Abstract
Background
Several biomarkers developed from freshwater sentinel species are used to complement chemical assessment of freshwater ecosystems and improve the evaluation of their contamination levels. While active biomonitoring strategies allow to compare biological data in the field, the lack of assessment criteria to interpret biomarkers modulations restricts their application in freshwater biomonitoring surveys. For this purpose, the determination of threshold values emerges as a relevant approach to support the biomarker-based diagnosis within biomonitoring surveys. Based on the distribution of contamination baselines, a statistical approach was proposed to define threshold values of bioaccumulated concentrations in the sentinel species Gammarus fossarum. Although this methodology is limited to chemical databases, this approach could be adapted for defining threshold values at the biological level. This study aims to demonstrate the possibility of defining threshold values for different classes of biomarkers monitored in two freshwater sentinel species (Dreissena polymorpha, G. fossarum) and evidence the added value of such an approach in biomarker-based diagnosis.
Results
For this purpose, the study benefited from exceptional environmental biomarker datasets acquired from mussels and gammarids caged in hundred environmental sites, representative of the diversity of French water bodies. Thanks to these environmental datasets, inhibition and/or induction thresholds could be defined for 8 biochemical, cellular, and molecular biomarkers of D. polymorpha and 25 peptide biomarkers of G. fossarum, not influenced by confounding factors. Threshold values were then applied to another biomarker dataset acquired from mussels and gammarids caged upstream and downstream four wastewater treatment plants (WWTPs). Several biomarkers were modulated both downstream and upstream of WWTPs, indicating a potential contamination not related to WWTPs. In contrast, some biomarkers were impacted exclusively downstream of the WWTPs, highlighting an impact of WWTP discharges on the aquatic fauna.
Conclusions
This study demonstrates the possibility to define threshold values for a wide range of biomarkers (cellular, biochemical and molecular) monitored on different sentinel species. The definition of threshold values constitutes an attractive solution to support the application of biomarkers in active biomonitoring surveys and improve the biomarker-based diagnosis without the need of “reference sites”.
Similar content being viewed by others
Background
Thousands of pollutants are routinely discharged into the aquatic environment [1], among them, new synthetic substances referred as ‘emerging contaminants (ECs)’ have become a major source of concern given their adverse effects on the environment and human well-being [2, 3]. The importance of ecosystem services (biodiversity, drinking water treatment, recreation) provided by natural water bodies leads to strong societal expectations regarding water quality and the protection of this resource. In Europe, these statements led to the development of the Water Framework Directive (WFD, Directive 2000/60/EC) which requires the Member States to achieve good quality states of their freshwater ecosystems [4]. Based on the monitoring of 45 priority substances, the current WFD strategy is struggling to consider the impact of complex chemical cocktails found in aquatic environments [5,6,7]. In this regard, biological responses, called biomarkers, have been proposed as a promising alternative to current regulatory actions [8,9,10]. In particular, several studies have demonstrated the value of monitoring a comprehensive set of biomarkers in multiple sentinel species to reveal hot spots of pollution and evaluate its adverse effects on aquatic wildlife [11,12,13,14].
The Crustacean Gammarus fossarum and the Bivalve Dreissena polymorpha are two relevant sentinel species for tracking the pollution in freshwater ecosystems. Both species have a broad distribution over the European continent and populations of G. fossarum and D. polymorpha are found in most of the European countries [15, 16]. Many studies have shown that these organisms bioaccumulate different micropollutants in a dose- and time-dependent manner [17, 18]. Given their different biological (i.e., feeding behavior, reproduction) and ecological traits [19,20,21], these two species are likely to be affected differently by the pollution and are therefore complementary to better evaluate the exposure of aquatic wildlife to hazardous chemical contamination [14]. For these two species, a panel of biomarkers has been developed to assess the biological impacts of pollutants. Specifically, several biochemical, cellular and molecular biomarkers related to the energy metabolism, detoxification processes and immune response of D. polymorpha were found to be particularly sensitive to pollutants under laboratory and field exposure conditions [14, 22]. For G. fossarum, a set of protein biomarkers with putative key functional roles (i.e., homeostasis, osmoregulation, detoxification, molting) are quantified by multiple reaction monitoring (MRM) mass spectrometry (MS) [23, 24]. This new generation of molecular biomarkers is emerging as promising tools for revealing the biological impact of pollutants on G. fossarum [24]. Easy to handle, caging of both species is now well mastered and standardized, offering the possibility to obtain comparable biological data in real environmental conditions, by using calibrated organisms following an active biomonitoring strategy [14, 22, 25,26,27]. However, the lack of assessment criteria to interpret biomarkers modulations restricts biomarkers use in freshwater biomonitoring surveys. The determination of threshold values emerges as a relevant and operational strategy to support the application of biomarkers in biomonitoring surveys. This strategy consists in defining threshold values that take the natural variability into account; a value observed out of this threshold then reflects a significant perturbation. In marine biomonitoring, studies have initiated the determination of threshold values for the application of biomarkers. International Council for Exploration of the Sea (ICES) and the Oslo–Paris Commission (OSPAR) have developed a framework to propose an integrated assessment of contaminant impacts in coastal and offshore areas [28]. Background assessment criteria (BAC) are defined as the baseline biological condition in healthy organisms. Ecotoxicological assessment criteria (EAC) are defined as the level of biological response above which significant acute and long-term adverse biological effects are likely to occur. These threshold values were defined for a set of biochemical and cellular biomarkers developed from diverse marine sentinel species, and are already implemented in biomonitoring surveys to assess the biological effects of environmental pollutions [29, 30]. In freshwater biomonitoring, threshold values were defined for some biomarkers of G. fossarum (i.e., feeding behavior, acetylcholinesterase, digestive enzymes) [31,32,33,34] and the three-spined stickleback, Gasterosteus aculeatus [35]. Threshold values were also used to model the influence of confounding factors in biomarker modulations, such as feeding and reproductive behaviors of G. fossarum likely influenced by temperature changes [31, 34]. However, the methodologies applied for defining threshold values were limited to data collected from laboratory experiments or data collected from organisms caged in "reference sites", considered as lowly impacted by anthropogenic activities. Based on the Gaussian distribution of bioaccumulation baselines, Besse et al. [17] proposed a statistical approach to determine thresholds values of bioaccumulated concentrations of different metallic and organic pollutants in G. fossarum, without a priori regarding the status of sites (reference or contaminated). Once defined, these threshold values were used in active biomonitoring studies to detect when pollutants are abnormally bioaccumulated in gammarids and to characterize the contamination profiles of investigated environmental sites [17, 36]. The applicability of this methodology for defining threshold values of biomarker data was also discussed from a preliminary assay with Gammarus feeding rate inhibition records in a regional watershed [37].
Using two sentinel species employed in active biomonitoring surveys (D. polymorpha and G. fossarum), this study aims to propose a methodology for defining threshold values without a priori and how it can be implemented for different categories of biomarkers. For this purpose, the study benefited from exceptional environmental data sets, which comprised the measure of several biochemical, cellular and molecular biomarkers from mussels and gammarids caged in a hundred environmental sites representative of the diversity of French water bodies. After checking that biomarkers were not influenced by confounding factors (temperature, conductivity, pH and dissolved oxygen), the methodology of Besse et al. [17] was improved to define inhibition and/or induction thresholds specific to each biomarker tested. To further demonstrate the added value of this approach in biomonitoring surveys, threshold values were applied to a new dataset and used to support biomarker-based diagnosis from mussels and gammarids caged upstream and downstream of four wastewater treatment plants (WWTPs).
Materials and methods
Training datasets
To determine threshold values using a procedure based on the one proposed by Besse et al. [17], the main condition is to dispose of environmental data acquired on disconnected sites that integrate a diversity of environmental toxicity conditions. For this purpose, threshold values of mussels and gammarid biomarkers were established from data sets acquired at sites belonging to the monitoring network of French water agencies (Fig. 1). More specifically, these datasets were obtained from active biomonitoring campaigns conducted within two distinct research projects performed in 2017 and 2018: the "EQUAL" project for D. polymorpha and the "AFB48" project for G. fossarum. For each biomonitoring surveys, mussels and gammarids were caged for 2 months and 1 week, respectively, according to a standardized caging methodology described in previous studies [38]. For each investigated site, physicochemical parameters (pH, temperature, dissolved oxygen and conductivity) of water were monitored throughout the experiments.
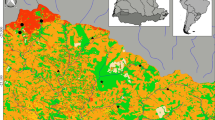
Distribution of caging sites monitored within the project “EQUAL” for D. polymorpha, and the project “AFB48” for G. fossarum. The blue triangles correspond to the 23 caging sites of zebra mussels (EQUAL) while red dots represent the 56 environmental sites where gammarids were caged (AFB48). The geographical boundaries on the map represent the boundaries of the six French regional environmental agencies: Adour Garonne (AG), Artois-Picardie (AP), Loire-Bretagne (LB), Rhin-Meuse (RM), Rhône Méditerranée-Corse (RMC) and Seine-Normandie (SN)
Within the “EQUAL” project, more than 200 mussels were caged in 23 sites monitored by the French water agency of the Rhin-Meuse basin (RM) (Fig. 1). In consultation with water operators, caging sites were selected: (i) to be representative of the diversity of the water bodies found in the Rhin-Meuse basin, and (ii) to consider different types of anthropogenic pressure, including agricultural and urban discharges. After 2 months of exposure, hemolymph and gland digestives from 10 mussels were sampled for cellular and molecular analysis, respectively, and 9 pools of 3 digestive glands were sampled for measuring energy reserves (lipids, glucose, glycogen and protein contents) and enzymatic assays (the remaining organisms were used in another study). In the AFB48 project, more than 250 male gammarids were caged for 7 days in 56 environmental stations: (i) distributed over the French territory, and (ii) monitored by the six regional French regional water agencies to implement WFD requirements (Fig. 1). After 7 days of exposure, 5 gammarids per site were randomly sampled, weighed, frozen in liquid nitrogen, and stored at – 80 °C for proteomic investigations. Remaining gammarids were used in another study for feeding assays and acetylcholinesterase (AChE) assays.
Case study
As a case study, threshold values were applied to a biomarker dataset obtained from the Interreg France-Wallonie-Vlaanderen program DIADeM (“Development of an Integrated Approach for the Diagnosis of the water quality of the Meuse River”). In this collaborative European project, zebra mussels and gammarids were caged upstream and downstream of several WWTPs located in four cities of the French–Belgian Meuse River basin (Fig. 2). More specifically, mussels were caged for two months at up- and downstream WWTPs of three cities (Namur, Charleville Mézières and Charleroi) while gammarids were caged up- and downstream WWTPs of Namur and Bouillon. As detailed before, mussels and gammarids were sampled after 2 months and one week of exposure, respectively, for biomarker investigations. For D. polymorpha, energy reserves and enzyme activities were analyzed from 9 pools of 3 digestive glands while 10 digestive glands and 10 hemolymph fractions were sampled for molecular and cellular analysis, respectively. For gammarids, 10 organisms per caging site were collected, weighed and frozen until proteomic investigations.
Environmental sites investigated within the DIADeM project. While mussels were caged up and downstream WWTPs of Namur, Charleroi and Charleville Mézières, gammarids were exclusively caged up- and downstream WWTPs of Namur and Bouillon (the figure modified from Catteau et al. [14])
Biomarker measurements
Biomarkers in Dreissena polymorpha
A panel of biochemical, cellular and molecular biomarkers related to the energy metabolism, immune system and general stress response was measured in different tissues of zebra mussels (Table 1). As described in Hani et al. [39], energy acquisition (Ea) was estimated by measuring proteins, lipids, carbohydrates (glycogen and glucose) contents in whole tissues of zebra mussels while energy consumption (Ec) was estimated by measuring the activity of the electron transport system (ETS). Then, an integrative biomarker (CEA: cellular energy allocation) was calculated, according to the following formula: [CEA = Ea/Ec] [40]. Still linked to the energy metabolism, several digestive enzyme activities (amylase, lipase, acid phosphatase “PAC”, alanine aminotransferase “ALAT”, lipase and lactate dehydrogenase “LDH”) were monitored in the digestive glands of mussels using a standardized protocol described in Hani et al. [39].
For the general stress response in mussels, both phenoloxidase (PO) activity and glutathione-s-transferase activity (GST) were monitored in digestive glands as described in previous studies [42, 43]. In addition to these biochemical biomarkers, gene expressions of the cytochrome C oxidase (Cox) and the mitochondrial ATP synthase (ATPase) were measured by RT-qPCR in the digestive glands of D. polymorpha as performed in Louis et al. [44]. Finally, after withdrawing hemolymph from the mussel posterior muscle, phagocytic efficiency of hemocytes was assessed by flow cytometry (Accuri™ C6 flow cytometer, Becton Dickinson) following the protocol described in Barjhoux et al. [41].
Protein biomarkers in Gammarus fossarum
A total of 32 peptides related to 23 proteins involved in reproduction, osmoregulation, molting processes and general stress response (Additional file 1: Table S1) were quantified by dynamic MRM (dMRM) in male gammarids using a standardized protocol validated by Faugere et al. [45]. Briefly, after protein extraction and digestion, 32 heavy-labeled peptides were spiked in samples with the same quantities. Samples were then purified and concentrated by solid-phase extraction (SPE) prior to LC–MS/MS analysis. Peptide solutions were analyzed using an Agilent 1290 Infinity II High Performance Liquid Chromatography (HPLC) system coupled to an Agilent 6495B triple quadrupole mass spectrometer (Agilent Technologies, Waldbronn, Germany) in dMRM mode with the same parameters as detailed by Faugere et al. [45].
Quantitative data of targeted peptides were estimated after automatic and manual integration of chromatographic peak areas using MassHunter Quantitative Analysis software (version B.09.00; Agilent Technologies). Peptides were quantified thanks to their most intense and the least interfered MRM transition. Peptide concentrations in gammarid samples were calculated using the endogenous/labeled peptide peak area ratio, since labeled peptides were spiked in samples at the same concentration (4 µg/mL). Except for the definition of threshold values, peptides quantified below their limit of quantification (LOQ) were replaced by LOQ concentrations defined in previous studies for each targeted peptide.
Statistical approach
Correlation between biomarkers and confounding factors
Pearson's correlation tests were performed to evaluate the effects of confounding factors (temperature, conductivity, pH and dissolved oxygen levels) on biomarker modulations. Thanks to the R package “ggcorrplot”, correlation matrices were performed to pinpoint the most correlated variables. Then, correlation details between biomarkers and confounding factors were inspected visually using scatter plots. Two variables were considered correlated with a correlation coefficient (R) greater than |0.5| and a p value lower than 0.05 according to Pearson’s correlation tests.
Definition of threshold values
Determination of biomarker threshold values was conducted using mussel and gammarid biomarker datasets obtained from the “EQUAL” and “AFB48” projects, respectively. For each biomarker not influenced by confounding factors and measured in at least 70% of samples, threshold values were statistically defined using a methodology adapted from Besse et al. [17] (only values higher than the LOQ were retained). This methodology assumes that the natural variability of a biomarker, for organisms living in a “healthy” environment without pollution, combined with noise would be characterized by a Gaussian distribution. The methodology was initially developed to determine bioaccumulation thresholds: for a given substance, all bioaccumulation data observed in organisms exposed at different sites were sorted from the lowest to the highest value. A Shapiro–Wilk test was applied on the overall dataset to test if the distribution is Gaussian. If not, the highest value was removed from the dataset and the normality tested again. This iterative process was stopped when the dataset was distributed according to a Gaussian law. The threshold value was thus defined by the Gaussian distribution obtained at the end as the 95th percentile (Fig. 3). Using a home-made R script, this original approach was improved by adding bootstrapping and adapted to biomarkers that could be inhibited by a pollutant (and for which an inhibition threshold should be defined) and to biomarkers that could be induced and inhibited (and for which both induction and inhibition thresholds should be defined) (Table 1).
Pipeline for the determination of threshold values of D. polymorpha and G. fossarum biomarkers. For well-measured biomarkers, and not influenced by confounding factors, induction (green) and/or inhibition (red) thresholds were defined according to the methodology adapted from Besse et al. [17]
For biomarkers that are only induced, the methodology used to define an induction threshold was: (i) from the complete initial dataset, a random sample of n = 30 values was realized, sorted from the lowest to the highest value and the approach proposed by Besse et al. [17] described above was applied to obtain an induction threshold value; (ii) this procedure was repeated z = 1000 times to deduce, from the z data samples, z threshold values; and (iii) finally, the estimated threshold value corresponded to the mean of the z threshold values.
For biomarkers that are only inhibited, the methodology used to define an inhibition threshold was: (i) from the complete initial dataset, a random sample of n = 30 values was realized, sorted here from the highest to the lowest value. Then, a Shapiro–Wilk test was realized and if the Gaussian distribution was not verified, it was in this case the lowest value that was removed from the data. The normality was tested again and the iterative process was run until obtaining a Gaussian distribution from which an inhibition threshold was defined as the 95th percentile; (ii) as before, this procedure was repeated z = 1000 times to deduce, from the z data samples, z inhibition threshold values; and (iii) finally, the inhibition threshold value retained corresponded to the mean of the z threshold values.
For biomarkers that could be induced and inhibited, we used the two methods previously described to separately estimate the inhibition and the induction thresholds. The issue here is to define a methodology to separate the “inhibition dataset” to the induction one. From the whole data sorted from the lowest to the highest value, we defined: (i) the “induction dataset” by eliminating the lowest values constituted by the q = 35 first percentiles; and (ii) the “inhibition dataset” by eliminating the highest values constituted by the q = 35 last percentiles. Then, the methodology described above to determine an induction threshold was applied on the induction dataset and the one used to determine an inhibition threshold was applied on the inhibition dataset.
Note that we tested different values for n, z and q and those retained for all datasets ensure the robustness of the results. The distribution of biomarker data and their defined threshold values (inhibition and/or induction thresholds) are illustrated in Additional file 5: Fig. S1.
Identification of modulated biomarkers using threshold values
Threshold values were applied to biomarker datasets obtained from the DIADeM project to reveal biomarkers affected in mussels and gammarids caged up- and down-stream several WWTPs. Thanks to these thresholds, a biomarker was considered impacted within a specific environmental site when more than 50% of values were observed below or above its inhibition and/or induction thresholds, respectively.
Results and discussion
Definition of threshold values specific to D. polymorpha and G. fossarum biomarkers
The definition of threshold values requires data representative of spatial and temporal variations encountered in the natural environment. In this study, active biomonitoring surveys conducted at the Rhine-Meuse basin (EQUAL) and the national (AFB48) scales provided a large number of environmental data regarding the biological responses of D. polymorpha and G. fossarum. More specifically the panel of biochemical, cellular and molecular biomarkers measured in mussels and gammarids caged in sites representative of the diversity of French water bodies, constitutes an ideal data set to demonstrate the possibility of estimating relevant threshold values. Caging experiments performed within the “EQUAL” project resulted in the acquisition of more than 180 data of biomarkers responses acquired from zebra mussels transplanted in 23 environmental sites (Additional file 2: Table S2). In the AFB48 project, 32 peptides related to 21 proteins were quantified by dMRM in hundreds of male gammarids caged in 55 environmental sites distributed over the French territory, resulting in the acquisition of more than 270 recorded data per peptide (Additional file 3: Table S3).
Using Pearson’s correlation tests, we first made sure that biomarker fluctuations were not influenced by temperature, conductivity, pH and dO2 levels found in freshwater environments (Additional file 6: Fig. S2 and Additional file 7: Fig. S3). No linear correlation was observed between gammarid peptides and confounding factors. Correlation coefficients (R) between peptide concentrations and confounding factors ranged from − 0.16 to 0.31. The peptide “GIDIIGDAFEADR” and the temperature were the most correlated variables with an R of 0.31. However, using scatterplot visualization, no clear correlation was observed (Additional file 6: Fig. S2). Higher correlations were observed between mussel biomarkers and confounding factors with R values ranged from − 0.59 to 0.54 (Additional file 7: Fig. S3). More specifically, several biomarkers linked to energy metabolism were found correlated with some confounding factors with R values greater than |0.5| and a p value lower than 0.05 according to Pearson’s correlation tests. It included LDH activity, positively correlated to the temperature with an R of 0.54 as well as glycogen content, ETS and PAC activity negatively correlated to dO2 levels with R values lower than − 0.5. Scatter plots showed that extreme values of glycogen content, ETS and LDH activity were observed at extreme levels of temperature and dO2, supporting the hypothesis that these biomarkers are influenced by confounding factors. The methodology of Besse et al. [17] is not adapted for defining threshold values of biomarker measurements correlated with confounding environmental factors. This is because the resulting inflated variability of biomarker levels impedes a clear discrimination of a statistical group of reference data from marginal levels due to the sole influence of contamination. Such a challenge first requires to better characterize the impact of confounding factors in biomarker modulation through laboratory-controlled experiments [31, 35]. Then normalized data can be analyzed by the methodology based on the Gaussian hypothesis, as exemplified by feeding rate data [37]. For our demonstration exercise, threshold values were not defined for mussel biomarkers suspected of being influenced by confounding factors, which included biomarkers related to the energy reserves, LDH and PAC activities (Fig. 3, Additional file 4: Table S4).
For other biomarkers, thresholds were defined using the methodology adapted from Besse et al. [17], which relies on the normal distribution of data to statistically define threshold values (Fig. 3). Contaminants can positively or negatively affect biological responses of sentinel species, therefore, induction and/or inhibition thresholds were defined depending on the nature of tested biomarkers and their implication in response to environmental pollution (Fig. 3). Overall, threshold values could be established for 8 biochemical, cellular and molecular biomarkers monitored in D. polymorpha (Fig. 3, Additional file 4: Table S4) and 25 peptide biomarkers quantified in G. fossarum (Fig. 3, Additional file 1: Table S1). More specifically, inhibition thresholds could be defined for 4 biomarkers of D. polymorpha and 25 peptides of G. fossarum, with 3–13% of data observed under the inhibition thresholds (Fig. 3). Induction thresholds were defined for 7 biomarkers of D. polymorpha and 24 peptides of G. fossarum, with 7–27% of data measured above the induction thresholds (Fig. 3).
For D. polymorpha, phagocytosis efficiency of hemocytes included exclusively inhibition threshold (Table 1), since previous ecotoxicological studies have shown that chemical pollutants can induce immunotoxic effects on wildlife species by decreasing their phagocytosis capacity [46]. Furthermore, according to Le Guernic et al. [47], a decrease in beads phagocytosis activities of D. polymorpha hemocytes may reflect the stimulation of the immune response during microbial infection. Conversely, only induction thresholds were defined for gene expressions (Cox, ATPase) and PO enzymatic activities monitored in the digestive glands of D. polymorpha (Table 1). The Cox and ATPase genes are key molecular players in cellular energy production and are generally induced during cellular stress to supply the energy needed to cope with pollutants [14, 44, 48]. Involved in detoxification, antioxidant defense and immune processes, PO enzymes are emerging as valuable biomarkers for tracking environmental pollution. Several laboratory and field studies observed an increase in PO activities when aquatic invertebrates were exposed to pollutants, including in D. polymorpha [49,50,51]. Finally, both inhibition and induction thresholds were set for digestive enzyme activities (lipase, amylase, ASAT) monitored in the digestive gland of D. polymorpha. The activities of digestive enzymes depend on different parameters (intrinsic hydrolytic properties, rates of synthesis, rates of secretion for enzymes for extracellular activity) on which a pollutant is likely to have an effect by interacting directly or indirectly with the protein. As pinpointed by previous research studies, pollutants found in the environment may promote the activation of digestive enzymes while many other pollutants impaired by inhibiting the activity of digestive enzymes [52].
Under stressful conditions, the synthesis of specific proteins can be boosted to support an appropriate biological response to pollutants. In contrast, pollutants and other cellular stress can damage proteins, leading to their degradation. Thus, both inhibition and induction thresholds were defined for 24 peptides well detected and quantified in gammarid samples (Fig. 3). These ready-to-use peptide biomarkers were linked to several vital functions of G. fossarum, including osmoregulation, molting processes, and general stress response, likely impacted by the environmental contamination [53, 54]. In contrast, thresholds could not be defined for 7 peptides detected below the LOQ in more than 70% of gammarids (Additional file 1: Table S1) since the methodology proposed here requires a minimum amount of data for defining a Gaussian distribution and then determine threshold values. Not surprisingly, most of these peptides were related to female yolk proteins, such as vitellogenin-like proteins (Additional file 1: Table S1). Given their implication in vitellogenesis processes, VTGs are normally found in female organisms. However, their detection in male gammarids may indicate a stressful situation induce by environmental pollution since VTGs are also believed to play roles in antioxidant and immune defenses [55]. Thus, the simple presence of peptides related to VTGs in male gammarids could be indicative of pollution-induced stress and LOQs established for these peptides could be set as induction thresholds. Other peptides detected at low levels in male gammarids were related to three proteins involved in molting processes (Cytochrome P450, JHE-like carboxylesterase) and general stress response (hemocytin). Although these peptides may be present in too low abundance in the samples, they may not respond sensitively to MRM instruments or may be subject to post-translational modifications, rendering them undetectable [56]. For example, in this study, two peptides (“APILEGYFSK”, “VPAILESFPGR”) of a PO-like protein were quantified in G. fossarum by MRM. While the peptide “APILEGYFSK” is located in the central part of the PO protein, the peptide "VPAILESFPGR" is found at the end of the N-terminal domain, which is actually considered as the cleavage site of pro-PO enzymes [57] (Additional file 8: Fig. S4). Therefore, the quantification of the peptide "APILEGYFSK" may reflect the true abundance of PO in the sample whereas the depletion of the peptide “VPAILESFPGR” may indicate the cleavage and activation of PO enzymes. For this reason, both induction and inhibition thresholds were proposed for the peptide “APILEGYFSK”, but only inhibition threshold was defined for the peptide “VPAILESFPGR”.
Biomarker-based diagnosis using threshold values
Once defined, the threshold values were used to assess biomarker modulations in mussels and/or gammarids caged upstream and downstream of 4 WWTPs (DIADeM project). Several biomarkers were impacted in zebra mussels caged in Namur, Charleroi and Charleville Mézières (Fig. 4). Out of the 8 biomarkers with defined threshold values, 6 biomarkers were considered modulated with more than 50% of data below the inhibition or above the induction thresholds in at least one environmental site. A higher number of biomarkers were considered impacted in mussels caged at Charleroi compared to other cities, with four biomarkers (PO, amylase, ASAT activities and Cox gene expression) observed above induction thresholds in mussels caged upstream and downstream the WWTP (Fig. 4). In all sites, some biomarkers were modulated in mussels caged downstream the WWTPs but not in mussels caged upstream the WWTPs, highlighting the adverse effects of WWTP discharges on aquatic wildlife. For example, Cox gene expression and amylase activity were measured above the induction thresholds only in mussels caged downstream of WWTPs of Charleville Mézière and Namur, respectively (Fig. 4). Similarly, phagocytosis efficiency was observed below the inhibition threshold exclusively in mussels caged downstream the WWTP of Charleroi, revealing a potential microbial or immunotoxic pollution released by the WWTP [58, 59] (Fig. 4). Several biomarkers were also considered impacted in mussels caged upstream the WWTPs, indicating a potential contamination not related to the discharges of WWTPs. For example, ASAT and lipase activities were observed above the induction thresholds in more than 50% of mussels caged upstream and downstream the WWTP of Namur (Fig. 4). At Charleville Mézière, the amylase activity of mussels was modulated in mussels caged upstream the WWTP while no modulation was observed in mussels caged downstream the WWTP (Fig. 4).
Phenoloxidase activity (A) Cox gene expression (B), amylase activity (C), ASAT activity (D), lipase activity (E) and phagocytosis efficiency (F) measured in zebra mussels caged up- and down-stream the WWTPs of Charleroi, Charleville Mézières and Namur. Red lines represent the inhibition thresholds while green lines represent induction thresholds defined for each biomarker
Among the 26 peptide biomarkers quantified in G. fossarum, only one peptide was observed out of threshold values and was considered as impacted. More specifically, the peptide "VPAILESFPGR" was found at concentrations below the inhibition threshold in 50% and 90% of gammarids caged upstream the WWTP of Bouillon and downstream the WWTP of Namur, respectively (Fig. 5). As detailed before two peptides from the cleavage site and the core part of a PO enzyme were targeted by MRM. The core peptide “APILEGYFSK” was found in similar concentrations among caged gammarids (Fig. 5), suggesting that PO is present in the same quantity. As discussed before, the modulated peptide “VPAILESFPGR” is located at the cleavage site of the pro-PO and its loss of detection in MRM assays may indicate the cleavage of Pro-PO to PO. Thus, the decrease of "VPAILESFPGR" concentrations observed in gammarids caged upstream of Bouillon-WWTP and downstream of Namur-WWTP suggests an activation of PO enzymes in these sites.
Concentrations of the PO-related peptides VPAILESFPGR (A) and APILEGYFSK (B) quantified in gammarids caged up- and down-stream the WWTPs of Bouillon and Namur. Red lines represent the inhibition thresholds defined for the two peptides while green line represents the induction threshold for APILEGYFSK
Overall, these results demonstrate the relevance of implementing threshold values for the application of biomarkers in freshwater biomonitoring surveys. The implementation of threshold values, defined from calibrated organisms, allowed the comparison of sites disconnected from each other without the need for a related a priori reference site. In contrast to the classical upstream/downstream impact study approaches, the implementation of threshold values improves the diagnosis of water quality by offering the possibility to assess the quality of upstream sites and making it possible to better characterize the impact of discharges at specific sites. Indeed, without the use of threshold values, the degraded quality at the upstream sites of the Namur and Charleroi WWTPs (ASAT, lipase and amylase) could not have been detected and their use as reference sites to investigate the contamination downstream of the WWTPs would lead to misinterpretations. This statement is consistent with the observations made by Chaumot et al. [37] on AChE activity measured in caged gammarids. Similarly, in female gammarids caged in situ, the definition and application of threshold values provided the opportunity to discriminate molt modulations related to the presence of contaminants from those related to temperature variations among investigated sites [34]. The results acquired on mussels and gammarids were not comparable due to the different biomarkers tested and the different exposure times. However, given the large number of biomarkers impacted in mussels caged both up- and downstream of the WWTP, biomarker investigation suggests that Charleroi sites are the most impacted ones by human activities. This hypothesis agrees with the chemical contamination profiles since Catteau et al. [14], which showed that the highest concentrations of pharmaceutical and domestic pollutants were found in Charleroi site. The biomarker-based diagnostic using thresholds also demonstrates the adverse impacts of WWTP effluents on freshwater invertebrates. In particular, biomarkers linked to the energy metabolism (amylase activity) for mussels and the general stress response (PO activity) for gammarids were modulated downstream the WWTP of Namur while no modulation was noted in animals caged upstream the WWTP.
Conclusions
Thanks to several environmental datasets representative of French water bodies, this study demonstrates the possibility of proposing threshold values for a wide range of biochemical, cellular and molecular biomarkers measured in two freshwater sentinel species, D. polymorpha and G. fossarum. Apart from the biomarkers suspected of being influenced by abiotic confounders, inhibition and/or induction thresholds were defined according to existing literature knowledge regarding their modulation profiles against pollutants. Subsequently, the application of these threshold values proved to be particularly relevant for diagnosing the impact of pollution in upstream and downstream several WWTPs of the Rhin-Meuse basin. Without the need for "reference sites", the biomarkers found abnormally modulated in caged animals made it possible to identify environmental sites impacted by environmental pollution and evaluate the effect of WWTPs on the biology of freshwater invertebrates.
For mussel biomarkers suspected sensitive to temperature and dO2, future investigations will be required to integrate the influence of these confounding factors into the definition of threshold values. Furthermore, it should be mentioned that threshold values defined in this study should be validated and/or refined by further investigations. Indeed, much more data you have along the gradient of possible modulation for a biomarker, much more relevant and precise will be the thresholds. Once validated, thresholds defined in situ by the methodology proposed in this study will offer the possibility to compare biological data collected from a multitude of environmental sites and sampling dates. Finally, the threshold values specific to each biomarker could be used by water operators to improve and facilitate the diagnosis of environmental pollution within freshwater biomonitoring surveys.
Availability of data and materials
Not applicable.
Abbreviations
- AChE:
-
Acetylcholinesterase
- ALAT:
-
Alanine aminotransferase
- ATPase:
-
ATP synthase
- BAC:
-
Background assessment criteria
- Cox:
-
Cytochrome C oxidase
- dMRM:
-
Dynamic multiple reaction monitoring
- Ea:
-
Energy acquisition
- EAC:
-
Ecotoxicological assessment criteria
- Ec:
-
Energy consumption
- ECs:
-
Emerging contaminants
- ETS:
-
Electron transport system
- GST:
-
Glutathione S-transferase
- HPLC:
-
High-performance liquid chromatography
- ICES:
-
International Council for Exploration of the Sea
- LDH:
-
Lipase and lactate dehydrogenase
- LOQ:
-
Limit of quantification
- MRM:
-
Multiple reaction monitoring
- MS:
-
Mass spectrometry
- OSPAR:
-
Oslo–Paris Commission
- PAC:
-
Acid phosphatase
- PO:
-
Phenoloxidase
- SPE:
-
Solid-phase extraction
- WFD:
-
Water framework directive
- WWTPs:
-
Wastewater treatment plants
References
Schwarzenbach RP, Escher BI, Fenner K et al (2006) The challenge of micropollutants in aquatic systems. Science 313:1072–1077. https://doi.org/10.1126/science.1127291
Sauvé S, Desrosiers M (2014) A review of what is an emerging contaminant. Chem Cent J 8:15. https://doi.org/10.1186/1752-153X-8-15
Sivaranjanee R, Kumar PS (2021) A review on remedial measures for effective separation of emerging contaminants from wastewater. Environ Technol Innov 23:101741. https://doi.org/10.1016/j.eti.2021.101741
Mostert E (2003) The European water framework directive and water management research. Phys Chem Earth Parts ABC 28:523–527. https://doi.org/10.1016/S1474-7065(03)00089-5
Roberta B, Benedetta P, Silvia Q (2014) An ecotoxicological approach to assess the environmental quality of freshwater basins: a possible implementation of the EU water framework directive? Environments 1:92–106. https://doi.org/10.3390/environments1010092
Altenburger R, Ait-Aissa S, Antczak P et al (2015) Future water quality monitoring—adapting tools to deal with mixtures of pollutants in water resource management. Sci Total Environ 512–513:540–551. https://doi.org/10.1016/j.scitotenv.2014.12.057
Kortenkamp A, Faust M, Backhaus T et al (2019) Mixture risks threaten water quality: the European Collaborative Project SOLUTIONS recommends changes to the WFD and better coordination across all pieces of European chemicals legislation to improve protection from exposure of the aquatic environment to multiple pollutants. Environ Sci Eur 31:69. https://doi.org/10.1186/s12302-019-0245-6
Gonzalez P, Pierron F (2015) Omics in aquatic ecotoxicology: the ultimate response to biological questions? In: Amiard-Triquet C, Amiard J-C, Mouneyrac C (eds) Aquatic ecotoxicology. Academic Press, Cambridge, pp 183–203
Milinkovitch T, Geffard O, Geffard A et al (2019) Biomarkers as tools for monitoring within the water framework directive context: concept, opinions and advancement of expertise. Environ Sci Pollut Res 26:32759–32763. https://doi.org/10.1007/s11356-019-06434-x
Wernersson A-S, Carere M, Maggi C et al (2015) The European technical report on aquatic effect-based monitoring tools under the water framework directive. Environ Sci Eur 27:7. https://doi.org/10.1186/s12302-015-0039-4
Schmitz M, Deutschmann B, Markert N et al (2022) Demonstration of an aggregated biomarker response approach to assess the impact of point and diffuse contaminant sources in feral fish in a small river case study. Sci Total Environ 804:150020. https://doi.org/10.1016/j.scitotenv.2021.150020
Catteau A, Bado-Nilles A, Beaudouin R et al (2021) Water quality of the Meuse watershed assessment using a multi-biomarker approach with caged three-spined stickleback (Gasterosteus aculeatus L.). Ecotoxicol Environ Saf 208:111407. https://doi.org/10.1016/j.ecoenv.2020.111407
Lomartire S, Marques JC, Gonçalves AMM (2021) Biomarkers based tools to assess environmental and chemical stressors in aquatic systems. Ecol Indic 122:107207. https://doi.org/10.1016/j.ecolind.2020.107207
Catteau A, Porcher J-M, Bado-Nilles A et al (2021) Interest of a multispecies approach in active biomonitoring: application in the Meuse watershed. Sci Total Environ. https://doi.org/10.1016/j.scitotenv.2021.152148
Stepien C, Grigorovich I, Gray M et al (2013) Evolutionary, biogeographic, and population genetic relationships of Dreissenid mussels, with revision of component taxa. In: Nalepa TF, Schloesser DW (eds) Quagga and zebra mussels, 2nd edn. CRC Press, Boca Raton, pp 403–444
Wattier R, Mamos T, Copilaş-Ciocianu D et al (2020) Continental-scale patterns of hyper-cryptic diversity within the freshwater model taxon Gammarus fossarum (Crustacea, Amphipoda). Sci Rep 10:16536. https://doi.org/10.1038/s41598-020-73739-0
Besse J-P, Coquery M, Lopes C et al (2013) Caged Gammarus fossarum (Crustacea) as a robust tool for the characterization of bioavailable contamination levels in continental waters: towards the determination of threshold values. Water Res 47:650–660. https://doi.org/10.1016/j.watres.2012.10.024
Bervoets L, Voets J, Covaci A et al (2005) Use of transplanted zebra mussels (Dreissena polymorpha) to assess the bioavailability of microcontaminants in Flemish surface waters. Environ Sci Technol 39:1492–1505. https://doi.org/10.1021/es049048t
Nalepa TF, Schloesser DW (1992) Zebra mussels biology, impacts, and control. CRC Press, Boca Raton
Devin S, Beisel J-N (2007) Biological and ecological characteristics of invasive species: a gammarid study. Biol Invasions 9:13–24. https://doi.org/10.1007/s10530-006-9001-0
Binelli A, Della Torre C, Magni S, Parlini M (2015) Does zebra mussel (Dreissena polymorpha) represent the freshwater counterpart of Mytilus in ecotoxicological studies? A critical review. Environ Pollut Barking Essex 196:386–403. https://doi.org/10.1016/j.envpol.2014.10.023
Ladeiro MP, Barjhoux I, Bigot-Clivot A et al (2017) Mussel as a tool to define continental watershed quality. IntechOpen, London
Charnot A, Gouveia D, Armengaud J et al (2017) Multiplexed assay for protein quantitation in the invertebrate Gammarus fossarum by liquid chromatography coupled to tandem mass spectrometry. Anal Bioanal Chem 409:3969–3991. https://doi.org/10.1007/s00216-017-0348-0
Gouveia D, Chaumot A, Charnot A et al (2017) Assessing the relevance of a multiplexed methodology for proteomic biomarker measurement in the invertebrate species Gammarus fossarum: a physiological and ecotoxicological study. Aquat Toxicol 190:199–209. https://doi.org/10.1016/j.aquatox.2017.07.007
Scheil V, Triebskorn R, Köhler H-R (2008) Cellular and stress protein responses to the UV filter 3-benzylidene camphor in the amphipod crustacean Gammarus fossarum (Koch 1835). Arch Environ Contam Toxicol 54:684–689. https://doi.org/10.1007/s00244-007-9072-7
Binelli A, Cogni D, Parolini M, Provini A (2010) Multi-biomarker approach to investigate the state of contamination of the R. Lambro/R. Po confluence (Italy) by zebra mussel (Dreissena polymorpha). Chemosphere 79:518–528. https://doi.org/10.1016/j.chemosphere.2010.02.033
Serdar O (2019) The effect of dimethoate pesticide on some biochemical biomarkers in Gammarus pulex. Environ Sci Pollut Res 26:21905–21914. https://doi.org/10.1007/s11356-019-04629-w
Vethaak AD, Davies IM, Thain JE et al (2017) Integrated indicator framework and methodology for monitoring and assessment of hazardous substances and their effects in the marine environment. Mar Environ Res 124:11–20. https://doi.org/10.1016/j.marenvres.2015.09.010
Giltrap M, Ronan J, Bignell JP et al (2017) Integration of biological effects, fish histopathology and contaminant measurements for the assessment of fish health: a pilot application in Irish marine waters. Mar Environ Res 129:113–132. https://doi.org/10.1016/j.marenvres.2017.04.004
Martínez-Gómez C, Fernández B, Robinson CD et al (2017) Assessing environmental quality status by integrating chemical and biological effect data: the Cartagena coastal zone as a case. Mar Environ Res 124:106–117. https://doi.org/10.1016/j.marenvres.2016.04.008
Coulaud R, Geffard O, Xuereb B et al (2011) In situ feeding assay with Gammarus fossarum (Crustacea): modelling the influence of confounding factors to improve water quality biomonitoring. Water Res 45:6417–6429. https://doi.org/10.1016/j.watres.2011.09.035
Xuereb B, Chaumot A, Mons R et al (2009) Acetylcholinesterase activity in Gammarus fossarum (Crustacea Amphipoda) Intrinsic variability, reference levels, and a reliable tool for field surveys. Aquat Toxicol Amst Neth 93:225–233. https://doi.org/10.1016/j.aquatox.2009.05.006
Charron L, Geffard O, Chaumot A et al (2013) Effect of water quality and confounding factors on digestive enzyme activities in Gammarus fossarum. Environ Sci Pollut Res Int 20:9044–9056. https://doi.org/10.1007/s11356-013-1921-5
Lopes C, Chaumot A, Xuereb B et al (2020) In situ reproductive bioassay with caged Gammarus fossarum (Crustacea): part 2—evaluating the relevance of using a molt cycle temperature-dependent model as a reference to assess toxicity in freshwater monitoring. Environ Toxicol Chem 39:678–691. https://doi.org/10.1002/etc.4656
Marchand A, Tebby C, Beaudouin R et al (2019) Modelling the effect of season, sex, and body size on the three-spined stickleback, Gasterosteus aculeatus, cellular innate immunomarkers: a proposition of laboratory reference ranges. Sci Total Environ 648:337–349. https://doi.org/10.1016/j.scitotenv.2018.07.381
Babut M, Ferrari BJD, Jame P et al (2020) Monitoring priority substances in biota under the Water Framework Directive: how effective is a tiered approach based on caged invertebrates? A proof-of-concept study targeting PFOS in French rivers. Environ Sci Eur 32:131. https://doi.org/10.1186/s12302-020-00416-4
Chaumot A, Geffard O, Armengaud J, Maltby L (2015) Chapter 11—gammarids as reference species for freshwater monitoring. In: Amiard-Triquet C, Amiard J-C, Mouneyrac C (eds) Aquatic ecotoxicology. Academic Press, Cambridge, pp 253–280
Alric B, Geffard O, Chandesris A et al (2019) Multisubstance indicators based on caged Gammarus bioaccumulation reveal the influence of chemical contamination on stream macroinvertebrate abundances across France. Environ Sci Technol 53:5906–5915. https://doi.org/10.1021/acs.est.9b01271
Hani YMI, Prud’Homme SM, Nuzillard J-M et al (2021) 1H-NMR metabolomics profiling of zebra mussel (Dreissena polymorpha): a field-scale monitoring tool in ecotoxicological studies. Environ Pollut Barking Essex 1987(270):116048. https://doi.org/10.1016/j.envpol.2020.116048
Verslycke T, Vercauteren J, Devos C et al (2003) Cellular energy allocation in the estuarine mysid shrimp Neomysis integer (Crustacea: Mysidacea) following tributyltin exposure. J Exp Mar Biol Ecol 288:167–179. https://doi.org/10.1016/S0022-0981(03)00006-6
Barjhoux I, Rioult D, Geffard A, Palos Ladeiro M (2020) A new protocol for the simultaneous flow cytometric analysis of cytotoxicity and immunotoxicity on zebra mussel (Dreissena polymorpha) hemocytes. Fish Shellfish Immunol 98:224–235. https://doi.org/10.1016/j.fsi.2019.12.092
Janssens L, Stoks R (2014) Non-pathogenic aquatic bacteria activate the immune system and increase predation risk in damselfly larvae. Freshw Biol 59:417–426. https://doi.org/10.1111/fwb.12274
Habig WH, Pabst MJ, Jakoby WB (1976) Glutathione S-transferase AA from rat liver. Arch Biochem Biophys 175:710–716. https://doi.org/10.1016/0003-9861(76)90563-4
Louis F, Rocher B, Barjhoux I et al (2020) Seasonal monitoring of cellular energy metabolism in a sentinel species, Dreissena polymorpha (bivalve): effect of global change? Sci Total Environ 725:138450. https://doi.org/10.1016/j.scitotenv.2020.138450
Faugere J, Gouveia D, Ayciriex S et al (2020) High-multiplexed monitoring of protein biomarkers in the sentinel Gammarus fossarum by targeted scout-MRM assay, a new vision for ecotoxicoproteomics. J Proteomics 226:103901. https://doi.org/10.1016/j.jprot.2020.103901
Fournier M, Cyr D, Blakley B et al (2000) Phagocytosis as a biomarker of immunotoxicity in wildlife species exposed to environmental xenobiotics1. Am Zool 40:412–420. https://doi.org/10.1093/icb/40.3.412
Le Guernic A, Geffard A, Le Foll F, Palos Ladeiro M (2020) Comparison of viability and phagocytic responses of hemocytes withdrawn from the bivalves Mytilus edulis and Dreissena polymorpha, and exposed to human parasitic protozoa. Int J Parasitol 50:75–83. https://doi.org/10.1016/j.ijpara.2019.10.005
Achard-Joris M, Gonzalez P, Marie V et al (2006) Cytochrome c oxidase subunit I gene is up-regulated by cadmium in freshwater and marine bivalves. Biometals 19:237–244. https://doi.org/10.1007/s10534-005-5671-9
Wheelock C, Phillips B, Anderson B et al (2008) Applications of carboxylesterase activity in environmental monitoring and toxicity identification evaluations (TIEs). Rev Environ Contam Toxicol 195:117–178. https://doi.org/10.1007/978-0-387-77030-7_5
Solé M, Rivera-Ingraham G, Freitas R (2018) The use of carboxylesterases as biomarkers of pesticide exposure in bivalves: a methodological approach. Comp Biochem Physiol Part C Toxicol Pharmacol 212:18–24. https://doi.org/10.1016/j.cbpc.2018.06.002
Luna-Acosta A, Breitwieser M, Renault T, Thomas-Guyon H (2017) Recent findings on phenoloxidases in bivalves. Mar Pollut Bull 122:5–16. https://doi.org/10.1016/j.marpolbul.2017.06.031
Dedourge-Geffard O, Palais F, Geffard A, Amiard-Triquet C (2013) Origin of energy metabolism impairments. In: Amiard-Triquet C, Amiard J-C, Rainbow PS (eds) Ecological biomarkers. CRC Press, Boca Raton
Dayras P, Charmantier G, Chaumot A et al (2017) Osmoregulatory responses to cadmium in reference and historically metal contaminated Gammarus fossarum (Crustacea, Amphipoda) populations. Chemosphere 180:412–422. https://doi.org/10.1016/j.chemosphere.2017.04.016
Gouveia D, Chaumot A, Charnot A et al (2017) Ecotoxico-proteomics for aquatic environmental monitoring: first in situ application of a new proteomics-based multibiomarker assay using caged amphipods. Environ Sci Technol 51:13417–13426. https://doi.org/10.1021/acs.est.7b03736
Sun C, Zhang S (2015) Immune-relevant and antioxidant activities of vitellogenin and yolk proteins in fish. Nutrients 7:8818–8829. https://doi.org/10.3390/nu7105432
Silva AMN, Vitorino R, Domingues MRM et al (2013) Post-translational modifications and mass spectrometry detection. Free Radic Biol Med 65:925–941. https://doi.org/10.1016/j.freeradbiomed.2013.08.184
Sirikharin R, Söderhäll K, Söderhäll I (2020) The N-terminal peptide generated after activation of prophenoloxidase affects crayfish hematopoiesis. Dev Comp Immunol 108:103687. https://doi.org/10.1016/j.dci.2020.103687
Gagné F, André C, Cejka P et al (2008) Immunotoxic effects on freshwater mussels of a primary-treated wastewater before and after ozonation: a pilot plant study. Ecotoxicol Environ Saf 69:366–373. https://doi.org/10.1016/j.ecoenv.2007.10.027
Akaishi FM, St-Jean SD, Bishay F et al (2007) Immunological responses, histopathological finding and disease resistance of blue mussel (Mytilus edulis) exposed to treated and untreated municipal wastewater. Aquat Toxicol 82:1–14. https://doi.org/10.1016/j.aquatox.2007.01.008
Acknowledgements
The authors thank the mobile environmental flow cytometry core facility (MOBICYTE platform) for the flow cytometry analyses. This work benefitted from the French GDR “Aquatic Ecotoxicology” framework which aims at fostering stimulating scientific discussions and collaborations for more integrative approaches. The authors also thank Rémi Recoura-Massaquant for his help in mapping the caging sites.
Funding
The results of this study were obtained within the scope of EQUAL, a project funded by the Agence de l’Eau Rhin-Meuse and within the framework of the DIADEM project (“Development of an integrated approach for the diagnosis of the water quality of the RiverMeuse”) and was supported by the Interreg France-Wallonia-Vlaanderen Program (European Regional Development Fund-ERDF). This study also received financial support from the program "OFB action DCE-biomarqueurs”. This work has been supported by the APPROve project funded by the ANR (ANR-18-CE34-0013-01).
Author information
Authors and Affiliations
Contributions
ML, CL, AG, OG, DD and AC contributed to the conception and design of the study. MPL, OD, ED, IBO, IBA, PN, AE, MN participated in data acquisition, ML and CL processed and interpreted the data. ML and OG drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Description of G. fossarum peptide biomarkers and threshold values defined for each peptide measured within the project "AFB48". "-" means that thresholds were not determined.
Additional file 2: Table S2.
Data of biomarkers measured in mussels caged in situ within the project Equal and Diadem.
Additional file 3: Table S3.
Data of peptide biomarkers quantified in gammarids caged in situ within the project AFB48 and Diadem.
Additional file 4: Table S4.
Threshold values defined for biomarkers measured in D. polymorpha within the project "EQUAL". "-" means that thresholds could not be determined.
Additional file 5: Figure S1.
Distribution of biomarker data used for defining threshold values. Red lines represent inhibition threshold while green lines represent induction thresholds.

Additional file 6: Figure S2.
Pearson correlations between confounding factors and peptide concentrations measured in gammarids. Figure A shows the Pearson correlation matrix where the green boxed cells highlight the peptides most correlated with confounding factors. Correlation between these variables were further investigated using scatter plot visualizations (B, C, D, E).

Additional file 7: Figure S3.
Pearson correlations between confounding factors and mussel biomarkers. Figure A shows the Pearson correlation matrix where the green boxed cells highlight the biomarkers correlated with an R coefficient greater than 0.5 or less than -0.5 between biomarker and confounding factors. Correlation between these variables were further investigated using scatter plot visualizations (B, C, D, E).

Additional file 8: Figure S4.
Targeted peptides of the gammarid phenoloxidase. The figure describes where targeted peptides are located within the protein sequence and functional domains of the phenoloxidase. Protein sequences and domain annotations were analyzed with interproscan tools.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Leprêtre, M., Geffard, A., Palos Ladeiro, M. et al. Determination of biomarkers threshold values and illustration of their use for the diagnostic in large-scale freshwater biomonitoring surveys. Environ Sci Eur 34, 115 (2022). https://doi.org/10.1186/s12302-022-00692-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12302-022-00692-2