The genetic distance among population pairs ranged from 0 to 0.075 (Table 2). The genetic distance was approximately zero among population pairs from Korea (K), Eastern China (CN) and Inner Mongolia (CIM), and it was highest between the populations from Japan (J) and Iran (I) (Table 2).
Maximum likelihood (ML) tree based on the COI gene (618 bp) of mtDNA of seven populations of Amphitetranychus viennensis and two populations of A. quercivorus (Ehara & Gotoh). Tetranychus kanzawai Kishida was used as the outgroup. Bootstrap values based on 1000 replications are indicated at the nodes. Operational taxonomic unit is indicated by the population (abbreviation refers to Table 1) with accession number in brackets
Endosymbiont infections
No infection with Wolbachia, Cardinium, Spiroplasma or Rickettsia was detected in any of the seven populations of A. viennensis used in the experiments.
Reproductive isolation and genetic distance
Relative production of daughters, sons, unhatched eggs and dead offspring varied among combinations of crosses (Fig. 2; Additional file 1: Table S1). Two reproductive isolation patterns were found in the cross experiments. One is that offspring sex ratio was more male-biased as females mated with more genetically distant males (females: Inner Mongolia (CIM), Eastern China (CN), and Korea (K) populations). The other is that there was no clear relationship between offspring sex ratio and genetic distance, but viability of F1 hybrids was lower as females mated with more genetically distant males (females: France (F), Turkey (T), Iran (I) and Japan (J) populations).
Relative proportions of male offspring, unhatched eggs, dead offspring and female offspring in intra- and interpopulation crosses for females from population France (a), Turkey (b), Iran (c), Inner Mongolia (d), Eastern China (e), Korea (f) and Japan (g). In each population, crossed males are shown in order of genetic distance (GD) arrangement of their population with that of the female. For the abbreviation of population names, see Table 1. N shows the number of pairs in the cross experiments
Relative production of daughters, sons, unhatched eggs and dead offspring also varied among combinations of backcrosses (Fig. 3; Additional file 1: Table S1). In most backcrosses, there were no or very few (in backcrosses between France (F) and Turkey (T)) male offspring, indicating that hybrid males were hardly viable. There were male offspring in the crosses among Inner Mongolia (CIM, China), Eastern China (CN) and Korea (K), but these populations were not genetically different (Table 2).
Relative proportions of male offspring, unhatched eggs, dead offspring and female offspring in backcrosses using female hybrids produced from France × Turkey (a), France × Iran (b), France × Japan (c), Turkey × Iran (d), Inner Mongolia × Korea (e), Inner Mongolia × Eastern China (f), Inner Mongolia × Japan (g), Eastern China × Korea (h), and Eastern China × Japan (i). For the abbreviation of population names in males, see Table 1. Hybrid females are indicated by the mothers’ abbreviation followed by the fathers’ abbreviation. For example, TF stands for hybrid females from the cross between Turkey (female) × France (male), and FT stands for hybrid females from the cross between France (female) × Turkey (male). N shows the number of pairs in the cross experiments
Prezygotic isolation (post-mating fertilization)
Due to the arrhenotokous reproduction mode of this mite species, which means females develop from fertilized eggs and males develop from unfertilized eggs, we can assess several measures of prezygotic and postzygotic reproductive isolation by analyzing the produced offspring by sex. Specifically, prezygotic isolation due to lack of egg fertilization leads to overproduction of haploid males developing from the unfertilized eggs. We used the ratio of male offspring to egg (#sons/#eggs) in the analysis of prezygotic reproductive isolation: lack of egg fertilization in interpopulation crosses leads to higher ratio of males compared to the control cross. We tested the effects of genetic distance, female populations and the interaction on the ratio of male offspring to egg. The effect of genetic distance on the ratio of male offspring to egg was different among female populations (quasibinomial GLM; Genetic distance × Female population, F6,35 = 7.025, P < 0.001; Fig. 4a). Therefore, we reanalyzed the effect of genetic distance in each population separately (Fig. 4a; Additional file 2: Table S2). Genetic distance significantly affected the ratio of male offspring to egg in the populations from Korea (K; F1,5 = 45.048, P < 0.01), Inner Mongolia (CIM; F1,5 = 42.190, P < 0.01) and Eastern China (CN; F1,5 = 35.610, P < 0.01), however, not in the populations from Iran (I; F1,5 = 1.169, P = 0.329), Turkey (T; F1,5 = 0.232, P = 0.650), France (F; F1,5 = 0.0286, P = 0.872), and Japan (J; F1,5 = 0.523, P = 0.502).
Relationship of prezygotic isolation (a), postzygotic isolation due to female hybrid inviability (b) and total reproductive isolation (sum of prezygotic and female hybrid inviability) (c) with genetic distance. For evaluation of each reproductive isolation, see text. Colored lines show the model prediction of the relationship between reproductive barrier and genetic distance in each female population (Additional file 2: Table S2), where the data is analyzed with genetic distance, female populations and the interaction. When genetic distance did not have a significant effect on reproductive barrier in each female population, the line is absent. Black bold lines show the model predictions where the models were reconstructed only with genetic distance (without consideration of female population differences) to estimate the genetic distance for which reproductive isolation is nearly complete (Table 3). For the abbreviation of population names, see Table 1
Postzygotic isolation—hybrid female inviability
Postzygotic isolation due to genetic incompatibilities in the hybrid diploid genome (after egg fertilization, i.e., hybrid female inviability) leads to higher mortality among (diploid) offspring. In the analysis of postzygotic reproductive isolation due to hybrid female inviability, we used offspring mortality among diploid offspring as [(#unhatched-eggs + #dead juveniles)/(#eggs − #sons)], because mortality among unfertilized, male offspring was negligibly low (Additional file 1: Table S1). The diploid offspring mortality varied among combinations of crosses (Fig. 4b). In the statistical model, the interaction between genetic distance and population was not significant (quasibinomial GLM; F6,35 = 0.671, P = 0.674), therefore, the interaction term was removed from the model. Genetic distance significantly affected the diploid offspring mortality (F1,47 = 30.031 P < 0.001; Fig. 4b), and there was also significant variation among populations (F6,41 = 2.460, P < 0.05; Fig. 4b).
Total reproductive isolation in parent-population crosses
In the analysis of total reproductive isolation in parent population crosses, we used the sum of these two values by values obtained by subtracting viable diploid offspring ratio from [1 − (#daughter/#eggs)]. [1 − (#daughter/#eggs)] in parent-population crosses varied among combinations of crosses (Fig. 4c). The interaction between genetic distance and population in the statistical model was not significant (quasibinomial GLM; F6,35 = 1.367, P = 0.255), therefore, the interaction term was removed from the model. Genetic distance significantly affected [1—(#daughter/#eggs)] (F1,47 = 33.888, P < 0.001; Fig. 3c), and there was also significant variation among populations (F6,41 = 3.006, P < 0.05; Fig. 4c).
Postzygotic barrier—hybrid female sterility
In the analysis of cumulative hybrid female sterility, we used the fraction of dead offspring in the backcrosses: [(#unhatched-eggs + #dead juveniles)/#eggs] (note this is a slight overestimation due to background mortality in the experiment as evident in the control crosses, this background mortality is typically < 5% and comparable among the populations, see Additional file 1: Table S1). F1 hybrid females from crosses of Iran (I) × Turkey (T), Iran (I) × France (F), Japan (J) × Inner Mongolia (CIM, China) did not produce viable sons at all (Fig. 4; Additional file 1: Table S1). Viable sons were produced only from F1 hybrid females from crosses among Inner Mongolia (CIM, China), Eastern China (CN) and Korea (K), for which genetic distances were approximately zero. Genetic distance in parental populations has a significant effect on fraction of dead offspring in the backcrosses (quasibinomial GLM; F1,37 = 127.381, P < 0.001). As the genetic distance increased, so fraction of dead offspring in the backcrosses increased (Fig. 5).
Relationship between degree of postzygotic isolation due to female hybrid sterility, based on the backcrosses, and genetic distance. The bold line shows the model prediction for all 16 types of hybrid female together (Table 3). Note that there are 8 estimates of female hybrid sterility from backcrosses between populations with zero genetic distance (CIM, CN, and K)
Genetic distance for which reproductive isolation is nearly complete
Based on the GLMs (Table 3), prezygotic isolation, hybrid female inviability and total reproductive isolation in the parental population crosses (sum of prezygotic isolation and hybrid female inviability) were estimated to be nearly complete (90.0–99.9% barrier) at genetic distances of 0.475–0.657, 0.150–0.209 and 0.145–0.210, respectively. Hybrid female sterility was estimated to be nearly complete at a genetic distance of 0.108–0.151 (90.0–99.9% barrier), suggesting that hybrid female sterility evolves earlier than other stages of reproductive barrier.
Discussion
In this study, we investigated the relationships of prezygotic reproductive isolation, hybrid female inviability and hybrid female sterility with genetic distance by using seven populations of a haplodiploid spider mite, A. viennensis. We found that intensities of reproductive isolation vary among cross combinations, and the intensity has a positive relationship with genetic distance in all stages of reproductive isolation, as reported in other taxa [28,29,30,31,32,33,34,35,36,37,38] including a haplodiploid spider mite, S. miscanthi species group [9].
Total reproductive isolation in parent population crosses (sum of prezygotic reproductive isolation and hybrid female inviability) was nearly complete at a genetic distance of 0.145–0.210 (99.0–99.9% reproductive isolation) based on an mtDNA genetic marker (COI). This is similar to that in the haplodiploid spider mite, S. miscanthi species group (0.152–0.210) [9]. Hence, this genetic distance may be an estimate of the minimum genetic distance in mtDNA required for speciation in spider mites. COI is an important DNA barcoding tool for molecular identification of species in animals including spider mites [41,42,43]. The minimum genetic distance in COI required for speciation in spider mites is possibly useful as the threshold value of identification in spider mites. Besides, the minimum genetic distance may be useful to estimate reproductive isolation status between closely related species and to find presence of cryptic species in spider mites.
Prezygotic reproductive isolation seemed to evolve much slower than hybrid female inviability in A. viennensis (genetic distance for 99.0–99.9% isolation: 0.457–0.657 in prezygotic isolation and 0.150–0.209 in hybrid female inviability), and also than prezygotic isolation in the S. miscanthi species group (genetic distance for 99.0–99.9% isolation: 0.190–0.258) [9]. These differences were possibly caused by geographic relationships of populations used in the cross experiments. If the diverging groups contact frequently, the prezygotic reproductive barrier can be reinforced by natural selection to prevent maladaptive hybridization [1, 2]. The geographic distribution of A. viennensis populations used in this study is completely allopatric whereas populations of S. miscanthi species group used in [9] are parapatric, caused by secondary contact, and semi-allopatric. Besides, the geographic scale is also different between this and the previous studies: A. viennensis was collected from the Palaearctic region ranging from France to Japan, whereas S. miscanthi species group was collected from Japan and surrounding countries [9]. Given that A. viennensis populations experience more isolation-by-distance, reinforcement of prezygotic reproductive isolation would possibly not occur in A. viennensis, resulting in the pattern of slow evolution of prezygotic reproductive isolation. However, the isolation-by-distance might also have caused higher genetic diversity among populations in A. viennensis, which might have promoted genetic incompatibility, and therefore a stronger postzygotic reproductive barrier.
The reproductive isolation pattern including the strength of prezygotic reproductive barrier showed two patterns among female populations in A. viennensis. In the crosses with females from Inner Mongolia (CIM), Eastern China (CN), and Korea (K), prezygotic reproductive barrier was found and stronger as the genetic distance was larger. However, in the crosses with females from France (F), Turkey (T), Iran (I) and Japan (J), the relationship between prezygotic barrier and genetic distance was not clear, but viability of F1 hybrids was lower as genetic distance was larger. The presence of these two patterns of reproductive isolation in A. viennensis can be useful to investigate whether prezygotic reproductive isolation in spider mites is often associated with reinforcement or whether other mechanisms affect prezygotic reproductive isolation. It can be thought that Inner Mongolia (CIM), Eastern China (CN) and Korea (K) are in geographical locations where contact with other genetically different groups frequently occur. Otherwise, as we mentioned previously, A. viennensis is thought to originate from Eastern Asia [40], although the molecular phylogenetic tree and the bootstrap values in this study are not sufficient to confirm this (Fig. 1). It can be thought that whether a population is ancestral or derived affects the frequency of contact with other genetically different populations or directly affects the reproductive isolation patterns in spider mites. Prezygotic reproductive isolation is often found between conspecific strains and between closely related species in spider mites [18, 22, 44,45,46,47]. For understanding the speciation pattern in spider mites, and also in haplodiploids in general, it would be important to figure out the evolutionary mechanisms of prezygotic reproductive isolation, for which further studies are necessary.
In this study, we found a positive relationship between reproductive isolation and genetic distance, showing that accumulation of genetic changes caused by natural selection and genetic drift is important in the evolution of reproductive isolation in the haplodiploid spider mite A. viennensis. Yet, we often found asymmetries in reproductive isolation in this study. For example, hybrid females were partly produced from Iran (female) × France (male), but not at all from France (female) × Iran (male) (Additional file 1: Table S1). Hybrid females from Turkey (female) × Iran (male) produced viable female offspring, but those from Iran (female) × Turkey (male) produced no viable offspring (Additional file 1: Table S1). Endosymbiont infection was not detected in the populations used in this study, although we could not reject the possibility that currently unknown endosymbionts, i.e. not Wolbachia, Cardinium, Spiroplasma or Rickettsia, infest and control the reproduction of A. viennensis. Therefore, the asymmetries are likely to be caused by cytonuclear interaction, for example, by a negative interaction between mitochondrial genes and nuclear genes [10, 48]. Similar asymmetries in reproductive isolation, not apparently caused by endosymbiont infections, were also found in the spider mite S. miscanthi species group [9]. In addition, cytoplasmic interaction was detected in another haplodiploid spider mite, Tetranychus evansi [10]. The importance of cytonuclear interactions in postzygotic reproductive isolation was pointed out in studies on a haplodiploid wasp [13, 49]. Cytonuclear interactions possibly have an important role, and not only the view point of nuclear-nuclear interactions, but also the view point of cytonuclear interactions is necessary for understanding evolutionary mechanisms of reproductive isolation in haplodiploid animals.
Conclusions
In this study, we focus on a haplodiploid spider mite and investigated the evolutionary mechanism of reproductive isolation among populations within the species A. viennensis. We found that the degree of reproductive isolation varies among cross combinations in two distinct patterns, and that the reproductive isolation has a positive relationship with genetic distance based on COI in any stage of isolation (prezygotic, postzygotic-hybrid inviability and postzygotic-hybrid fertility). We estimated the genetic distances which complete the reproductive barrier, and discussed about the importance of reinforcement of prezygotic reproductive barrier and the importance of cytonuclear interaction in haplodiploid animals. The findings are significant for understanding evolutionary patterns of reproductive isolation in haplodiploid animals.
Methods
Mites and host plants
A list of populations used in the present study is provided in Table 1. To establish laboratory populations, the collected mites were reared on leaves of Yoshino cherry, Prunus × yedoensis Matsum., which were placed on a water-soaked sponge in Petri dishes (9 cm in diameter) under constant climatic conditions (25 ± 1℃, 60–70% relative humidity and16:8 h light:dark photoperiod). We placed the leaves underside up and the perimeter was covered with water-soaked tissue paper. In winter, these populations were kept as diapause females, which were reared from eggs under 15 ± 1 °C and 8:16 light:dark photoperiod. Diapause females were put onto black paper and set into glass vials, which were kept in a refrigerator (ca. 5 °C) for five months from December to April under darkness. We collected Tetranychus kanzawai Kishida from Japan (city: Miyakojima, latitude-longitude: 24°45ʹN–125°23ʹE, host plant: Benincasa hispida (Thunb.) Cogn., date: January 31, 2008), and used it as the outgroup of phylogenetic analysis of A. viennensis.
DNA preparation and sequencing
In measures of genetic distance among populations and the phylogenetic analyses, we used the cytochrome c oxidase subunit I gene (COI) of mitochondrial DNA (mtDNA). DNA was extracted from a single female mite from each population by using PrepMan Ultra Sample Preparation Reagent (Thermo Fisher Scientific Inc.). To amplify the fragment of mtCOI region, PCR was carried out using primers given in Additional file 3: Table S3 [50, 51] in a 36 µl reaction mixture containing 0.5 ul of DNA sample, 3.6 µl of 10 × Ex Taq buffer (20 mM mg2 + plus, Takara Bio Inc.), 0.14 µl of TaKaRa Ex Taq (5U/µl, Takara Bio Inc.), 2.88 µl of dNTP mix (2.5 mM each, Takara Bio Inc.), 0.72 µl of each primer (10 pmol/ul each) and 27.44 µl of ddH2O. PCR cycling conditions were 3 min at 94 °C, followed by 35 cycles of 1 min at 94 °C, 1 min at 51 °C and 1.5 min at 72 °C, and a final extension at 72 °C for 10 min. In some samples, the fragment was not amplified. Therefore, in these samples, we carried out PCR by decreasing the annealing temperature or increasing the number of cycles. PCR products were purified using MinElute PCR Purification Kit (QIAGEN). The purified products were sequenced using ABI BigDye Terminator ver. 3 Cycle Sequencing Kit (Applied Biosystems) and ABI3130xlGenetic Analyzer (Applied Biosystems).
Phylogenetic analyses and genetic distance measurements
Obtained sequences of the COI (618 bp, GenBank accession numbers: LC579422-LC579429) for A. viennensis and T. kanzawai and the COI sequences for A. quercivorus from previously published data (accession numbers: AB981238 and LC435686) [52] were aligned using CLUSTAL W in MEGA X [50]. A maximum likelihood (ML) tree of the aligned COI sequences was constructed with MEGA X [53]. As the substitution model for the ML tree, we used the Tamura 3-parameter model in which non-uniformity of evolutionary rates among sites is modeled by using a discrete Gamma distribution, because the model performed better than other models according to the Bayesian Information Criteria (BICs) in ML fits of 24 different nucleotide substitution models. Reliability of trees was evaluated by the bootstrap test (N = 1000).
Kimura 2-parameter genetic distances [54] were calculated among the populations using MEGA X [53]. We did not correct the data for their phylogenetic independence in the genetic distances as other papers did [28, 29, 38], since we also focus on asymmetries in reproductive incompatibility among the reciprocal combinations.
Endosymbiont infections
To detect the presence of Wolbachia, Cardinium, Spiroplasma and Rickettsia in the mites, we carried out PCR assay for these endosymbionts using primers given in Additional file 3: Table S3 [55,56,57,58,59]. DNA was extracted from five female mites from each population, by homogenizing them in a 1.5 ml microtube with 18 µl of STE buffer (100 mM NaCl, 10 mM Tris–HCl, 1mH EDTA, pH 8.0) and 2 µl proteinase K, and incubating them at 55℃ for 30 min and 95℃ for 3 min. PCR was carried out in a 20 µl reaction mixture containing 1.0 µl of DNA sample, 2 µl of 10 × NH4 Reaction buffer (Nippon Genetics Co., Ltd), 1 µl of 50 mM MgCl2 Solution, 0.2 µl of BIOTAQ DNA Polymerase (5 U/µl, Nippon Genetics Co., Ltd), 0.4 µl of dNTP mix (10 mM each), 1 µl of each primer (10 pmol/µl each) and 13.4 µl of ddH2O. PCR cycling conditions were 3 min at 95 °C, followed by 36 cycles of 30 s at 95 °C, 30 s at 52 °C and 30 s at 72 °C, and a final extension at 72 °C for 5 min. To be sure, we carried out this check for endosymbiont infections twice by using the same DNA template from each population (i.e., two technical replicates per DNA sample). We used DNA of Wolbachia-infected Panonychus mori Yokoyama (Toyama, voucher specimen no. 665), Cardinium-infected Tetranychus urticae (Koch) (red form, Nagano, no. 171), Spiroplasma-infected Tetranychus truncatus Ehara (Inner Mongolia, no. 199) and Rickettsia-infected Nephotettrix cincticeps (Uhler) [60] as positive controls of Wolbachia, Cardinium, Spiroplasma and Rickettsia infection, respectively. Distilled water was used as negative control.
Cross experiments
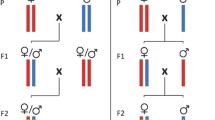
We carried out cross experiment among the seven populations of A. viennensis in all combinations and both directions (42 combinations). As controls, intra-population crosses were carried out in each population (7 controls). The number of replicates in each cross combination ranged between 12 and 30 (Additional file 1: Table S1). We also carried out 16 different backcrosses by using F1 hybrids obtained from the following cross experiments: F (female) × T (male), F (female) × I (male), F (female) × J (male), T (female) × F (male), T (female) × I (male), I (female) × F (male), I (female) × T (male), CIM (female) × CN (male), CIM (female) × K (male), CN (female) × CIM (male), CN (female) × K (male), K (female) × CIM (male), K (female) × CN (male), J (female) × F (male), J (female) × CIM (male) and J (female) × CN (male). In the other 26 crosses we obtained few hybrid females, therefore we did not carry out backcrosses with those. The number of replicates in each backcross combination was 10 to 32 (Additional file 1: Table S1).
The cross experiments were carried out under the same conditions as the mite rearings. A leaf of Yoshino cherry was placed onto wet sponge in a Petri dish (9 cm in diameter) in the mite rearing. To make the area unified, a square (4 × 4 cm) was created by using strings of water-soaked tissue paper. A female in the last molt before adulthood (teleiochrysalis stage) and an adult male were collected from the mite culture, and placed on the leaf arena. The female and male were allowed to mate and oviposit for five days, then they were removed from the leaf arena. The number of eggs, the offspring survival and the gender of offspring were checked and recorded.
Statistical analyses
Analyses were carried out with the statistical package R version 3.6.2 [61]. We analyzed the ratio of male offspring to egg (#sons/#eggs), offspring mortality among diploid offspring [(#unhatched-eggs + #dead juveniles) / (#eggs − #sons)] and the sum of these two values by values obtained by subtracting viable diploid offspring ratio from [1 − (#daughter / #eggs)] by using generalized linear models (GLMs) with genetic distance, female population and their interaction on these three variables. We applied a quasibinomial distribution as the error distribution to account for overdispersion. For models where the interaction did not have a significant effect, we reanalyzed the effects of genetic distance and female population by removing the interaction term. For models where the interaction did have a significant effect, we reanalyzed the effect of genetic distance in each female population separately. We analyzed the relationship between the fraction of dead offspring in the backcrosses and genetic distance between parent populations by using a GLM. We applied a quasibinomial distribution as the error distribution to account for overdispersion. In the analysis, we included intra-population crosses as the controls. To estimate genetic distance for which reproductive barrier is nearly complete, we reconstructed the quasibinomial GLMs only with genetic distance, and calculated the genetic distances for 99.0% and 99.9% reproductive barriers complete by using the models. In these analyses, we used the R packages stats and MASS [61, 62].
Availability of data and materials
Most data generated or analyzed during this study are included in this published article and its supplementary information files. The sequence data used in the analysis are available in GeneBank (accession numbers: AB981238 and LC435686).
Abbreviations
- F:
-
France
- T:
-
Turkey
- I:
-
Iran
- CIM:
-
Inner Mongolia, China
- CN:
-
Eastern China
- K:
-
Korea
- J:
-
Japan
- ML:
-
Maximum likelihood
References
Howard DJ, Berlocher SH, editors. Endless forms: species and speciation. New York: Oxford University Press; 1998.
Coyne JA, Orr HA. Speciation. 1st ed. Sunderland, Mass: Sinauer Associates, Inc.; 2004.
de la Filia AG, Bain SA, Ross L. Haplodiploidy and the reproductive ecology of Arthropods. Curr Opin Insect Sci. 2015;9:36–43.
Price CSC, Kim CH, Gronlund CJ, Coyne JA. Cryptic reproductive isolation in the Drosophila simulans species complex. Evolution. 2001;55:81–92.
Nosil P, Crespi BJ. Ecological divergence promotes the evolution of cryptic reproductive isolation. Proc R Soc B Biol Sci. 2006;273:991–7.
Sato Y, Saito Y, Mori K. Reproductive isolation between populations showing different aggression in a subsocial spider mite, Schizotetranychus miscanthi Saito (Acari: Tetranychidae). Appl Entomol Zool. 2000;35:605–10.
Sato Y, Saito Y, Mori K. Patterns of reproductive isolation between two groups of Schizotetranychus miscanthi Saito (Acari: Tetranychidae) showing different male aggression traits. Appl Entomol Zool. 2000;35:611–8.
Sato Y, Breeuwer JAJ, Egas M, Sabelis MW. Incomplete premating and postmating reproductive barriers between two parapatric populations of a social spider mite. Exp Appl Acarol. 2015;65:277–91.
Sato Y, Sakamoto H, Gotoh T, Saito Y, Chao J-T, Egas M, et al. Patterns of reproductive isolation in a haplodiploid—strong postmating, prezygotic barriers among three forms of a social spider mite. J Evol Biol. 2018. https://doi.org/10.1111/jeb.13270.
Knegt B, Potter T, Pearson NA, Sato Y, Staudacher H, Schimmel BCJ, et al. Detection of genetic incompatibilities in non-model systems using simple genetic markers: hybrid breakdown in the haplodiploid spider mite Tetranychus evansi. Heredity. 2017;118:311–21.
Nouhaud P, Blanckaert A, Bank C, Kulmuni J. Understanding admixture: haplodiploidy to the rescue. Trends Ecol Evol. 2020;35:34–42.
Koevoets T, Beukeboom LW. Genetics of postzygotic isolation and Haldane’s rule in haplodiploids. Heredity. 2009;102:16–23.
Beukeboom LW, Koevoets T, Morales HE, Ferber S, van de Zande L. Hybrid incompatibilities are affected by dominance and dosage in the haplodiploid wasp Nasonia. Front Genet. 2015. https://doi.org/10.3389/fgene.2015.00140.
Lohse K, Ross L. What haplodiploids can teach us about hybridization and speciation. Mol Ecol. 2015;24:5075–7.
Patten MM, Carioscia SA, Linnen CR. Biased introgression of mitochondrial and nuclear genes: a comparison of diploid and haplodiploid systems. Mol Ecol. 2015;24:5200–10.
Helle W, Sabelis MW. Spider mites: their biology, natural enemies and control, vol. 1A. Amsterdam: Elsevier; 1985.
Fujimoto H, Hiramatsu T, Takafuji A. Reproductive interference between Panonychus mori Yokoyama and P. citri (McGREGOR) (Acari: Tetranychidae) in peach orchards. Appl Entomol Zool. 1996;31:59–65.
Sato Y, Alba JM, Sabelis MW. Testing for reproductive interference in the population dynamics of two congeneric species of herbivorous mites. Heredity. 2014;113:495–502.
Chae Y, Yokoyama N, Ito K, Fukuda T, Arakawa R, Zhang Y-X, et al. Reproductive isolation between Stigmaeopsis celarius and its sibling species sympatrically inhabiting bamboo (Pleioblastus spp.) plants. Exp Appl Acarol. 2015;66:11–23.
Gomi K, Gotoh T. Host plant preference and genetic compatibility of the kanzawa spider mite, Tetranychus kanzawai KISHIDA (Acari: Tetranychidae). Appl Entomol Zool. 1996;31:417–25.
Navajas M, Gutierrez J, Lagnel J, Fauvel G, Gotoh T. DNA sequences and cross-breeding experiments in the hawthorn spider mite Amphitetranychus viennensis reveal high genetic differentiation between Japanese and French populations. Entomol Exp Appl. 1999;90:113–22.
Navajas M, Tsagkarakov A, Lagnel J, Perrot-Minnot M-J. Genetic differentiation in Tetranychus urticae (Acari: Tetranychidae): polymorphism, host races or sibling species? Exp Appl Acarol. 2000;24:365–76.
Uesugi R, Goka K, Osakabe M. Development of genetic differentiation and postzygotic isolation in experimental metapopulations of spider mites. Exp Appl Acarol. 2003;31:161.
Gotoh T, Noda H, Fujita T, Iwadate K, Higo Y, Saito S, et al. Wolbachia and nuclear–nuclear interactions contribute to reproductive incompatibility in the spider mite Panonychus mori (Acari: Tetranychidae). Heredity. 2005;94:237–46.
Gotoh T, Noda H, Ito S. Cardinium symbionts cause cytoplasmic incompatibility in spider mites. Heredity. 2007;98:13–20.
Ros VID, Breeuwer JAJ. The effects of, and interactions between, Cardinium and Wolbachia in the doubly infected spider mite Bryobia sarothamni. Heredity. 2009;102:413–22.
Zhu L-Y, Zhang K-J, Zhang Y-K, Ge C, Gotoh T, Hong X-Y. Wolbachia strengthens Cardinium-induced cytoplasmic incompatibility in the spider mite Tetranychus piercei McGregor. Curr Microbiol. 2012;65:516–23.
Coyne JA, Orr HA. Patterns of speciation in Drosophila. Evolution. 1989;43:362–81.
Coyne JA, Orr HA. Patterns of speciation in Drosophila revisited. Evolution. 1997;51:295–303.
Sasa MM, Chippindale PT, Johnson NA. Patterns of postzygotic isolation in frogs. Evolution. 1998;52:1811–20.
Presgraves DC. Patterns of postzygotic isolation in Lepidoptera. Evolution. 2002;56:1168–83.
Price TD, Bouvier MM. The evolution of F1 postzygotic incompatibilities in birds. Evolution. 2002;56:2083–9.
Moyle LC, Olson MS, Tiffin P. Patterns of reproductive isolation in three angiosperm genera. Evolution. 2004;58:1195–208.
Scopece G, Musacchio A, Widmer A, Cozzolino S. Patterns of reproductive isolation in mediterranean deceptive orchids. Evolution. 2007;61:2623–42.
Malone JH, Fontenot BE. Patterns of reproductive isolation in toads. PLoS ONE. 2008;3: e3900.
Yukilevich R. Asymmetrical patterns of speciation uniquely support reinforcement in Drosophila. Evolution. 2012;66:1430–46.
Turelli M, Lipkowitz JR, Brandvain Y. On the Coyne and Orr-igin of species: effects of intrinsic postzygotic isolation, ecological differentiation, X chromosome size, and sympatry on Drosophila speciation. Evolution. 2014;68:1176–87.
Sánchez-Guillén RA, Córdoba-Aguilar A, Cordero-Rivera A, Wellenreuther M. Genetic divergence predicts reproductive isolation in damselflies. J Evol Biol. 2014;27:76–87.
Ehara S, Gotoh T. A new Tetranychus closely related to 7. viennensis Zacher (Acari: Tetranychidae). Int J Acarol. 1990;16:55–8.
Bolland HR. World catalogue of the spider mite family: Leiden. Boston: Brill; 1998.
Hebert PDN, Cywinska A, Ball SL, de Waard JR. Biological identifications through DNA barcodes. Proc R Soc B. 2003;270:313–21.
Hinomoto N, Tran DP, Pham AT, Le Ngoc TB, Tajima R, Ohashi K, Osakabe M, Takafuji A. Identification of spider mites (Acari: Tetranychidae) by DNA sequences: a case study in Northern Vietnam. Int J Acarol. 2007;33:53–60.
Matsuda T, Hinomoto N, Singh RN, Gotoh T. Molecular-based identification and phylogeny of Oligonychus species (Acari: Tetranychidae). J Econ Entomol. 2012;105:1043–50.
Takafuji A, Fujimoto H. Reproductive compatibility between populations of the citrus red mite, Panonychus citri (McGregor) (Acarina: Tetranychidae). Res Popul Ecol. 1985;27:361–72.
Gotoh T, Takayama K. Developmental characteristics, genetic compatibility and esterase zymograms in three strains of the hawthorn spider mite, Tetranychus viennensis Zacher (Acari: Tetranychidae). J Acarol Soc Jpn. 1992;1:45–60.
Gotoh T, Abe T, Kurihara A, Suzuki M. Genetic incompatibility in local populations of the spider mite, Tetranychus quercivorus Ehara et Gotoh (Acari: Tetranychidae). Appl Entomol Zool. 1995;30:361–8.
Perrot-Minnot MJ, Migeon A, Navajas M. Intergenomic interactions affect female reproduction: evidence from introgression and inbreeding depression in a haplodiploid mite. Heredity. 2004;93:551–528.
Hill GE. Mitonuclear ecology. Mol Biol Evol. 2015;32:1917–27.
Niehuis O, Judson AK, Gadau J. Cytonuclear genic incompatibilities cause increased mortality in male F2 hybrids of Nasonia giraulti and N. vitripennis. Genetics. 2008;178:413–26.
Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am. 1994;87:651–701.
Gotoh T, Araki R, Boubou A, Migeon A, Ferragut F, Navajas M. Evidence of co-specificity between Tetranychus evansi and Tetranychus takafujii (Acari: Prostigmata, Tetranychidae): comments on taxonomic and agricultural aspects. Int J Acarol. 2009;35:485–501.
Arabuli T, Negm MW, Matsuda T, Kitashima Y, Abramishvili T, Akimov IA, et al. Morphological identification of Amphitetranychus species (Acari: Tetranychidae) with crossbreeding, esterase zymograms and DNA barcode data. PLoS ONE. 2019;14: e0221951.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9.
Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20.
Webb L, Carl M, Malloy DC, Dasch GA, Azad AF. Detection of murine typhus infection in fleas by using the polymerase chain reaction. J Clin Microbiol. 1990;28:530–4.
Jeyaprakash A, Hoy MA. Long PCR improves Wolbachia DNA amplification: wsp sequences found in 76% of sixty-three arthropod species. Insect Mol Biol. 2000;9:393–405.
Tsuchida T, Koga R, Shibao H, Matsumoto T, Fukatsu T. Diversity and geographic distribution of secondary endosymbiotic bacteria in natural populations of the pea aphid Acyrthosiphon pisum. Mol Ecol. 2002;11:2123–35.
Morimoto S, Kurtti TJ, Noda H. In vitro cultivation and antibiotic susceptibility of a cytophaga-like intracellular symbiote isolated from the tick Ixodes scapularis. Curr Microbiol. 2006;52:324–9.
Nakamura Y, Kawai S, Yukuhiro F, Ito S, Gotoh T, Kisimoto R, et al. Prevalence of Cardinium bacteria in planthoppers and spider mites and taxonomic revision of “Candidatus Cardinium hertigii” based on detection of a new Cardinium group from biting midges. Appl Environ Microbiol. 2009;75:6757–63.
Noda H, Watanabe K, Kawai S, Yukuhiro F, Miyoshi T, Tomizawa M, et al. Bacteriome-associated endosymbionts of the green rice leafhopper Nephotettix cincticeps (Hemiptera: Cicadellidae). Appl Entomol Zool. 2012;47:217–25.
R Core Team. R: a language and environment for statistical computing. Vienna: Foundation for Statistical Computing; 2020.
Venables WN, Ripley BD. Modern applied statistics with S. 4th ed. New York: Springer-Verlag; 2002. https://doi.org/10.1007/978-0-387-21706-2.
Acknowledgements
We thank Drs. M. Kawashima (Andong National University), Z.-Q. Li (Inner Monglia Academy of Agricultural Sciences), X.-Y. Hong (Nanjing Agricultural University), A. Saboori (University of Tehran), I. Kasap (Çanakkale Onsekiz Mart University) and M. Navajas (INRA) for providing samples, and Ms. N. Nishizawa, Ms. C. Fukumoto and Mr. C. Yoshioka (Ibaraki University) for supporting DNA experiments.
Funding
The work was supported in part by JSPS KAKENHI Grant number 17H03775 (Grant-in-Aid for Scientific Research B to TG).
Author information
Authors and Affiliations
Contributions
SF and TG carried out the experiments, TG secured funding, TM analyzed phylogenetic data and deposited DNA data, YS analyzed data of cross experiments and phylogenetic data, YS and ME wrote the manuscript, and all authors contributed to revision of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Spider mites used in this study (Table 1) were collected by cooperation of Drs. M. Kawashima in Andong National Univerity, Z.-Q. Li in Inner Mongolia Academy of Agricultural Sciences, X.-Y. Hong in Nanjing Agricultural University, A. Saboori in University of Tehran, I. Kasap in Çanakkale Onsekiz Mart University and M. Navajas in National Institute of Agricultural Research (INRA, France). The mites were imported to Japan with the authorization of the Ministry of Agriculture, Forestry and Fisheries of Japan.
Consent for publication
Not applicable.
Competing interest
The authors have no conflict of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Number of eggs laid during the first five days of the oviposition period, hatchability, survival rate of immature stages, female ratio of offspring and female offspring per ovipositing female in crosses.
Additional file 2: Table S2.
Quasibinomial generalized linear models used in the lines in each female population in Fig. 4a.
Additional file 3: Table S3.
Primers used in genetic analyses of Amphitetranychus viennensis and in PCR assay for endosymbiont infection.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Sato, Y., Fujiwara, S., Egas, M. et al. Patterns of reproductive isolation in a haplodiploid mite, Amphitetranychus viennensis: prezygotic isolation, hybrid inviability and hybrid sterility. BMC Ecol Evo 21, 177 (2021). https://doi.org/10.1186/s12862-021-01896-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12862-021-01896-5