Abstract
Background
Many plant pathogen secretory proteins are known to be elicitors or pathogenic factors,which play an important role in the host-pathogen interaction process. Bioinformatics approaches make possible the large scale prediction and analysis of secretory proteins from the Puccinia helianthi transcriptome. The internet-based software SignalP v4.1, TargetP v1.01, Big-PI predictor, TMHMM v2.0 and ProtComp v9.0 were utilized to predict the signal peptides and the signal peptide-dependent secreted proteins among the 35,286 ORFs of the P. helianthi transcriptome.
Results
908 ORFs (accounting for 2.6% of the total proteins) were identified as putative secretory proteins containing signal peptides. The length of the majority of proteins ranged from 51 to 300 amino acids (aa), while the signal peptides were from 18 to 20 aa long. Signal peptidase I (SpI) cleavage sites were found in 463 of these putative secretory signal peptides. 55 proteins contained the lipoprotein signal peptide recognition site of signal peptidase II (SpII). Out of 908 secretory proteins, 581 (63.8%) have functions related to signal recognition and transduction, metabolism, transport and catabolism. Additionally, 143 putative secretory proteins were categorized into 27 functional groups based on Gene Ontology terms, including 14 groups in biological process, seven in cellular component, and six in molecular function. Gene ontology analysis of the secretory proteins revealed an enrichment of hydrolase activity. Pathway associations were established for 82 (9.0%) secretory proteins. A number of cell wall degrading enzymes and three homologous proteins specific to Phytophthora sojae effectors were also identified, which may be involved in the pathogenicity of the sunflower rust pathogen.
Conclusions
This investigation proposes a new approach for identifying elicitors and pathogenic factors. The eventual identification and characterization of 908 extracellularly secreted proteins will advance our understanding of the molecular mechanisms of interactions between sunflower and rust pathogen and will enhance our ability to intervene in disease states.
Similar content being viewed by others
Background
Sunflower rust, caused by Puccinia helianthi Schw., is a widespread disease of sunflower (Helianthus annuus L.) throughout the world and may cause significant yield losses and loss of seed quality. P. helianthi is an obligate pathogen and completes its life cycle on sunflower. Although P. helianthi is a pathogen of great economic importance, little is known about the molecular mechanisms involved in its pathogenicity and host specificity.
Pathogen secretory proteins and host plant defense interactions involve complex signal exchanges at the plant surface and at the interface between the pathogen and the host [1, 2]. Plant pathogens are endowed with a special ability to interfere with physiological, biochemical, and morphological processes of the host plants through a diverse array of extracellular effectors. These are present or active at the intercellular interface or delivered inside the host cell to reach their cellular target and facilitate infection or trigger defense responses [3–5]. Thus, genes encoding extracellular proteins have a higher probability of being involved in virulence.
Many Avr genes encoding secreted proteins were identified from haustoria-forming pathogens, such as AvrL567, AvrM, AvrP4, and AvrP123 in flax rust caused by Melampsora lini [6, 7], AvrPi-ta and AvrPiz-t in rice blast Magnaporthe grisea [8, 9], Avr1b-1 in stem and root rot of soybean Phytophthora sojae [10], Avr3a in potato late blight P. infestans [11], and ATR13 and ATR1 in downy mildew of Arabidopsis caused by Hyaloperonospora parasitica [12, 13]; all of which exhibit pathogenic functions during pathogen infection. In addition, some cell wall degrading enzymes (CWDEs) produced by pathogens are secretory proteins, such as the wood Xylanase Xyn22 and Xyn33 of M. grisea [14], and pectinlyase Pmr6 of Erysiphe cichoracearum [15]. Some virulence-related proteins, such as Gas1 and Gas2 (expressed specifically at the appressorium formation stage) [16], hydrophobic protein Mpg1 [17], tetraspanin-like protein Pls1 [18] and chitin binding protein Cbp1 of rice blast [19] are in the same category.
Amino terminal signal peptides are responsible for transporting the virulent factors [20]. The N-terminal signal peptides can be classified into four types based on recognition sequences of signal peptidases. The first class is composed of “typical” signal peptides, which are cleaved by one of the various type I SPases of Bacillus subtilis [21–23] and most secretory proteins with this signal peptide are secreted into the extracellular environment. This group also includes signal peptides with a so-called twin-arginine motif (RR-motif) that are transported via the twin-arginine translocation pathway (Tat pathway). In bacteria, the Tat translocase is found in the cytoplasmic membrane and exports proteins to the cell envelope or to the extracellular space [24]. The second class of signal peptides are lipoproteins cleaved by the lipoprotein-specific (type II) SPase of B. subtilis (Lsp) [25, 26]. Secretory proteins with the aforementioned signal peptides are transported via the general secretion pathway (Sec-pathway) [27]. The third class constitutes prepilin-like proteins cleaved by the prepilin-specific SPase ComC and the fourth class of signal peptides consists of ribosomally synthesized bacteriocin and pheromone [28, 29]. These signal peptides lack a hydrophobic H-domain and they can be removed from the mature protein by a subunit of the ABC transporter or by specific SPases.
With the development of molecular biology, large scale genome and transcriptome sequencing has been used as an effective method for discovering gene expression profiles and novel genes. Several computer-based prediction algorithms have been used to predict the secretomes from many microbial species, such as Candida albicans [30], P. infestans [31, 32], Saccaromyces cerevisiae [33], Agrobacterium tumefaciens [34], Fusarium graminearum [35], Neurospora crassa [36], Verticillium dahliae [37], Aspergillus oryzae [38], Puccinia striiformis f. sp. tritici [39], and Colletotrichum graminicola [40]. These predicted secretomes provide a basis for further investigations using wet-lab procedures or more in-depth computational comparisons of relevant data sets.
An examination of the pathogenesis-related secretome of P. helianthi is important for understanding the molecular mechanism of pathogen-host interaction. Here, we generated a high-throughput transcriptome analysis of proteins containing a signal peptide. We analyzed a total of 35,286 ORFs of the P. helianthi transcriptome using SignalP v4.1, TMHMM v2.0, TargetP v1.1, TatP v1.0 and big-PI predictor bioinformatics tools to identify secretory proteins.
Methods
Isolates and culture conditions
Rust-infected sunflower leaves were collected in paper bags seperately, air dried at room temperature for 24 h and then spores from mature uredial pustules were brushed off the leaves and stored at 4–5 °C. The collected inocula were inoculated on universal susceptible line 7350. After 10–15 days urediospores of a single pustule were used inoculating two weeks old susceptible plants to produce purified isolates. Subsequently, fresh urediniospores of each isolate were collected from rusted leaves by flicking leaves against parchment paper, and then fresh spores were dried for 3 days in a desiccator and stored individually in the refrigerator at 80 °C below zero. In this experiment, the transcriptome data were obtained from P. helianthi isolate SY.
Puccinia helianthi transcriptomic data sets
We constructed a P. helianthi reference transcriptome for different growing stage urediniospores (0 h fresh urediniospores, 4, and 8 h germinated spores). The cDNA library was sequenced on the Illumina HiSeq™ 2500. For the assembly library, raw reads were filtered to remove those containing an adapter and reads with more than 5% unknown nucleotides. Low quality reads were also removed, in which the percentage of low Q-value (≤10) bases was more than 20%. Clean reads were de novo assembled by the Trinity Program yielding 59,409 transcripts with a mean size of 1394 bp. Sequence data has been uploaded to the Short Read Archive (https://www.ncbi.nlm.nih.gov/sra) of the National Center for Biotechnology Information (NCBI); accession number SRP059519. The secretory proteins were predicted according to the N-terminal amino acid sequences of 35,286 ORFs (Additional file 1).
Prediction and validation of excretory/secretory (ES) proteins
ORFs fulfilling the following four criteria were defined as the computational secretome: (a) the ORF contains an N-terminal signal peptide; (b) the ORF has no transmembrane domains; (c) the ORF has no GPI-anchor site; and (d) the sequence does not contain the localization signal, which may target mitochondria or other intracellular organelles.
Table 1 summarizes the bioinformatic tools used in this study. SignalP v4.1, TMHMM v2.0, TargetP v1.1, ProtComp v9.0 and big-PI predictor tools were employed to identify expected secretory proteins of P. helianthi. SignalP predicts classical secretory proteins in eukaryotes and a truncation protein sequence at 70 amino acids as filters. The standard was L = −918.235-123.455* (Mean S score) +1983.44* (HMM score) and L > 0 for predicting signal peptide proteins. TargetP allowed the prediction of mitochondrial proteins with a cut-off of 0.95 for mitochondrial proteins and 0.90 for proteins in other locations. Transmembrane proteins were predicted with TMHMM (version 2.0) with default options. The putative proteins generated from the transcriptome were initially analyzed by SignalP to predict classical secretory proteins on the basis of a D-score greater than 0.5. The proteins identified were then analyzed with TMHMM to screen for classical secretory proteins without transmembrane segments. Proteins that passed the first two steps were then evaluated by TargetP to identify mitochondrial proteins. Once mitochondrial proteins were identified, the remaining secretory proteins were examined and their sub-cellular localization was predicted with Protcomp. Those assigned to extracellular (secreted) categories were considered pathogenic secretory proteins.
Analysis of signal peptide sequences
In order to further examine the length of signal peptide sequences, the secretory proteins obtained from the previous step were analyzed using custom Perl script. Lipoprotein signal peptide prediction was done with LipoP v1.0, which was able to distinguish among lipoproteins (SPaseII-cleaved proteins), SPaseI-cleaved proteins, cytoplasmic proteins, and transmembrane proteins [41]. Signal peptides with an RR-motif were selected by TatP v1.0 and homology prediction of those signal peptide sequences was evaluated following alignment by Clustal Omega.
ES proteins annotation
Predicted ES proteins were annotated with InterProScan and gene ontology (GO) terms for protein domain and family classification [42]. GO term enrichment analysis was performed using the DAVID bioinformatics resource [43]. KAAS (KEGG Automatic Annotation Server) performed functional annotation by BLAST search against the manually curated KEGG database [44] and provided insight into BRITE functional hierarchies and KEGG pathway maps [45]. The ES proteins were independently assessed for homology matches against NCBI’s non-redundant protein database and for orthologs against the Cluster of Orthologous Groups of proteins (COG) database using BLAST with permissive (E-value: 1e-10) search strategies. Finally, the ES proteins were predicted to have pathogenic function by BLAST analysis of the Pathogen Host Interaction (PHI) database (identity > 25, E-value: 1e-10).
Results
ES protein prediction from the transcriptome data set of P. helianthi
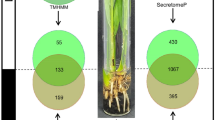
A total of 2,350 (6.7%) out of 35,286 ORFs were predicted as classical secretory proteins with SignalP. According to TMHMM v2.0 tool prediction, 149 (6.3%) proteins have two or more transmembrane helices, 422 (18.0%) proteins have one transmembrane helix, and 1,779 proteins lack transmembrane helices, accounting for 75.7% among 2350 proteins with N-terminal signal peptides. The remaining 1,779 proteins without transmembrane helices were queried with big-PI Predictor yielding 22 potential GPI-anchored proteins that may not be extracellularly secreted and 1,757 non GPI-anchored proteins ORFs.
TargetP v1.1 software was used to predict mitochondrial proteins. Among 1,757 proteins, 1,676 (95.4%) proteins had extracellular targeting signals, 68 (3.9%) proteins contained mitochondria targeting signals and 15 proteins (0.9%) contained other targeting signals.
The application of ProtComp v9.0 to the remaining 1,676 ORFs yielded a total of 908 ORFs (54.2%) as ES proteins (Additional file 2) and the remaining 768 proteins were predicted to be transported to the mitochondria (11.3%), cell membrane (14.9%), nucleus (3.8%), golgi (2.9%), cytoplasm (3.0%), endoplasmic reticulum (4.4%), lysosome (2.9%), peroxisome (1.3%) and vacuole (1.6%).
ORF length of the secretory proteins from P. helianthi
To examine the ORF length of the predicted secretory proteins from P. helianthi, 35,286 P. helianthi ORFs were analyzed by bioinformatics tools and 908 (2.6%) ORFs were identified as secretory proteins. Among them, 728 proteins contained the complete ORF. The longest protein was 1001 amino acids (aa) and the shortest one was 34 aa. The length of most secretory proteins (79.8% of the total identified proteins with a complete ORF) was between 51 and 300 aa. Within this group, 41.0% of them were 101–200 aa long. Thus, we suggest most secretory proteins probably fall in the shorter length range (Fig. 1).
Characteristics of signal peptides of predicted secretory proteins in P. helianthi
The analysis of the signal peptides of 908 predicted secretory proteins reveals the length of the signal peptide ranges from 10 to 34 aa (mean = 21 aa) and most signal peptides (35.8%) ranged from 18 to 20 aa. Signal peptides with 19 aa length, however, were the most abundant, accounting for 13.7% (Fig. 2). The alignment of all 908 signal peptide sequences was done by Clustal Omega. The homology among the signal peptide sequences was low with the highest similarity (66.7%) observed between signal peptide sequence KU994941 and KU994981. No protein with an RR-motif signal peptide was found by TatP v1.0 while 463 proteins contained secretory pathway signal peptides cleavable by SpaseI, and 55 proteins harbored lipoprotein signal peptides cleavable by SpaseII. N-terminal transmembrane helices were found in 30 proteins and 360 of them could be localized to cytoplasmic organelles. Thus, most of the secretory proteins were determined to be secreted through the general secretion pathway (Sec-pathway).
Amino acid composition of signal peptides of predicted secretory proteins in P. helianthi
The distribution of 20 amino acids in the signal peptide was statistically analyzed and the frequencies of amino acid residues in a descending order were: L - S - T - R - A - I - C - V - F - E - K - M - G - N - Q - P - Y - H - W - D. Hydrophobic amino acid leucine (L) showed an appearance rate of 16.1%, followed by serine (S) as 10.8% (Fig. 3). The occurrence of the negatively charged hydrophilic amino acid aspartate (D) is the lowest, accounting for 0.5%.
In general, the C-terminal region of signal peptides contains an enzyme recognition site. Based on this cleavage site, the amino acids of negative direction were named as −1, −2, and −3; those of positive direction were named as +1, +2, and +3. Between protein cutting locus positions −3 and +3, valine (V) is most likely to occupy the position −3 at a frequency of 26.7%. The frequency of serine (S) being at position −2 is 16.5%, alanine (A) has a 49.1% chance to be at position −1, while 12.9% of the time glutamine (Q) is found in position +1 (Table 2). Interestingly, it was found that most amino acids were widely used in the range of cleavage site −3 to +3 position in sunflower rust but no H, K, or Y was observed at position −1. This indicates amino acids near the cleavage site are highly polymorphic in sunflower rust.
Annotation of excretory/secretory (ES) of P. helianthi
All ES proteins identified were searched for sequence homology against our non-redundant dataset using BLAST. It was found that 581 (64.0%) computationally predicted ES proteins shared similarities with known proteins. A total of 143 ES proteins could be annotated in Gene Ontology (GO) and were classified into 27 functional groups, including 14 groups in biological process, seven in cellular component, and six in molecular function (Fig. 4). Within biological process, “metabolic process” (GO: 0008152) with 63 ES proteins and “cellular process” (GO: 0009987) with 26 ES proteins were predominant. In the category of cellular component, the three main groups were “extracellular region” (GO: 0005576, 19 ES proteins), “cell” (GO: 0005623, 18 ES proteins), and “cell part” (GO: 0044464, 18 ES proteins). The categories “catalytic activity” (GO: 0003824) and “binding” (GO: 0005488) were most common in molecular function, represented by 63 and 37 ES proteins, respectively.
ES proteins were subjected to GO enrichment analysis. The 10 top significant enriched GO terms are shown in Table 3. The hydrolase activity, hydrolyzing O-glycosyl compounds (GO:0004553), hydrolase activity (GO:0016787), hydrolase activity, acting on glycosyl bonds (GO:0016798), carbohydrate metabolic process (GO:0005975), peptidase activity, acting on L-amino acid peptides (GO:0070011), extracellular region (GO:0005576), peptidase activity (GO:0008233), serine-type endopeptidase activity (GO:0004252), serine-type peptidase activity (GO:0008236) and serine hydrolase activity (GO:0017171) are significantly enriched. These proteins included glycoside hydrolase, glucoamylase, phosphatase, phosphoesterase, lipase, cysteine peptidase, peptidase, cysteine-rich secretory protein, etc. Pathway associations were established for 82 (9.0%) ES proteins with the majority belonging to metabolism. The predicted ES protein dataset is comprised of important biological molecules, including enzymes, the spliceosome and the ribosome (Table 4).
Function prediction of predicted secretory proteins in P. helianthi
Out of 908 secretory proteins queried against our non-redundant dataset using BLAST, 581 had functional descriptions, of which 279 had clear functional descriptions and 302 were predicted as hypothetical, conserved hypothetical, uncharacterized, or unnamed proteins. The querying of 908 secretory proteins against the COG database was performed for functional classification (Fig. 5). A total of 80 proteins could be assigned to the COG classification, of which 26 (32.5%) potentially participated in the transport and metabolism of carbohydrates (G; Fig. 5), followed by 23.8% involved in post-translational modifications, protein turnover, and molecular chaperones (O; Fig. 5). Proteins participating in inorganic ion transport and metabolism; replication, recombination and repair; transcription; amino acid transport and metabolism accounted for only 1.3%, respectively (P, L, K, E; Fig. 5). 188 out of the 908 proteins had annotations based on InterPro, of which 62 (33.0%) were hydrolases, including 19 peptidases, 15 glycoside hydrolases, seven esterases, five phosphatases, four each ribonuleases, and polysaccharide deacetylases, three each alpha/beta hydrolases, and glucanases (Table 5).
COG classifications of predicted secretory proteins in the transcriptome of Puccinia helianthi. All 80 putative proteins showing significant homology to those in the COG database were functionally classified into 14 families. Note: P, Inorganic ion transport and metabolism; L, Replication, recombination and repair; K, Transcription; E, Amino acid transport and metabolism; C, Energy production and transformation; U, Intracellular trafficking, secretion, and vesicular transport; S, Function unknown; M, Biosynthesis of cell and outer membrane; J, Translation, ribosomal structure and biogenesis; Q, The biosynthesis of secondary metabolites, transport and catabolism; R, General function prediction; G, The transport and metabolism of carbohydrates; O, Post-translational modification, protein turnover and molecular chaperones; I, Lipoid metabolism
Peptidase, glycoside hydrolase, pectinesterase, polysaccharide deacetylase, pectate lyase and glucanosyltransferase were found possibly to be related to cell wall degradation. Nine proteins contained an MD-2-related lipid-recognition (ML) domain, six contained a lipocalin/cytosolic fatty-acid binding domain, and three contained a tyrosinase copper-binding domain. Six were annotated as lipocalin, four as the proteinase inhibitor I25 cystatin, four as apolipoprotein, three each as ribosomal protein, one as thaumatin, and two were annotated as the cysteine-rich allergen V5/Tpx-1-related secretory protein. The functions of most predicted secretory proteins are still unknown.
Blasting PHI yielded a total of 43 secretory proteins that could be correlated to pathogenicity (Tables 6 and 7). Of these, three secretory proteins (KU994907, KU994919 and KU994955) were predicted to be similar to an effector (plant avirulence determinant, Phibase accesstion ID: PHI: 653, PHI: 653 and PHI: 652, respectively) of P. sojae (Table 7).
Discussion
Protein is the major functional component of living organisms. Many pathogenic microbes can secrete proteins into host cells to promote their infection process [46]. Therefore, analysis of secretory proteins in the pathogen genome or transcriptome will help reveal pathogenic mechanisms. According to the signal peptide hypothesis [47], secretory protein destination is determined by its signal peptide. The signal peptide will be cleaved off when the protein reaches its destination. A free online program, SignalP, has been developed that accurately identifies eukaryotic signal peptides [48, 49]. An analysis of 47 known secretory protein and 47 other proteins of C. albicans by SignalP v2.0 showed that the putative results obtained were credible [30].
Signal peptides structures from various proteins commonly contain a positively charged N-region, a hydrophobic H-region and a neutral polar C-region. In the C-terminal region, helix breaking proline and glycine residues and small uncharged residues which are often found at the positions −3 and −1 determine the signal peptide cleavage site [50]. In P. helianthi, valine was observed more frequently (26.7%) at position −3, alanine was most likely to be at position −1 (49.1%), while histidine, lysine, tyrosine were not observed at this position. This indicates amino acids at −3 and −1 positions are relatively conserved, which might guarantee the recognition accuracy of signal peptidases.
Numerous algorithms are freely available for the prediction of protein structures, functions and interactions. Analyses of entire S. cerevisiae genome databases have included identification of GPI-anchored proteins [51], a prediction of protein sub-cellular localization [52] and a prediction of the “typical” secretory protein with Internet-based software SignalP v3.0, TargetP v1.01, Big-PI predictor and TMHMM v2.0 [33]. Bioinformatics approaches made the large scale prediction and analysis of ES proteins of Helminths possible, which included a comprehensive BLAST analysis to annotate the function of the ES proteins [53]. Thus, one approach to rapidly analyze the entire P. helianthi transcriptome and to predict its secretome is to utilize a wide range of appropriate and efficient bioinformatics tools.
After screening 35,286 ORFs of transcriptome data, 908 (2.6%) were predicted as secretory proteins. These putative secretory proteins were small proteins. Up to 79.8% of these secretory proteins were between 51 and 300 aa with signal peptide length between 18 and 20 aa. The short length of amino acids in secretory proteins is likely due to the reference genome of P. helianthi is not available and the unavoidable limitations of de novo transcriptome reconstruction. In signal peptides, the frequency of leucine (L), a hydrophobic amino acid, reached 16.1%. Abundant hydrophobic amino acids may be relevant to the secretion of secretory proteins and their subsequent destination. Most of the amino acids in signal peptides were aliphatic, which are mostly neutral amino acids or hydroxyl or sulfur amino containing amino acids. These amino acids may be important for physiochemical properties of the secretory proteins, which can make the signal peptide cross the plasma membrane easier and enhance signal guidance function. Prediction result showed most of the signal peptides of 908 putative secretory proteins were cleaved by SpI. The majority of the secretory proteins in P. helianthi are likely transported via the general secretory pathway. Furthermore, no signal peptide contained the RR-motif, which may indicate the Tat pathway does not exist or has minor roles in P. helianthi.
Signal peptides can guide the secretory proteins to subcellular locations, and play a key role in the process of metabolism. Signal peptide sequence analysis of all 908 secretory proteins showed sequence similarity is low, which indicates higher sequence variability, consistent with previous reports [34]. The low conservation might contribute to accurate positioning and specific metabolic functions of individual secretory proteins.
Among the 908 secretory proteins, most with functional descriptions are proteins responsible for transport and metabolism of carbohydrates, which is similar to previous research on Bradyrhizobium japonicum [54] and Rhizobium etli [55]. This implies a great deal of materials needed for rust pathogen development and infection may involve sugars, inorganic salt, and organic small molecules, which can be used as cofactors and to meet pathogen energy requirements. Our GO enrichment analysis indicated that hydrolase activity, carbohydrate metabolic process, peptidase activity were significantly enriched in the putative secretory proteins. It suggests rust pathogen P. helianthi can secrete various types of extracellular hydrolases which may include nucleases that can degrade the genetic material of the host plants and interfere with the host genetic metabolism. Additional hydrolase enzymes may be responsible for cell wall degradation; thereby making the host conducive to rust pathogen colonization by destroying the host cell structure and accelerating the process of infection. In addition, the secretory proteins also contain relatively unique serine proteases and similar proteins. In fungi, serine proteases are closely linked with pathogen infection and are often used to degrade the host plant proteins [56]. This suggests serine proteases may also be associated with the rust infection process. Cysteine peptidases (CPs) play important roles in facilitating the survival and growth of mammalian parasites [57]. CPs found in the sunflower rust pathogen, in turn, could also be associated with virulence to the host. In addition, two cysteine-rich secretory proteins identified as calcium chelating serine proteases [58] could be candidate effectors of this pathogen [59]. Three proteins similar to effectors of P. sojae were also found that might be similarly correlated with the pathogenicity of P. helianthi. These candidate proteins may provide more insight into common pathogenesis pathways utilized by both P. sojae and P. helianthi but more experimental evidence is necessary to confirm the biological roles of P. helianthi effectors.
Proteins containing the conserved ML domain are involved in lipid recognition or metabolism and are particularly important for the recognition of pathogen-related processes such as lipopolysaccharide (LPS) binding and signaling [60]. LPS and glycoproteins have been detected in the neck region of haustoria [61]. Proteins containing the ML domain in P. helianthi may, therefore, play a role in the recognition of host lipid-related products.
The thaumatin protein is considered a model pathogen-response protein domain for pathogenesis-related (PR) proteins involved in systematically acquired resistance and stress responses in plants, although their precise role is unknown [62]. Thaumatin-like secreted proteins of rust fungi may alter the plant-signalling pathway and have also been reported in the Melampsora secretome [63]. Future research into the role of thaumatin in sunflower rust infection will provide a better understanding of general and specific mechanisms of thaumatin-mediated resistance and pathogenesis.
Among these 908 secretory proteins in P. helianthi, the majority of them were unclassified due to rust fungi are biotrophic species and require specific genes in their life. The similar results were reported in wheat rust fungus P. striiformis f. sp. tritici [64, 65].
Conclusion
In this study, various open source bioinformatics tools were used to predict and analyze ES proteins from P. helianthi transcriptome. Out of 35,286 ORFs of transcriptome data, 908 (2.6%) were predicted as secretory proteins and most were short proteins. A BLAST analysis was used to annotate the function of the ES proteins and provided further evidence for some proteins as candidates participating in the infection process of P. helianthi. Blasting PHI yielded a total of 43 secretory proteins that could be involved in pathogenicity and three secretory proteins were predicted to be similar to the effectors of P. sojae. Therefore, this investigation provides a novel approach for identifying elicitors and pathogenic factors. It also establishes a sound foundation for understanding the structures and functions of the pathogenic factors of P. helianthi. In conclusion, our data can be used as a candidate gene resource for further computational or wet lab research to unveil the molecular mechanisms underlying the interaction between sunflower and P. helianthi.
Abbreviations
- aa:
-
Amino acid
- COG:
-
Cluster of Orthologous Groups of proteins
- CPs:
-
Cysteine peptidases
- CWDEs:
-
Cell wall degrading enzymes
- ES:
-
Excretory/secretory
- GO:
-
Gene ontology
- KAAS:
-
KEGG Automatic Annotation Server
- LPS:
-
Lipopolysaccharide
- Lsp:
-
Lipoprotein-specific SPase
- ML:
-
MD-2-related lipid-recognition
- PHI:
-
Pathogen host interaction
- PR:
-
Pathogenesis-related
- RR-motif:
-
Twin-arginine motif
- Sec-pathway:
-
Secretion pathway
- SpI:
-
Signal peptidase I
- SpII:
-
Signal peptidase II
- Tat:
-
Twin-arginine translocation
References
Parniske M. Intracellular accommodation of microbes by plants: a common developmental program for symbiosis and disease? Curr Opin Plant Biol. 2000;3(4):320–8.
Hahn M, Mendgen K. Signal and nutrient exchange at biotrophic plant-fungus interfaces. Curr Opin Plant Biol. 2001;4(4):322–7.
Collmer A, Badel JL, Charkowski AO, Deng WL, Fouts DE, Ramos AR, Rehm AH, Anderson DM, Schneewind O, van Dijk K, Alfano JR. Pseudomonas syringae Hrp type III secretion system and effector proteins. Proc Natl Acad Sci. 2000;97(16):8770–7.
Kjemtrup S, Nimchuk Z, Dangl JL. Effector proteins of phytopathogenic bacteria: bifunctional signals in virulence and host recognition. Curr Opin Microbiol. 2000;3(1):73–8.
Staskawicz BJ, Mudgett MB, Dangl JL, Galan JE. Common and contrasting themes of plant and animal diseases. Science. 2001;292(5525):2285–9.
Catanzariti AM, Dodds PN, Lawrence GJ, Ayliffe MA, Ellis JG. Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell. 2006;18(1):243–56.
Dodds PN, Lawrence GJ, Catanzariti AM, Ayliffe MA, Ellis JG. The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell. 2004;16(3):755–68.
Orbach MJ, Farrall L, Sweigard JA, Chrmley FG, Valent B. A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell. 2000;12(11):2019–32.
Li W, Wang B, Wu J, Lu G, Hu Y, Zhang X, Zhang Z, Zhao Q, Feng Q, Zhang H, Wang Z, Wang G, Han B, Wang Z, Zhou B. The Magnaporthe oryzae avirulence gene AvrPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t. Mol Plant-Microbe Interact. 2009;22(4):411–20.
Shan W, Cao M, Leung D, Tyler BM. The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol Plant-Microbe Interact. 2004;17(4):394–403.
Armstrong MR, Whisson SC, Pritchard L, Bos JI, Venter E, Avrova AO, Rehmany AP, Böhme U, Brooks K, Cherevach I, Hamlin N, White B, Fraser A, Lord A, Quail MA, Churcher C, Hall N, Berriman M, Huang S, Kamoun S, Beynon JL, Birch PR. An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc Natl Acad Sci U S A. 2005;102:7766–71.
Allen RL, Bittner-Eddy PD, Grenvitte-Briggs LJ, Meitz JC, Rehmany AP, Rose LE, Beynon JL. Host-parasite coevolutionary conflict between Arabidopsis and downy mildew. Science. 2004;306(5703):1957–60.
Rehmany AP, Gordon A, Rose LE, Allen RL, Armstrong MR, Whisson SC, Kamoun S, Tyler BM, Birch PR, Beynon JL. Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell. 2005;17(6):1839–50.
Wu SC, Kauffmann S, Darvill AG, Albersheim P. Purification, cloning and characterization of two xylanases from Magnaporthe grisea, the rice blast fungus. Mol Plant-Microbe Interact. 1995;8(4):506–14.
Vogel JP, Raab TK, Schiff C, Somerville SC. PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell. 2002;14(9):2095–106.
Xue C, Park G, Choi W, Zheng L, Dean RA, Xu JR. Two novel fungal virulence genes specifically expressed in appressoria of the rice blast fungus. Plant Cell. 2002;14(9):2107–19.
Talbot NJ, Ebbole DJ, Hamer JE. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell. 1993;5(11):1575–90.
Clergeot PH, Gourgues M, Cots J, Laurans F, Latorse MP, Pepin R, Tharreau D, Notteghem JL, Lebrun MH. PLS1, a gene encoding a tetraspanin-like protein, is required for penetraion of rice leaf by the fungal pathogen Magnaporthe grisea. Proc Natl Acad Sci U S A. 2001;98(12):6963–8.
Kamakura T, Yamaguchi S, Saitoh K, Teraoka T, Yamaguchi I. A novel gene, CBP1, encoding a putative extracellular chitin-binding protein, may play an important role in the hydrophobic surface sensing of Magnaporthe grisea during appressorium differentiation. Mol Plant-Microbe Interact. 2002;15(5):437–44.
Rapoport TA. Transport of proteins across the endoplasmic reticulum membrane. Science. 1992;258(5084):931–6.
Tjalsma H, Noback MA, Bron S, Venema G, Yamane K, van Dijl JM. Bacillus subtilis contains four closely related typeIsignal peptidases with overlapping substrate specificities: constitutive and temporally controlled expression of different sip genes. J Biol Chem. 1997;272(41):25983–92.
Tjalsma H, Bolhuis A, van Roosmalen ML, Wiegert T, Schumann W, Broekhuizen CP, Quax W, Venema G, Bron S, van Dijl JM. Functional analysis of the secretory precursor processing machinery of Bacillus subtilis: identification of a eubacterial homolog of archaeal and eukaryotic signal peptidases. Genes Dev. 1998;12(15):2318–31.
Tjalsma H, van den Dolder J, Meijer WJ, Venema G, Bron S, van Dijl JM. The plasmid-encoded typeIsignal peptidase SipP can functionally replace the major signal peptidases SipS and SipT of Bacillus subtilis. J Bacteriol. 1999;181(8):2448–54.
Sargent F, Berks BC, Palmer T. Pathfinders and trailblazers: a prokaryotic targeting system for transport of folded proteins. FEMS Microbiol Lett. 2006;254(2):198–207.
Pra´gai Z, Tjalsma H, Bolhuis A, van Dijl JM, Venema G, Bron S. The signal peptidase II (lsp) gene of Bacillus subtilis. Microbiology. 1997;143(4):1327–33.
Tjalsma H, Kontinen VP, Pra´gai Z, Wu H, Meima R, Venema G, Bron S, Sarvas M, van Dijl JM. The role of lipoprotein processing by signal peptidaseII in the Grampositive eubacterium Bacillus subtilis: signal peptidasIIis required for the efficient secretion of a-amylase, a non-lipoprotein. J Biol Chem. 1999;274(3):1698–707.
Natale P, Brüser T, Driessen AJM. Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane—Distinct translocases and mechanisms. Biochim Biophys Acta. 2008;1778(9):735–1756.
Paik SH, Chakicherla A, Hansen JN. Identification and characterization of the structural and transporter genes for, and the chemical and biological properties of, sublancin 168, a novel lantibiotic produced by Bacillus subtilis 168. J Biol Chem. 1998;273(36):23134–42.
Weiner JH, Bilous PT, Shaw GM, Lubitz SP, Frost L, Thomas GH, Cole JA, Turner RJ. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell. 1998;93(1):93–101.
Lee SA, Wormsley S, Kamoun S, Lee AF, Joiner K, Wong B. An analysis of the Candida albicans genome database for soluble secreted proteins using computer-based prediction algorithms. Yeast. 2003;20(7):595–610.
Torto TA, Li S, Styer A, Huitema E, Testa A, Gow NA, van West P, Kamoun S. EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora. Genome Res. 2003;13(7):1675–85.
Zhou XG, Hou SM, Chen DW, Tao N, Ding Y, Sun M, Zhang S. Genome-wide analysis of the secreted proteins of Phytophthora infestans. Hereditas. 2011;33(7):125–33.
Yang J, Li CY, Wang Y, Zhu Y, Li J, He X, Zhou X, Liu L, Ye Y. Computational analysis of signal peptide dependent secreted proteins in Saccharomyces cerevisiae. Agric Sci China. 2006;5(3):221–7.
Fan C, Li C, Zhao M, He Y. Analysis of signal peptides of the secreted proteins in Agrobacterium tumefaciens C58. Acta Microbiol Sin. 2005;45(4):561–6.
Yu QL, Ma L, Liu L, Yang J, Su Y, Wang Y, Zhu Y, Li C. Primary analysis of host-targeting-motif harbored secreted proteins in genome of Fusarium graminearum. Biotechnol Bull. 2008;18(1):160–5. 180.
Zhou X, Li C, Zhao Z, Su Y, Zhang S, Li J, Yang J, Liu L, Ye Y. Analysis of the secreted proteins encoded by genes in genoma of filamental fungus (Neurospora crassa). Hereditas. 2006;28(2):200–7.
Tian L, Chen JY, Chen XY, Wang J, Dai X. Prediction and analysis of Verticillium dahliae VdLs 17 secretome. Sci Agric Sin. 2011;44(15):3142–53.
Ren N, Li JX, Shen XK. Prediction and analysis of secretome in Aspergillus oryzae. J Anhui Agri Sci. 2010;38(25):13622–5.
Xue XD, Qu ZP, Wang XJ, Zhang Y, Li G, Huang L, Kang Z. Prediction secreted proteins from cDNA library of Puccinia striiformis f. sp. tritici. J Northwest A & F Univ (Nat Sci Ed). 2009;37(2):105–11.
Han CZ. Prediction for secreted proteins from Colletotrichum graminicola genome. Biotechnology. 2014;24(2):36–41.
Juncker AS, Willenbrock H, von Heijne G, Brunak S, Nielsen H, Krogh A. Prediction of lipoprotein signal peptides in gram-negative bacteria. Protein Sci. 2003;12(8):1652–62.
Zdobnov EM, Apweiler R. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17(9):847–8.
da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57.
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–5.
Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38(2):D355–60.
Ellis J, Catanzariti AM, Dodds P. The problem of how fungal and oomycete avirulence proteins enter plant cells. Trends Plant Sci. 2006;11(2):61–3.
Blobel G, Sabatini DD. Ribosome-membrane interaction in eukaryotic cells. Biomembranes. 1971;2:193–5.
Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10(1):1–6.
Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–6.
Von Heijne G. Protein targeting signals. Curr Opin Cell Biol. 1990;2(4):604–8.
Caro LH, Tettelin H, Vossen JH, Ram AF, den EH v, Klis FM. In silicio identification of glycosyl-phosphatidylinositol-anchored plasmamembrane and cell wall proteins of Saccharomyces cerevisiae. Yeast. 1997;13(15):1477–89.
Kumar A, Agarwal S, Heyman JA, Matson S, Heidtman M, Piccirillo S, Umansky L, Drawid A, Jansen R, Liu Y, Cheung K, Miller P, Gerstein M, Roeder GS, Snyder M. Subcellular localization of the yeast proteome. Genes Dev. 2002;16(6):707–19.
Garg G, Ranganathan S. Helminth secretome database (HSD): a collection of helminth excretory/secretory proteins predicted from expressed sequence tags (ESTs). BMC Genomics. 2012;13 Suppl 7:22–30.
Hempel J, Zehner S, Gottfert M, Patschkowski T. Analysis of the secretome of the soybean symbiont Bradyrhizobium japonicum. J Biotechnol. 2009;140(2):51–8.
Zhang W, Ma J, Pan W, Lei T. Genome-wide identification and analyses of the classical secreted proteins of Rhizobium etli CFN42. Genomics Appl Biol. 2014;33(5):961–9.
Monod M, Capoccia S, Léchenne B, Zaugg C, Holdom M, Jousson O. Secreted proteases from pathogenic fungi. Int J Med Microbiol. 2002;292(5–6):405–19.
Mottram JC, Coombs GH, Alexander J. Cysteine peptidases as virulence factors of Leishmania. Curr Opin Microbiol. 2004;7(4):375–81.
Milne TJ, Abbenante G, Tyndall JD, Halliday J, Lewis RJ. Isolation and characterization of a cone snail protease with homology to CRISP proteins of the pathogenesis-related protein superfamily. J Biol Chem. 2003;278(33):31105–10.
Stergiopoulos I, de Wit PJ. Fungal effector proteins. Annu Rev Phytopathol. 2009;47:233–63.
Inohara N, Nunez G. ML- a conserved domain involved in innate immunity and lipid metabolism. Trends Biochem Sci. 2002;27(5):219–21.
Larous L, Kameli A, Losel DM. Ultrastructural observations on Puccinia methae infections. J Plant Pathol. 2008;90(2):185–90.
Liu JJ, Sturrock R, Ekramoddoullah AK. The superfamily of thaumatin-like proteins: its origin, evolution, and expression towards biological function. Plant Cell Rep. 2010;29(5):419–36.
Joly DL, Feau N, Tanguay P, Hamelin RC. Comparative analysis of secreted protein evolution using expressed sequence tags from four poplar leaf rusts (Melampsora spp.). BMC Genomics. 2010;11(28):422.
Ling P, Wang M, Chen X, Campbell KG. Construction and characterization of a full-length cDNA library for the wheat stripe rust pathogen (Puccinia striiformis f. sp. tritici). BMC Genomics. 2007;8(1):145.
Yin C, Chen X, Wang X, Han Q, Kang Z, Hulbert SH. Generation and analysis of expression sequence tags from haustoria of the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. BMC Genomics. 2009;10(1):626.
Acknowledgements
Authors are grateful to Dr. C. S. Karigar who provided writing assistance and revised critically.
Funding
This work was supported by the National Natural Science Foundation of China (grant number 31360422) and China Agricultural Research System (grant number CARS-16). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files. Transcriptome sequence data has been uploaded to the Short Read Archive (https://www.ncbi.nlm.nih.gov/sra) of NCBI; accession number SRP059519.
Authors’ contributions
LJ conceived the study and drafted the manuscript. XF analyzed data and revised the manuscript. DD and WJ performed the prediction and analysis and also participated in manuscript preparation. All authors have read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1:
Dataset of 35286 ORFs of the Puccinia helianthi transcriptome (XLSX 4749 kb)
Additional file 2:
Dataset of 908 putative secretory proteins (XLSX 129 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Jing, L., Guo, D., Hu, W. et al. The prediction of a pathogenesis-related secretome of Puccinia helianthi through high-throughput transcriptome analysis. BMC Bioinformatics 18, 166 (2017). https://doi.org/10.1186/s12859-017-1577-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12859-017-1577-0