Abstract
Background
Canarian Black (CB) pigs belong to an autochthonous and endangered breed, which is spread throughout the Canarian archipelago. It is commonly accepted that they represent a relic of the pig populations that were bred by the Berbers in North Africa over millennia. It is important to note that the geographic isolation of the Canary Islands has preserved this genetic legacy intact from foreign introgressions until the Spanish conquest of the archipelago in the 15th century. Ten years ago, it was demonstrated that, in CB pigs, the frequency of the Asian A2 cytochrome-b haplogroup reached 73%. The current work aimed at investigating whether this observation is explained by either a recent or an ancient introgression of CB pigs with Far Eastern pigs.
Results
Genetic analyses of 23 ancient samples from pre-hispanic Canarian pigs (420 to 2500 years before present) showed that Near Eastern and Far Eastern genetic signatures were totally absent in the primitive Canarian pre-hispanic pigs. Indeed, the haplotypes detected in these pigs were closely related to those of North African and European wild boars.
Conclusions
Our results demonstrate that the high frequency of the Far Eastern mitochondrial cytochrome B A2 haplotype in modern Canarian Black pigs probably corresponds to a relatively recent introgression with British breeds.
Similar content being viewed by others
Findings
The only living representative of the domestic swine that was introduced into the Canary Islands 3000 YBP (years before present) is the Canarian Black (CB) pig, which in the 1980’s was very near extinction. However, an active conservation program has allowed the population to reach a census of a few hundred individuals [1,2]. Analysis of mitochondrial genome variation in modern CB pigs revealed that the Far Eastern A2 cytochrome-b (MT-CYB) haplogroup reached a high frequency in this breed (up to 73%) [3]. Far Eastern haplotypes are absent in local European breeds, such as the Iberian, Mangalitza, Majorcan Black or Basque pigs [4]. Clop et al. [3] proposed two alternative explanations for the presence of Far Eastern haplotypes in the mitochondrial genome of CB pigs i.e. either introgression of CB pigs with improved British breeds that had been extensively hybridized with Chinese sows in the 18th and 19th centuries to select for pigs with earlier reproductive maturity and increased fatness [5], or conversely, a more ancient introduction as a consequence of the settlement of Berber tribes in the Canary Islands. Although this latter interpretation is less likely, Far Eastern haplotypes are very common in East African breeds such as the Mukota breed [4] and it is possible that these haplotypes may have diffused westwards, as is the case for indicine alleles in cattle [6].
Obviously, these two alternative scenarios cannot be ascertained with mitochondrial data from modern CB samples. Thus, we decided to survey the mitochondrial variation of 23 ancient pig samples that cover a large period of time (from ≈ 420 to 2500 YBP) and represent 11 pre-Hispanic archeological sites across four of the seven Canary Islands (Figure 1) and [see Additional file 1; Additional file 2: Table S1]. Total DNA was extracted in laboratories that are dedicated to the analysis of ancient DNA at the Institute of Evolutionary Biology and University of Pompeu Fabra in Barcelona by applying proteinase-K digestion followed by phenol-chloroform precipitation and a column concentration (Amicon), as described elsewhere [7]. Extracts from skin samples were subsequently purified with a gene clean silica method using a DNA extraction Kit (Fermentas, USA). To the best of our knowledge, no previous work on modern pigs had ever been conducted at this laboratory and standard precautions for experiments involving ancient DNA samples were followed (see Appendix).
Pig specific primers [see Additional file 2: Table S2]) were designed to amplify two non-overlapping sequences of 89 (PCR1) and 77 bp (PCR2) corresponding to the MT-CYB gene. Sequence PCR1 contains the diagnostic single nucleotide polymorphisms (SNPs) that are located at positions 15036, 15038, 15041, 15044 and 15045 and differentiate European (E1, E2, E3 and E4) from Asian (A1, A2, A3, A4) haplogroups [3,8] and sequence PCR2 contains two SNPs that discriminate between Near Eastern vs Western (Europe and North Africa) haplotypes [4]. Each fragment was amplified using a two-step PCR protocol [9]. Amplified products were purified with a gene clean silica method using the DNA Extraction Kit (Fermentas, USA) and cloned using the Topo TA cloning kit (Invitrogen, The Netherlands). White colonies were subjected to 30 cycles of PCR with M13 universal primers and subsequently sequenced with an Applied BioSystems 3100 DNA sequencer, at the Servei de Seqüenciació of the Universitat Pompeu Fabra (Barcelona).
Amplicon sequences of PCR1 and PCR2 were obtained from 21 of 23 and 8 of 10 specimens of pre-Hispanic Canary pigs, respectively. PCR1 was analyzed in all 23 ancient pig samples collected. For PCR2, we excluded the samples for which PCR1 was not amplified and also most of the samples of mummified pig skin because, after purification and amplifications, the amount of DNA extracted was not sufficient [see Additional file 2: Table S1]. Although the high temperatures of the climatic conditions of the Canary Islands do not favor DNA preservation, the success rates of amplification (91.3% and 80%, respectively) were very high [10,11]. For identical temperature and environmental settings, preservation of ancient DNA is highly correlated with sample age [12]. Two of the samples for which no successful amplification was achieved (Buenavista 3 and Lanzarote 12, both from Lanzarote) were among the four oldest materials in the assemblage. The third sample for which amplification failed (Guadayaque11/3) originated from a leather skin that was used as shroud on the mummies. Possibly, the presence of inhibitor substances used during the embalming process may have prevented DNA amplification.
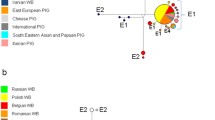
All PCR1 DNA sequences obtained from the 21 ancient pig samples from the Canary Islands belonged to a single haplogroup, i.e. MT-CYB haplogroup E1 (Figure 2) and [see Additional file 2: Table S1, Additional file 3: Figure S1]. Given that haplogroup E1 is represented by distinct haplotypes that segregate in wild boars from the Near East, Europe and North Africa, it does not allow us to conclude on the geographical origin of the primitive Canarian pre-hispanic pigs. Interestingly, the eight ancient pig samples that provided PCR2 amplicons harboured Western haplotypes [see Additional file 3: Figure S2]. It is important to note that Western MT-CYB variants have negligible frequencies in Near Eastern wild boars and they probably reflect introgression with Western pigs or feralization of domestic pigs [4,13]. This result agrees well with data on the autosomal nuclear genome: based on 60K SNP genotypes of Near Eastern and European pigs and wild boars, Manunza et al. [14] demonstrated that there was no Near Eastern footprint in extant Canarian pigs. The absence of such signatures in the eight pre-hispanic Canarian samples that varied in age from ≈ 960 to 2500 years suggests that, at the beginning of the first millennium BC, the Near East signature was absent, or at low frequency, in domestic pig populations from western North Africa.
Comparison of frequencies of mitochondrial MT-CYB haplogroups in the ancient Canarian pigs (described in this study) and modern pigs from Africa and the Canary Islands and wild boar from Near East and Europe described previously by Ramirez et al. [ 4 ].
Our data not only provide a first glimpse on the mitochondrial gene pool of pigs that probably have a Berber ancestry, but also help to solve a puzzling finding reported by Clop et al. [3] 10 years ago i.e. the high frequency of the Far Eastern A2 MT-CYB haplotype in CB pigs. Our results on ancient DNA strongly support the former hypothesis i.e. the complete absence of Far Eastern haplotypes in the dataset of ancient pig samples (from 11 locations across four of the seven Canary Islands) suggests that native CB pigs did not carry Far Eastern alleles. According to García-Martín and Capote [2], pigmentation and ear morphology and size of Canarian pigs resemble those observed in the Berkshire pig breed. Interestingly, the A2 haplotype segregates at high frequencies (around 35%) in this British local breed [8], which provides evidence for a recent Berkshire introduction. From an historical point of view, crossbreeding of CB pigs with British and Iberian breeds has been widely documented [15,16].
Certain insights about the origin of modern domestic breeds can only be obtained through the analysis of ancient DNA [13,17–19]. In this study, partial sequencing of the MT-CYB sequence in pig samples from Canarian archaeological assemblages has allowed us to demonstrate that the presence of Asian MT-CYB alleles in CB pigs was the result of a recent introgression event (probably with British breeds). However, ascertainment bias is a problem for proper interpretation of results that are exclusively based on maternal data. Analyses of ancient samples based on whole-genome sequencing may contribute to better understand the process of domestication and breed formation.
References
Robert A, Zamorano MJ, Ginés R, Argüello A, Delgado JV, López JL. Origin and standing of Canary Black Pig. Arch Zootec. 2000;49:291–6.
Garcia-Martin M, Capote J. El Cerdo Negro Canario. Cabildo Insular de La Palma. 1982. p. 21.
Clop A, Amills M, Noguera JL, Fernandez A, Capote J, Ramón MM, et al. Estimating the frequency of Asian cytochrome B haplotypes in standard. Genet Sel Evol. 2004;36:97–104.
Ramírez O, Ojeda A, Tomàs A, Gallardo D, Huang LS, Folch JM, et al. Integrating Y-chromosome, mitochondrial, and autosomal data to analyze the origin of pig breeds. Mol Biol Evol. 2009;26:2061–72.
Giuffra E, Kijas JMH, Amarger V, Carlborg O, Jeon JT, Andersson L. The origin of the domestic pig: independent domestication and subsequent introgression. Genetics. 2000;154:1785–91.
Hanotte O, Bradley DG, Ochieng JW, Verjee Y, Hill EW, Rege JEO. African pastoralism : genetic imprints of origins and migrations. Science. 2002;296:336–9.
Lalueza-Fox C, Römpler H, Caramelli D, Stäubert C, Catalano G, Hughes D, et al. A melanocortin 1 receptor allele suggests varying pigmentation among Neanderthals. Science. 2007;318:1453–5.
Fang M, Andersson L. Mitochondrial diversity in European and Chinese pigs is consistent with population expansions that occurred prior to domestication. Proc Biol Sci. 2006;273:1803–10.
Krause J, Dear PH, Pollack JL, Slatkin M, Spriggs H, Barnes I, et al. Multiplex amplification of the mammoth mitochondrial genome and the evolution of Elephantidae. Nature. 2006;439:724–7.
Ramírez O, Gigli E, Bover P, Alcover JA, Bertranpetit J, Castresana J, et al. Paleogenomics in a temperate environment: shotgun sequencing from an extinct Mediterranean caprine. PLoS One. 2009;4:e5670.
Ramirez O, Illera JC, Rando JC, Gonzalez-Solis J, Alcover JA, Lalueza-Fox C. Ancient DNA of the extinct lava shearwater (Puffinus olsoni) from the Canary Islands reveals incipient differentiation within the P. puffinus complex. PLoS One. 2010;5:e16072.
Poinar HN. The genetic secrets some fossils hold. Acc Chem Res. 2002;35:676–84.
Larson G, Albarella U, Dobney K, Rowley-Conwy P, Schibler J, Tresset A, et al. Ancient DNA, pig domestication, and the spread of the Neolithic into Europe. Proc Natl Acad Sci U S A. 2007;104:15276–81.
Manunza A, Zidi A, Yeghoyan S, Balteanu VA, Carsai TC, Scherbakov O, et al. A high throughput genotyping approach reveals distinctive autosomal genetic signatures for European and Near Eastern wild boar. PLoS One. 2013;8:e55891.
López-Fernández JL, Arguello A, Capote J, Darmanim N. Contribution to the study of Black Canary Pig. Arch Zootec. 1992;41:531–6.
Martínez A, Quiroz J, Marques J, Delgado J. Study of the genetic diversity of the Canary Black pig using DNA microsatellites. Arch Zootec. 2007;56:425–8.
Krause-Kyora B, Makarewicz C, Evin A, Flink LG, Dobney K, Larson G, et al. Use of domesticated pigs by Mesolithic hunter-gatherers in northwestern Europe. Nat Commun. 2013;4:2348.
Thalmann O, Shapiro B, Cui P, Schuenemann VJ, Sawyer SK, Greenfield DL, et al. Complete mitochondrial genomes of ancient canids suggest a European origin of domestic dogs. Science. 2013;342:871–4.
Groenen MAM, Archibald AL, Uenishi H, Tuggle CK, Takeuchi Y, et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature. 2012;491:393–8.
Lalueza-Fox C, Rosas A, Estalrrich A, Gigli E, Campos PF, García-Tabernero A, et al. Genetic evidence for patrilocal mating behavior among Neandertal groups. Proc Natl Acad Sci U S A. 2011;108:250–3.
Cooper A, Poinar HN. Ancient DNA: do it right or not at all. Science. 2000;289:1139.
Pääbo S, Poinar H, Serre D, Jaenicke-Després V, Hebler J, Rohland N, et al. Genetic analyses from ancient DNA. Annu Rev Genet. 2004;38:645–79.
Gilbert MT, Bandelt HJ, Hofreiter M, Barnes I. Assessing ancient DNA studies. Trends Ecol Evol. 2005;20:541–4.
Del-Arco Aguilar MC, Del-Arco Aguilar MM, Atiénzar Armas E, Hopf M. Estudio de los restos vegetales de la cueva de Don Gaspar y algunas anotaciones sobre la agricultura prehistórica de Tenerife. Investigaciones Arqueológicas en Canarias. 1990;2:13–30.
Del-Arco Aguilar MC,González Hernández C, Del-Arco Aguilar MM, Atiénzar Armas E, Del-Arco Aguilar MJ, C Rosario Adrián. El Menceyato de Icod en el poblamiento de Tenerife: D. Gaspar, Las Palomas y Los Guanches. Sobre el poblamiento y las estrategias de alimentación vegetal entre los Guanches. Eres Arqueología Bioantropología. 2000;9:67–129.
Diego CL. La cueva de Los Cabezazos, en el Bco. del Agua de Dios (Tegueste, Tenerife). NAH Prehistoria. 1975;4:291–336.
Rosario Adrian MC, Valencia Afonso V, Martín Diaz MÁ. Plan especial Barranco del Agua de Dios, Tegueste (Tenerife). Canarias Arqueológica: arqueología-bioantropología. 2010;18:109–16.
Arco Aguilar Mª del C del. Resultados de un sondeo arqueológico en la Cueva de Los Guanches (Icod, Tenerife). El Museo Canario. 1984;46:45-90.
González-Antón R, Del-Arco Aguilar MC, Rodríguez Martín C et al. La necrópolis de Ucazme (Adeje, Tenerife). Estudio arqueológico, Bio y Paleopatológico. Eres Arqueología Bioantropología. 1995;6:29–42.
Momias Canarias”. El Omnibus. 1855. p. 12.
Atoche P, Rodríguez MD, Ramírez MA. El yacimiento arqueológico de “El Bebedero” (Teguise, Lanzarote). Universidad de La Laguna, Ayuntamiento de Teguise, Madrid: Resultados de la primera campaña de excavaciones. Secretariado de Publicaciones; 1989.
Atoche P. Resultados preliminares de la tercera campaña de excavaciones arqueológicas en “El Bebedero” (Teguise, Lanzarote 1990). Vegueta. 1995;96(2):29–44.
Atoche P. Estratigrafías, cronologías absolutas y periodización cultural de la Protohistoria de Lanzarote. Zephyrus. 2009;63:105–34.
Atoche P, Ramírez MA, Torres JD, Pérez S. Excavaciones arqueológicas en el yacimiento de Buenavista (Tiagua, Lanzarote): primera campaña, 2006. Canarias Arqueológica (Arqueología/Bioantropología). 2009;17:9–51.
Atoche P, Ramírez MA, Pérez S, Torres JD. Segunda campaña de excavaciones arqueológicas en el yacimiento de Buenavista (Tiagua, Lanzarote): Resultados preliminares. Canarias Arqueológica (Arqueología/Bioantropología). 2010;18:1–55.
Atoche P, Ramírez Rodriguez MA. Nuevas dataciones radiocarbónicas para la Protohistoria canaria: el yacimiento de Buenavista (Lanzarote). Anuario de Estudios Atlánticos. 2011;57:139–70.
Acknowledgements
IO has a pre-doctoral fellowship from the Basque Government (DEUI). OR is a post-doctoral researcher from the JAEDOC program co-funded by The European Social Fund (ESF). This work has been founded by the grant BFU2012-34157 to C.L-F from the MINECO, Spain.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JC, MCD-A, PA, TD, FJP, RG-A, MA, CL-F and OR contributed to the design of this research. OR, and IO performed the experimental analyses. IO and OR performed the data analysis. IO, MA, CL-F and OR wrote the manuscript. All authors read and approved the final manuscript.
Additional files
Additional file 2: Table S1.
Archaeological sites and age of Canarian pig samples used. This Table provides the ID, lab codes, archeological site and age of the Canarian pig ancient samples from which cytochrome B haplogroups (Fragment 1) and haplotypes (Fragment 2) were retrieved. List of primers used for amplification of the two 89 and 77 bp sequences of the MT-CYB gene. Table S2. provides the names and sequences of the primers used to amplify the two 89 and 77 bp sequences of the MT-CYB gene. Primers were designed with the Primer 3 software (http://bioinfo.ut.ee/primer3-0.4.0/).
Additional file 3: Figure S1.
Alignment of the 48-bp sequences (PCR1) of the MT-CYB gene fragment 1 obtained from DNA from 21 Canarian pig ancient samples. Description: The diagnostic SNPs that are located at positions 15036, 15038, 15041, 15044 and 15045 (indicated in red) differentiate European: E1 (GenBankID: KJ746666), E2 (EU531827) and E4 (GU211924) haplogroups and, also, Far Asian: A1 (KP257599), A2 (KM215171), A3 (AB015072), A4 (KJ746664) and A5 (GU135825) haplogroups. Figure S2. Alignment of the 37-bp sequences (PCR2) of the MT-CYB gene fragment 1 obtained from DNA from 8 Canarian pig ancient samples. Description: The two diagnostic SNPs (indicated in red) differentiate haplotypes of haplogroup E1 present in wild boar individuals from the Near East: H12 (GenBank ID: EU531818), H31 (EU531827), H52 (EU531832), H53 (EU531833), and H54 (EU531834); and from Europe: H1 (AY237496), H4 (AY237512), H15 (AB015072), H16 (EU531821), H17 (EF061501), H18 (AY237516), H24 (AF136542), H27 (EU531824), H32 (EU531828), H34 (EU531830), H65 (AM492593) and H66 (AM492620).
Appendix
Appendix
Sequencing of mtDNA
DNA was extracted following the method described in detail in [20]. Extraction procedures were performed in an isolated pre-PCR area, by adopting the standard precautions for ancient DNA studies [21,22]. Multiple extraction and amplification negative controls to monitor for contamination in the reagents were added to each PCR reaction. During the whole study, no amplification products were obtained in these blank PCR controls.
Amplification was carried out according to a two-step PCR protocol reported by [9]. Both PCR steps were carried out with 2 units AmpliTaq Gold (ABI, USA), 1X AmpliTaq Gold buffer (ABI, USA), 2.5 mM MgCl2 (ABI, USA), and 500 μM of each dNTP. In the first multiplex step, 150 nM of each primer pair and 5 μl of DNA extract were added to a final reaction volume of 20 μl. The first amplification step consisted in a 12 min activation step at 94°C, followed by 27 cycles at 94°C for 20 s, 50°C for 20 s, and 72°C for 20 s. Five μl of a 1 to 10 dilution of the primary amplification product were used as template for the second PCR step. Conditions were the same as for the first step, except that the standard primer concentration was increased to 1.5 μM and that 33 cycles were performed, followed by a final step of 12 min at 72°C. Blank and mock PCR controls were included in each amplification step to monitor against contamination.
PCR products were visualized under UV light and the appropriate bands were excised from a low-melting point agarose gel and purified with a silica-based method. Subsequently, the amplification products were cloned in bacteria (TOPO-TA cloning kit, Invitrogen). Resulting white colonies were picked, amplified with M13 universal primers and sequenced with an ABI3730 capillary sequencer (Applied Biosystems).
We followed the criteria for authenticity described by Gilbert et al. [23]: (i) amplifications of both mtDNA sequences of the Buenavista1 sample were replicated in an independent laboratory (at the Universitat Pompeu Fabra); (ii) for all the samples, mock extractions and PCR blank controls were carried out and there was no evidence of contamination; and (iii) about 30% of the fragments were replicated twice [see Additional file 3: Figures S1 and S2].
To assign the genotype of the diagnostic SNPs, we used the majority rules consensus. If, for one or a few clones, additional substitutions are found present in the sequence of one PCR but not in the sequence of another PCR from the same sample, it is reasonable to attribute them to DNA damage, and thus they were not considered in the analyses.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Olalde, I., Capote, J., Del-Arco, M.C. et al. Ancient DNA sheds light on the ancestry of pre-hispanic Canarian pigs. Genet Sel Evol 47, 40 (2015). https://doi.org/10.1186/s12711-015-0115-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12711-015-0115-7