Abstract
The “Green revolution” gene sd1 has been used widely in the breeding of modern rice varieties for over half a century. The application of this gene has increased rice yields and thereby supported a significant proportion of the global population. The use of a single gene, however, has raised concerns in the scientific community regarding its durability, especially given the bottleneck in genetic background and the need for large input of fertilizer. New dwarfing or semi-dwarfing genes are needed to alleviate our dependence on the sole “Green revolution” gene. In the past few years, several new dwarfing and semi-dwarfing genes as well as their mutants have been reported. Here, we provide an extensive review of the recent discoveries concerning newly identified genes that are potentially useful in rice breeding, including methods employed to create and effectively screen new rice mutants, the phenotypic characteristics of the new dwarfing and semi-dwarfing mutants, potential values of the new dwarfing and semi-dwarfing genes in rice breeding, and potential molecular mechanisms associated with the newly identified genes.
Similar content being viewed by others
Background
Dwarfism is one of the most valuable traits in crop breeding, as it increases lodging resistance, improves crop yields, facilitates mechanical harvesting, and reduces microbe infection in fields (Evans 1993; Tar'an et al. 2003; Li et al. 2011; Xin et al. 2014). During the past half-century, the application of the dwarfing gene Rht in wheat and the semidwarf-1 (sd1) gene in rice triggered the so-called ‘Green Revolution’ in agriculture, resulting in dramatically elevated crop yields. Because of the Green Revolution, humans have largely avoided worldwide starvation during the great population expansion from four to approximately eight billion (Evans 1998; Hedden 2003).
The rice sd1 gene, encoding a gibberellin 20-oxidase (OsGA20ox-2) (Ashikari et al. 2002; Sasaki et al. 2002; Spielmeyer et al. 2002), was originally derived from the Taiwanese variety Dee-Geo-Woo-Gen (DGWG) (Chandler 1968; Ferrero-Serrano et al. 2019). DGWG was first crossed with the Indonesian variety Peta, resulting in the semi-dwarf cultivar IR8. IR8 has produced dramatically higher yields per unit area and has formed the basis for the development of new high-yield, semi-dwarf rice cultivars since the 1960s (Institue 1967; Tomita 2009; Monna et al. 2002). At present, sd1 remains one of the most important genes widely deployed in the world because of its rich polymorphism in nature (Spielmeyer et al. 2002; Asano et al. 2007; Peng et al. 2021).
Such extensive and worldwide use of a single gene will ineluctably result in limitations and potential problems. The situation has created a bottleneck for developing new rice varieties (Asano et al. 2009). To date, the rice yield potential (Evans 1993) has stagnated after the initial great leap in the ‘Green Revolution’ and the development of hybrid technology (Peng et al. 2008; Siddiq and Vemireddy 2021). The genetic uniformity also makes rice vulnerable to genetic erosion and sudden outbreaks of crop pests, diseases, or other abiotic threats (Siddiq and Vemireddy 2021; Asano et al. 2009). In addition, the use of sd1 requires heavy utilization of nitrogen fertilizers to achieve high yields. Constant use of high levels of nitrogen fertilizers threatens the diversity and sustainability of ecosystems and agricultural systems. Widespread and extensive use of nitrogen fertilizers also results in a huge increase in production costs (Wang et al. 2021a; Li et al. 2021). Currently, the nitrogen input of rice with the sd1 gene has been decreased by regulation of GROWTH-REGULATING FACTOR 4 (GRF4) in the laboratory (Li et al. 2018), but the practical application of GRF4 in the field is still being developed. GRF4 could simultaneously increase rice storage capacity and decrease seed shattering, both of which are highly desirable in modern rice breeding (Sun et al. 2016). Furthermore, the increasing rice-growing area with cultivars containing sd1 evokes another problem that of genetic erosion (Frankel 1970; Khoury et al. 2022; Harlan 1975). Recently, gene editing based on the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein9 (Cas9) system has been used to edit SD1 in rice cultivars. This new method can relieve genetic erosion in modern rice varieties, but more investigation is needed before the CRISPR/Cas9 system can be applied in rice molecular breeding (Chen et al. 2019; Hu et al. 2019; Biswas et al. 2020).
To overcome these limitations and potential problems, we need to enrich the dwarfing (semi-dwarfing) gene pool and identify additional dwarf and semi-dwarf genes that are suitable for practical use in rice breeding (Asano et al. 2009; Liu et al. 2018). A great deal of effort has been put into searching for new rice dwarf (semi-dwarf) mutants through various methods, but to date limited progress has been achieved (Peng et al. 2021). Here, we summarize recent advances on new dwarf (semi-dwarf) mutants and genes in the literature. We focus on the methods used to identify new mutants, their phenotypic characteristics including cytological characteristics, the potential value in rice breeding, and potential molecular mechanisms associated with the newly identified genes.
Methods of Identifying New Dwarfing/Semi-dwarfing Mutants
Generation of artificial mutations is considered as one of the most effective methods to discover new genetic resources for practical breeding. Rice artificial mutants can be generated by chemical reagents or radiation. Mutants Slr1-d1, Slr1-d2, Slr1-d3, Slr1-d4, D-h, and Tid1 resulted from treatment of rice with MNU (Asano et al. 2009, Hirano et al. 2010, KOH and HEU 1993, Sunohara et al. 2009). Slr1-d5 was generated using ethyl methane sulfonic acid (EMS) (Zhang et al. 2016). Physical radiation has also been utilized to produce new rice mutants. KL908 resulted from γ-irradiation (Wei et al. 2006), and Y98149 and Sdd were obtained using low-energy nitrogen ions (Liu et al. 2008a, 2008b). The DMF-1 mutant was obtained by X-ray (Sunohara et al. 2006). Tissue culture is a conventional tool employed to induce mutations (Jiang and Ramachandran 2010). For example, 986083D originated from the anther culture of an autotetraploid indica/japonica hybrid (Qin et al. 2008), and SV14 was obtained through somaclonal variation of a thermo-sensitive male sterile line (Liu et al. 2018). Wild rice is a good source of natural mutants that are useful in breeding (Sakai and Itoh 2010; Zhao et al. 2018). Recently, gene editing using the CRISPR/Cas9 system has also been used for generating mutants and identifying useful genes. Application of this cutting-edge technology has created semi-dwarf mutants slr1-d7, slr1-d8, slr1-d9, slr1-d10, slr1-d11, and slr1-d12 (Jung et al. 2020). Although natural mutation occurs at low frequency in higher plants, large and advanced backcross populations have been employed to produce new rice mutants (Jiang and Ramachandran 2010), including the spontaneous mutants Slr1-d5, Shennong9816d, sdt, and LB4D (Fan et al. 2016; Zhao et al. 2015; Wu et al. 2018; Liang et al. 2011). Researchers have also used space mutagenesis to create new crop germplasm. For example, CHA-2 was screened from the progeny of an indica TXZ13 after space-induced mutations (Guo et al. 2011).
Phenotypic Dwarfism of the Newly Identified Mutants
The rice culm usually includes 4–6 elongated internodes in a mature plant. Dwarf and semi-dwarf plants show various reductions in the internodes. Internodes on rice plants are counted from the top to the base at the mature stage. According to internode elongation patterns, dwarfing rice plants are classified into six groups: N-type, dn-type, sh-type, dm-type, nl-type, and d6-type (Takeda 1974, 1977); these are schematically representated in Fig. 1A. N-type plants elongate normally between all internodes like wild-type plants. All internodes are uniformly reduced in dn-type mutants. Plants of the other four mutant types exhibit reduction in elongation of a specific internode or combinations of nodes. Specifically, sh-type mutants exhibit reduction in the first internode; dm-type mutants exhibit reduction in the second internode. For nl-type mutants, the fourth internode is relatively longer, while the first internode is much shortened. For the d6-type mutants, all of the internodes are shortened except for the first internode (Takeda 1977).
Internode elongation patterns of the newly identified mutants. A Schematic diagram of internode elongation patterns of rice stem (redrawn from Takeda 1977). I: the first internode, shown in white block; II: the second internode, shown in green block; III: the third internode, shown in orange block; IV: the fourth internode, shown in black block; V: the fifth internode, shown in grey block. B The newly identified mutants with different internode elongation patterns
As shown in Fig. 1B, newly identified dwarfing and semi-dwarfing mutants, including SV14, slr1, Y98149, sdt, D-h, KL908, 986083D, and LB4D, are dn-type, with all internodes shortened (Liu et al. 2018, 2008a; Asano et al. 2009; Zhang et al. 2016; Jung et al. 2020; Wu et al. 2018; Zhao et al. 2015; Piao et al. 2014; Wei et al. 2006; Qin et al. 2008). DMF-1 is the only identified mutant that is considered a dm-type, with a shortened second internode (Miura et al. 2009; Wu 1997) while Shennong9816d is considered a d6-type (Fan et al. 2016).
Cytological studies have revealed that internode length reduction in plants with a dwarfing or semi-dwarfing phenotype is due to either shortening of stem cell lengths, reduced numbers of cells, or a combination of both shortened cell lengths and reduced cell numbers (Liu et al. 2018; Jung et al. 2020; Sunohara et al. 2009; Piao et al. 2014; Wu et al. 2018; Liang et al. 2011; Zhou et al. 2013; Zhao et al. 2018). Interestingly, the stem cells of the mutant sdt are actually longer than those of the wild type, and the shortened internodes in this mutant are the result of exclusively reduced cell numbers. Based on these results, the functions of dwarfing-related genes may involve negative regulation of cell growth in the stem (Zhao et al. 2015).
Potential Use of the Newly Identified Dwarfing/Semi-dwarfing Genes
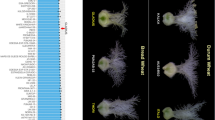
To date, more than 60 dwarf and semi-dwarf mutants have been identified from indica and japonica rice (Futsuhara and Kikuchi 1997; Gaur et al. 2020; Peng et al. 2021). We used both ‘dwarf’ and ‘semi-dwarf’ as the key words to search the Rice Annotation Project Database (https://rapdb.dna.affrc.go.jp/), and we found 66 dwarf and semi-dwarf genes, as shown in Fig. 2 (details in Additional file 1: Table S1). Although these genes are dispersed throughout the rice genome, chromosomes 1, 3, and 6 contain the most genes, with 15, 11, and 10 genes on these chromosomes, respectively. In addition, Ssi1, Dd7, Dx and LB4D were not shown in Fig. 2, because the gene loci have not been verified. Most of the genes identified from the mutants may not be suitable for breeding due to their unfavorable agronomic phenotypes, such as severe dwarf, low fertility, and short grain (Nagai et al. 2018; Wu et al. 2018).
Sixty-six dwarf and semi-dwarf genes obtained from the Rice Annotation Project Database (https://rapdb.dna.affrc.go.jp/). The genes in red are discussed in the following text. The chromosomal locations of these genes are determined using the program Map Gene 2 Chromosome V2 (MG2C) (Chao et al. 2021)
Rice mutant D-h with a semi-dwarf phenotype was obtained from the Korean japonica cultivar Hwachung after being treated with MNU (KOH and HEU, 1993). The mutant phenotype is linked with a 63-bp deletion in the d-h locus flanked by the two STS markers S3197-1 and D-h-3 on the short arm of chromosome 1 (Piao et al. 2014). The D-h gene was predicted to take part in an unknown GA signaling pathway that made the mutant insensitive to GA. The D-h mutant also exhibits both short compact panicles and small round grains, making it unsuitable for hybrid breeding (Piao et al. 2014). However, the dominant dwarf gene can facilitate rice breeding by avoiding using a recessive gene for both male and female parents (Piao et al. 2014; Asano et al. 2009).
X-ray irradiation was also used to induce mutations in the japonica cultivar Fujiminori to identify dwarf mutants. A new dominant semi-dwarf mutant named DMF-1 was created from irradiation (Yamaguchi 1976). A dominant gene named Ssi1 (Short second internode 1) was identified by this method. Ssi1 was mapped to a 7.3-cM region flanked by molecular markers M45 and M26 on the long arm of chromosome 1 (Wu 1997; Miura et al. 2009; Sunohara et al. 2006). A 1.3-Mbp genomic inversion around the Ssi1 locus reduced the second internode elongation in DMF-1 (Sunohara et al. 2006; Miura et al. 2009). The Ssi1 appears to be involved in brassinosteroid (BR) metabolism to reduce the rice plant height (Miura et al. 2009).
The newly identified SLR1 gene with the full name DELLA protein SLENDER RICE1 also functions in gibberellic acid (GA) metabolism. SLR1 acts as a repressor of GA signaling, as SLR1 degradation could trigger most of the GA-associated responses in rice (Hirano et al. 2010). Six alleles of natural Slr1-d mutants have been identified, namely, Slr1-d1, Slr1-d2, Slr1-d3, Slr1-d4, Slr1-d5, and Slr1-d6. These mutants resulted from gain-of-function mutations via amino acid substitutions. Slr1-d1, Slr1-d3, Slr1-d5, and Slr1-d6 have single amino acid substitutions in the TVHNP domain, Slr1-d2 in the DELLA domain, and Slr1-d4 in the GRAS domain (Wu et al. 2018; Hirano et al. 2010; Asano et al. 2009; Zhang et al. 2016). Slr1-d1 and Slr1-d3 are considered unsuitable for breeding due to their association with severe dwarf phenotype and low fertility, respectively (Asano et al. 2009). Rice lines with Slr1-d2 have shown suitable culm length, indicating that this gene may be useful for breeding (Asano et al. 2009). When Slr1-d5 was introduced into the indica variety 93–11, homozygous plants showed 30% reduction in plant height. Additionally, plants containing Slr1-d5 displayed large increases in the number of tillers that may contribute to higher yields (Zhang et al. 2016). When Slr1-d6 was introduced into the genetic background of 93–11, it reduced plant height by 37% but still maintained an acceptable seed setting rate for sterile line production (Wu et al. 2018). Slr1-d6 heterozygous plants resulted in a 25% increase in yield compared with sd1 semi-dwarf 93–11 (Wu et al. 2018). Thus, Slr1-d2, Slr1-d5, and Slr1-d6 hold great promise in rice breeding. Numerous studies on the application of these genes are currently underway (Asano et al. 2009; Zhang et al. 2016; Wu et al. 2018). More recently, allelic variants named as slr1-d7, slr1-d8, slr1-d9, slr1-d10, slr1-d11, and slr1-d12 were created by gene editing using the CRISPR/Cas9 system. All six mutants showed nucleotide mutations in the TVHYNP domain. Specifically, slr1-d7, slr1-d8, slr1-d9, and slr1-d10 were identified with bp deletions, while slr1-d11 and slr1-d12 had one bp insertion (Jung et al. 2020). After initial testing, rice lines containing slr1-d7 and slr1-d8 were considered potentially valuable in rice breeding as semi-dwarf germ resources (Jung et al. 2020).
One of the newly identified dwarf genes is the Shortened Basal Internodes (SBI) gene on chromosome 5. SBI is a semi-dominant gene encoding a gibberellin 2-oxidase that specifically controls the elongation of culm basal internodes by reducing the amounts of active gibberellins (GAs) in the tissue (Liu et al. 2018). SBI was identified from the cultivar SV14, a progeny of the indica male sterile line Zhu1S with somaclonal variation (Liu et al. 2002, 2018). Initial evaluation of over 94 varieties carrying SBI revealed strong lodging resistance in comparison with their parent lines. Among these evaluated SBI cultivars, Lingliangyou 22 and Lingliangyou 211 were the best based on their lodging resistance, yield, and grain quality. These two cultivars have been planted in over 366,000 ha in China (Liu et al. 2018).
An allelic variant of SEMIDWARF AND HIGH-TILLERING (SDT) was mapped to the long arm of chromosome 6 after a genetic analysis on one chromosome segment substitution line (CSSL) derived from a natural rice semi-dwarf and high-tillering (sdt) mutant (Zhao et al. 2015). The sdt allele is able to regulate the polyadenylation of the Osmi156h microRNA precursor. Thus, the sdt mutant exhibits both increased lodging resistance and tiller number, factors that may contribute to the harvest index and grain productivity (Zhao et al. 2015). When both sdt and sd1 were pyramided, the hybrid rice yield increased by about 20% (Zhao et al. 2015).
Rice mutant Y98149 has a semi-dwarf phenotype and was identified from the progeny of the japonica rice variety Y98148 (Wu et al. 2002; Liu et al. 2008b). The mutant phenotype is controlled by a new dominant gene designated as Sdd(t). Sdd(t) has been mapped to the short arm of chromosome 6 between two STS (sequence-tagged site) markers S9 and S13 (Liu et al. 2008a), and Sdd(t) was later found to be identical to Sdt97 (Tong et al. 2001; Tong et al. 2007). With a residual heterozygous line (RHL) population, this novel gene was mapped to the long arm of chromosome 6 flanked between STS marker N6 and SNP marker N16 (Tong et al. 2016). The Sdt97 gene can alter the transcription level of methyladenine glycosylase to regulate GA synthesis (Tong et al. 2007, 2016). Researchers have successfully combined Sdt97 and the photoperiod-sensitive male sterile gene pms3 to develop a series of semi-dwarf photoperiod-sensitive genic male-sterile (PSGMS) rice varieties, thereby providing a powerful genetic tool in two-line hybrid rice breeding systems (Tong et al. 2007, 2016).
Fan et al. (2016) identified a spontaneous mutant, Shennong9816d, from the rice japonica variety Shennong9816. The mutant had only 60.42% of the wild-type plant height. Genetic analysis demonstrated that a recessive gene osh15(t) near the short arm of chromosome 7 was responsible for the dwarfing phenotype. The osh15(t) gene is an allele of OSH15 that affects the architecture of internodes and leads to rice dwarfism (Sato et al. 1999). The Shennong9816d mutant showed decreased total number of grains per panicle, but increased tiller number compared with the wild type. Interestingly, the mutant Shennong9816d contains higher amounts of chlorophyll than the wild type.
Zhao et al. (2018) discovered a dominant dwarfing gene, Dd7, from crosses between the japonica cultivar Dianjingyou 1 (DJY1) and the African wild rice Oryza bathii. Dd7 is considered a potential valuable source for rice breeding based on its various favorable agronomic traits, such as number of grains per panicle, heading date, and 1000-grain weight. Dd7 was mapped to an 82-kb region on chromosome 7 with 14 open reading frames (ORFs). Dd7 is thought to be involved in abscisic acid (ABA) metabolism other than GA or BR biosynthesis/signaling pathways to regulate the rice dwarf phenotype.
Qin et al. identified a dominant dwarf mutant 986083D (japonica) from the progeny of the anther culture of an autotetraploid indica/japonica hybrid. A dominant dwarf gene named Dx was mapped onto the short arm of chromosome 8 flanked by the SSR markers RM6356 and RM6208. Dx altered the rice dwarf phenotype neither through GA metabolism or signaling nor by SL signaling (Qin et al. 2008). The molecular mechanism remains unknown. Plant heights of F1 crosses with 986083D displayed 40% to 53.5% reduction in a subtropical environment and with 37.5% to 48.2% reduction in a temperate environment (Qin et al. 2008). Dx may be a valuable material in rice breeding in the future (Qin et al. 2008).
Iwata et al. identified a mutant named KL908 in 1977 from irrated japonica variety NL8. Later, a dominant dwarfing gene, D-53, was identified from this mutant (Kinoshita 1984). D-53 was mapped onto the short arm of chromosome 11 and encodes a protein that belongs to the nucleoside triphosphate hydrolase superfamily with a double Clp-N motif-containing a P-loop (Wei et al. 2006; Jiang et al. 2013; Zhou et al. 2013). D-53 acts as a repressor in strigolactone (SL) signaling, and thus controls the dwarf phenotype in this mutant (Zhou et al. 2013; Jiang et al. 2013).
Sunohara and Kitano (2003) identified a mutant named Twisted dwarf 1 (Tid1) from the japonica cultivar Taichung 65 (T65) after being mutagenized with MNU. TID1 was mapped onto the short arm of chromosome 11 and encodes an α-tubulin protein. Mutated TID1 resulted in defective microtubules, leading to severe dwarfism and reduced right helical growth (Sunohara et al. 2009). Even though homozygous Tid1 mutants are lethal or sterile (Sunohara and Kitano 2003; Qin et al. 2008), the mutant may still be useful in practical rice breeding due to its semi-dominant characteristics.
Liang et al. (2011) idenfied a spontaneous dwarf mutant named LB4D from the progeny of wild rice backcrosses while constructing wild rice introgression lines. The LB4D mutant contains a single semi-dominant dwarf gene (LB4D) with a strong dwarfing effect in plants under different genetic backgrounds. LB4D reduced plant height in F1 plants from 27.9 to 38.1%. LB4D was mapped to a 46-kb region between markers Indel 4 and Indel G on the short arm of chromosome 11. The gene LB4D was supposed to be independent of GA biosynthesis and signaling pathways. Further studies on the molecular mechanism underlying rice height are needed. Based on its overall favorable characteristics, LB4D may be valuable for ‘two-line’ hybrid rice breeding.
At present, two genes, Sdt97/Sdd(t) and SBI from the mutants Y98148 and SV1, respectively, have begun to exhibit their breeding value. Mutant Y98149 reduced plant height by about 28% with a slightly shorter panicle length compared with the parental cultivar Y98148 (Liu et al. 2008a). Sdt97/Sdd(t) and pms3 have been combined to develop a series of semi-dwarf PSGMS rice varieties, and they are now available for a two-line hybrid rice breeding program (Tong et al. 2007, 2016). Heterozygous SV1 plants showed a slightly reduced seed setting rate and number of filled grains per panicle but greatly increased effective numbers of panicles per plant and 1000-grain weight, with overall improved yield (Liu et al. 2002). The SBI allele was introduced into various rice cultivars to breed more than 94 authorized new varieties that were confirmed as having enhanced lodging resistance as well as higher grain yield. Notably, two new varieties were considered as the elite rice varieties that are now planted in over 360,000 ha in China (Liu et al. 2018).
Genetic and Molecular Mechanisms of the Newly Identified Genes
Dominant and semi-dominant dwarfing genes are considered more valuable in hybrid crop breeding compared with recessive genes. For recessive genes such as sd1, lodging resistance only occurs when both parents carry the same recessive gene (Peng et al. 2021). For dominant or semi-dominant dwarfing genes, only one parent needs to have the gene for lodging resistance; this makes hybrid breeding much easier (Qin et al. 2008; Chen et al. 2018). The availability of newly identified dominant or semi-dominant dwarfing genes should reduce the problems associated with the extensively used recessive sd1 gene (Asano et al. 2009). Accordingly, researchers have focused on identifying dominant and semi-dominant dwarfing mutants during the past two decades. As a result, each the newly identified genes except one were dominant or semi-dominant (Liang et al. 2011; Sunohara et al. 2009, 2006; Qin et al. 2008; Wei et al. 2006; Piao et al. 2014; Zhao et al. 2015, 2018; Tong et al. 2016; Liu et al. 2008a, 2018; Asano et al. 2009).
The dominant or semi-dominant effects of these genes were unrelated with their capability to reduce plant height when they were introduced into various rice varieties. Zhu et al. reported that the plant height of the F1 generations from KL908 and other high rice varieties was close to or slightly greater than the midparent value, confirming that the dominant effect of D53 in the KL908 is quite weak (Zhu et al. 1995). The semi-dwarfing ability of Slr1-d6 has been demonstrated to be stronger than sd1 when they are in the same genetic background of indica 9311, while the culm length of Slr1-d6 heterozygote plants was similar to that of the 9311 with sd1 (Wu et al. 2018). Similarly, dominant Dd7 showed relatively weak ability to reduce the plant height in F1 hybrid plants (Zhao et al. 2018). The dominant reduction of plant height from Slr1-d5 homozygous plants was 30%, a value that was recognized as a mild level when indica 9311 was backcrossed with Slr1-d5 (Zhang et al. 2016). Another semi-dominant gene, LB4D, was able to reduce the plant height from 27.9 to 38.1% after being introduced into different rice varieties, showing a strong dwarfing effect (Liang et al. 2011). Dx was considered the strongest dominant dwarfism effect gene to date, since the plant height reduction of the F1 hybrids varied from 37.5 to 53.5% in different environments after crosses with various rice cultivars (Qin et al. 2008). As for the recessive dwarf gene osh15(t) in mutant Shennong9816d, the gene reduced plant height by 39.58% of compared with the wild type (Fan et al. 2016).
The newly identified dwarfing and semi-dwarfing genes usually affect other phenotypes in addition to plant height. Their pleiotropic characteristics make their usefulness in rice breeding more dependent on the balance between the positive contribution of dwarfism and the potential negative impact on other important agronomic traits. The genes with the most dramatic reduction in plant heights include Slr1-d1, Slr1-d2, Slr1-d3, Slr1-d4, Slr1-d5, Slr1-d8, KL908, Tid1-1, and Shennong9816d. These genes can reduce plant height by more than 50% compared to the wild types (Wu et al. 2018; Asano et al. 2009; Zhang et al. 2016; Hirano et al. 2010; Wei et al. 2006; Fan et al. 2016). Most of these genes, however, may not be useful in breeding unless their unfavorable agronomic characteristics can be overcome. For example, the culm lengths of the mutants Slr1-d1 and Slr1-d2 are too short for developing sterile lines for rice breeding, even though no other unfavorable phenotypes are associated with these genes (Asano et al. 2009). Fertility is too low for rice lines carrying the Slr1-d3 gene to be useful in breeding (Asano et al. 2009). Mutants carrying Slr1-d8 have shorter panicles with decreased grains per plant (Jung et al. 2020). Mutant KL908 had significantly decreased tillers, smaller panicles, and thinner stems (Wei et al. 2006). Homozygous plants with Tid1-1 are frequently lethal or sterile, and they are also too short (only 2 cm plant height) (Sunohara and Kitano 2003). Tid1-1 heterozygous plants exhibit much milder dwarfism but have significantly shorter roots (Sunohara et al. 2009). Shennong9816d plants have increased chlorophyll content and greater numbers of tillers. However, the lower 1000-grain weight together with reduced fertility and smaller grains mean that Shennong9816d is not useful in breeding without further improvement (Fan et al. 2016). Heights of the plants with the dominant gene D53 vary greatly once this gene is introduced into other rice varieties (Zhu et al. 1995; Qin et al. 2008). Therefore, by far the most promising gene for rice breeding among the genes with dramatic reduction in plant heights is the gene in the mutant Slr1-d5. Slr1-d5 homozygous lines resulted in 30% reduction in plant height when the indica variety 93–11 was backcrossed with mutant Slr1-d5 (Zhang et al. 2016).
The genes with milder dwarfism include Slr1-d6, Slr1-d7, Ssi1, and Dd7. These genes reduce plant height by around 10–20 cm, resulting in a favorable condition for plants to catch pollen during pollen shedding from female panicles for hybrid rice fertilization (Wu et al. 2018; Jung et al. 2020; Sunohara et al. 2006; Zhao et al. 2018). Slr1-d6 plants show no significant yield penalty. In fact, Slr1-d6 plants have significantly increased effective tillers per plant. However, this positive effect is partially offset by decreases in spikelets per panicle and 1000-grain weight during seed setting. The usefulness of Slr1-d6 in rice breeding comes from yield increase in heterogenous plants. When Slr1-d6 was introduced into rice variety 9311, heterozygous plants exhibited a 25% yield increase (Wu et al. 2018). Unlike Slr1-d6 that has shown potential usefulness in rice breeding, Slr1-d7 has several undesirable agronomic traits and needs significant improvement to have any practical value. Even though Slr1-d7 plants have thickened internodes (Jung et al. 2020) that makes it possible to support larger panicles with more grains, these plants have shorter panicles with reduced grains and withered leaves. A similar situation was found with plants carrying Slr1-d8 (Jung et al. 2020). Defects with Ssi1 plants include a high ratio of hull-cracked kernels that leads to low grain quality and reduced yield (Sunohara et al. 2006). The semi-dominant gene Ssi1 underlying the dm-type internode elongation pattern can be stably transferred to different genetic backgrounds, but only if it is in a homozygous status, a factor that further limits its usefulness in rice breeding. Dd7 from the wild rice Oryza barthii introduced into NIL produced no visible defects in agronomic traits except for the smaller number of grains per panicle that was compensated by the higher panicle number. Plants with Dd7 have overall improved grain yield (Zhao et al. 2018).
The LB4D and Dx genes produce effects that fall between genes causing dramatic plant height reduction and genes causing mild plant height reduction. Plant heights with LB4D or Dx also vary greatly under different genetic or environmental conditions. Heterozygous LB4D plants had 27.9–38.1% plant height reduction. Heterozygous Dx plants in various genetic backgrounds had 37.5–48.2% plant height reduction in a temperate environment and 40–53.5% reduction in a subtropical environment. LB4D decreased tiller number and average grain weight (Liang et al. 2011). Dx is a strongly dominant gene. Both homozygous and heterozygous Dx plants share similar phenotypes, including decreased fertility and increased tillers (Qin et al. 2008).
Most cloned dwarf (semi-dwarf) genes play important roles in the biosynthesis and/or response to three plant hormones, gibberellins (GAs), brassinosteriods (BRs), and strigolactones (SLs) (Zhang et al. 2016; Fan et al. 2016). GAs are a large family of natural tetracyclic diterpenoid carboxylic acids that function as regulators of plant growth and development. To date, 136 GAs have been isolated from higher plants, fungi, and bacteria (Hedden 2017). Rice mutants defective in GA metabolism or signaling pathways normally exhibit proportional dn-type shortened internodes (Zhang et al. 2016; Fleet and Sun 2005; Sakamoto et al. 2004; Wu et al. 2018). The gene named Shortened Basal Internodes (SBI) from mutant SV14 encodes a gibberellin 2-oxidase designated as GA2ox4. This enzyme converts active GA1, GA9, and GA20 into inactive GA8, GA51, and GA29 (Liu et al. 2018). As a result, the abundance of GA1 decreases and GA29 increases in the tissues of internodes of the mutant SV14, resulting in the dn-type dwarf (Liu et al. 2018). The GA signaling pathway includes GA receptor GA-INSENSITIVE DWARF1 (GID1) (Ueguchi-Tanaka et al. 2005), the F-box protein GA-INSENSITIVE DWARF2 (GID2) (Sasaki et al. 2003; Gomi et al. 2004), and DELLA protein SLENDER RICE1 (SLR1) (Ikeda et al. 2001). The DELLA protein SLR1 acts as a transcription regulator, suppressing the expression of intracellular GA-response genes in the absence of GA (Ogawa et al. 2000; Ikeda et al. 2001). In the presence of GAs, SLR1 binds with GID1 and stimulates the formation of a GA-GID1-DELLA complex that triggers the degradation of SLR1 via the SCFGID2-proteasome pathway, leading to transcriptional activation of GA-responsive genes (Sun and Gubler 2004; Eckardt 2007; Ikeda et al. 2001). Natural single amino acid substitutions or artificial deletion/insertion mutants in the functional motifs of DELLA protein led to similar dn-type dwarf slr1 mutants. The slr1 mutants were defective in GA signaling, resulting in dwarfism (Wu et al. 2018; Zhang et al. 2016; Jung et al. 2020; Asano et al. 2009). The d-h gene in the mutant D-h was predicted to participate in an unknown GA signaling pathway, since the expression levels of the genes in the GA biosynthetic and signaling pathways were conflicting (Piao et al. 2014). The exact mechanism of d-h mediated dwarfism remains unclear (Piao et al. 2014).
To investigate the relationship between GA and dwarfism, researchers also classified rice plants into four groups: N, T, D, and E based on two physiological processes: elongation of shoots and active α-amylase activity in the endosperm. Group N (normal type) usually includes normal cultivars and semidwarf mutants. The group N plants exhibited a slightly increased elongation of shoots and a much greater increase in α-amylase activity after exogenous GA3 treatment. Members of group T (Tan-ginbozu type) are often GA-deficient mutants that are highly responsive to exogenous GA3 in terms of elongation of shoots and production of α-amylase, even with minimum levels of endogenous GA treatment. Group D (Daikoku type) plants are GA-insensitive and are not responsive to exogenous GA3. Group E plants show responses to exogenous GA3 similar to group N plants (Mitsunaga et al. 1994). For example, mutant 986083D, a group D mutant, is moderately sensitive to exogenous GA3 based on the elongation of shoots as well as induction of α-amylase activity in the endosperm (Qin et al. 2008). Mutations in the functional motifs of DELLA protein SLR1 resulted in a set of slr1 mutants defective in GA signaling. Thus, these slr1 mutants are group D members (Wu et al. 2018; Zhang et al. 2016; Jung et al. 2020; Asano et al. 2009). The mutants Y98148 and LB4D exhibited similar responses to exogenous GA3 as their wild types, but the induction of α-amylase from half-seeds without embryos and the shoot length did not differ significantly from the corresponding wild types. Therefore, these mutants were considered as group E (Liang et al. 2011; Liu et al. 2008a). Mutant Shennong9816d is considered a group T member because it is sensitive to GA3 but deficient in GA synthesis (Fan et al. 2016). The reduced plant heights of Shennong9816d mutants are due to the inhibited elongation of the bottom internodes (d6-type dwarfism) (Fan et al. 2016).
BRs are a specific class of endogenous polyhydroxysteroids essential for plant growth and development (Li 2013). Defects in the BR biosynthesis or signaling pathway lead to mutants with pleiotropic aberrant phenotypes such as dwarfism, erect leaves, and small grains (Piao et al. 2014; Fan et al. 2016; Hong et al. 2002, 2005). Rice dm-type mutants are usually BR-deficient and -insensitive mutants (Hong et al. 2005; Hu et al. 2013; Zhang et al. 2014; Yamamuro et al. 2000). Key components in rice BR biosynthesis include OsDWARF, Dim/dwf1, D2/CYP90D2, SG4, CYP90B2/OsDWARF4, and D11/CYP724B1, as revealed from characterizing the mutants brassinosteroid-dependent 1 (brd1), BR-deficient dwarf2 (brd2), ebisu dwarf (dwarf2 or d2), small grain 4 (sg4), and osdwarf4-1, dwarf11, respectively (Hong et al. 2002, 2005, 2003; Shi et al. 2015; Sakamoto et al. 2006; Tanabe et al. 2005). The rice BR signaling pathway is overall similar to that of Arabidopsis, with conserved proteins such as BRI1, BAK1, GSK1, BZR1, and BIN2 (Zhang et al. 2014; Fàbregas and Caño-Delgado 2014; Tong and Chu 2012; Nakagawa et al. 2011). In rice plants, however, there is another G-protein signaling pathway involved in BR signaling via the U-box E3 ligase TAIHU DWARF1 (TUD1) that interacts with the rice Gα subunit D1 (Nakagawa et al. 2011; Hu et al. 2013; Oki et al. 2009a, 2009b). The D1-TUD1-mediated BR signaling pathway functions parallel to or overlapping with the typical BRI1 pathway in regulating the elongation of the second internode (Hu et al. 2013). The elongation of the second internode is also regulated by Ssi1 (Miura et al. 2009).
SLs are a class of root-derived terpenoid lactones involved in regulating plant development and growth features such as shoot architecture, stem secondary growth, seed germination, root development, leaf senescence, and photomorphogenesis as well as responding to environmental stresses such as low-phosphate and nitrate deficiency (Mashiguchi et al. 2021; Seto et al. 2012; Brewer et al. 2013). Biosynthesis of active SLs involves many proteins and enzymes, including D10, HTD1/D17, D27, and HTD2 (Arite et al. 2007; Lin et al. 2009; Zou et al. 2006; Wang et al. 2020; Liu et al. 2009). D14 is an α/β-fold hydrolase that serves as a receptor for active SLs (Arite et al. 2009; Gao et al. 2009; Hamiaux et al. 2012; Jiang et al. 2013; Hu et al. 2017). D3 is an F-box leucine-rich repeat protein that plays a central role in SL signaling (Ishikawa et al. 2005; Hu et al. 2017; Jiang et al. 2013; Zhou et al. 2013). D53 belongs to the superfamily of double Clp-N motif-containing, P-loop nucleoside triphosphate hydrolases and functions as a repressor of SL signaling (Zhou et al. 2013; Jiang et al. 2013). SLs trigger proteasome-mediated degradation of D53 in a D14- and D3-dependent manner (Jiang et al. 2013; Zhou et al. 2013). KL908 with a mutation in the gene D53 shares similar pleiotropism of phenotypes with other mutants d3, d10, d14, d17, and d27, including short stature, more tillers, and small panicles. These are considered as strigolactone-deficient or -insensitive mutants (Wei et al. 2006; Ishikawa et al. 2005; Gao et al. 2009).
Cytokinins are also regulators of dwarfism (Hirose et al. 2007; Wang et al. 2021b). One of the enzymes involved in cytokinin metabolism is OsCKX4, an oxidase/dehydrogenase involved in the degradation of cytokinins (Schmülling et al. 2003). Suppression of OsCKX4 gene expression represses cytokinin catabolism, leading to reduced plant height. The discovery of cytokinins’ involvement and the associated molecular mechanisms in rice dwarfism was drawn from characterizing the cytokinin-deficient mutant rice lateral branch (rlb). The mutant carries a T-DNA insertion in the rice homeobox gene OSH15. Thus, RLB is an allele of OSH15. The protein RLB recruits PRC2 (Polycomb repressive complex 2) to epigenetically repress the transcription of OsCKX4 by modifying H3K27me3, resulting in inhibition of OsCKX4 transcription (Wang et al. 2021b). The osh15(t) from the Shennong9816d mutant is another naturally allelic variation of OSH15, showing a 30 bp insertion in intron 2 and a 1 bp mutation in exon 5 (Fan et al. 2016).
Initial evidence has also shown that the hormone abscisic acid may also play a role in rice dwarfism. The plant height of a near-isogenic rice line, NIL-Dd7, was shortened in comparison with its parent lines, but NIL-Dd7 did not respond to either GA or BR treatments. Recently, Dd7 was mapped to a very short chromosomal fragment, and a candidate gene has been identified. The Dd7 candidate gene contains an ORF encoding 9-cis-epoxycarotenoid dioxygenase 1, a key regulator of ABA synthesis (Dong et al. 2015; Zhao et al. 2018). However, more work is needed to confirm the role of ABA in regulating plant height (Zhao et al. 2018).
In addition to plant hormones, there are likely other mechanisms involved in regulating rice dwarf phenotypes. For example, increased abundance of the OsmiR156h microRNA in a natural allelic variant of the sdt gene reduced plant height but increased tillers and grain yield (Zhao et al. 2015). Other reported genes and proteins that affect plant height include α-tubulin (Sunohara et al. 2009) and a methyladenine glycosylase (a DNA repair enzyme) (Tong et al. 2016).
Conclusion and Perspective
Rice is the staple food crop for more than half of the world's population and in more than 100 countries worldwide (Fukagawa and Ziska 2019). Rice breeding has experienced two quantum yield breakthroughs over the past three decades by introduction of the semi-dwarf gene sd1 and hybrid technology (Siddiq and Vemireddy 2021; Peng et al. 2008). After these significant advances, rice yield has been in stagnation. In contrast, global demand for rice is predicted to increase by more than 70% of the current level. Global climate change, a growing population, and decreasing farmable lands pose serious challenges to satisfying rice demand in the future (Godfray et al. 2010; Siddiq and Vemireddy 2021, Food and Nations 2009). Cultural changes in rice farming such as rice direct seeding (DSR) and adoption of more advanced equipment and machines also require adaptable rice cultivars such as more lodging resistant varieties (Farooq et al. 2011; Hafeez-ur-Rehman et al. 2019). Thus, identification and use of new dwarfism and semi-dwarfism genes are needed to guarantee future food supply.
Although more than 60 dwarfing and semi-dwarfing genes have been identified through various technologies, most of these genes have serious defects in other agronomic traits along with reduced plant heights (Futsuhara and Kikuchi 1997; Gaur et al. 2020; Peng et al. 2021; Nagai et al. 2018; Wu et al. 2018). These unfavorable agronomic traits prevent their usefulness in practical rice breeding. More effort must be put into reducing these unfavorable traits associated with dwarfism so that the newly identified genes can be useful for increasing rice yield and quality.
In the past, our effort has focused on finding mutants from wild rice or generation of artificial mutants by chemical treatments or radiation. These efforts have been shown to be productive for unearthing useful genes for crop breeding, and we need to continue to enlarge our rice artificial mutant libraries in order to discover more dwarfing and semi-dwarfing genes. Since most dwarfing genes are in the pathways of plant hormone metabolism, disruption of the expression of these genes or mutations in key residues for normal three-dimensional structures of the encoded proteins will inevitably affect the normal functions of these genes. Future efforts shall also focus on generating mutants with small structural changes so that the main functions of these genes are not dramatically affected, but their functions for controlling plant heights are modified, resulting in shorter rice plants. Such elegant engineering is of course difficult, but possible with recent dramatic advances in new technologies, including precise genome sequencing, accurate tools for prediction of three-dimensional structures such as FoldAlpha2, targeted mutagenesis, and genome editing.
Availability of Supporting data
All data and its supplementary information files of this study are included in this article.
References
Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J 51(6):1019–1029
Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J (2009) d14, a Strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol 50(8):1416–1424
Asano K, Takashi T, Miura K, Qian Q, Kitano H, Matsuoka M, Ashikari M (2007) Genetic and molecular analysis of utility of sd1 alleles in rice breeding. Breed Sci 57:53–58
Asano K, Hirano K, Ueguchi-Tanaka M, Angeles-Shim RB, Komura T, Satoh H, Kitano H, Matsuoka M, Ashikari M (2009) Isolation and characterization of dominant dwarf mutants, Slr1-d, in rice. Mol Genet Genom 281(2):223–231
Ashikari M, Sasaki A, Ueguchi-Tanaka M, Itoh H, Nishimura A, Datta S, Ishiyama K, Saito T, Kobayashi M, Khush GS, Kitano H, Matsuoka M (2002) Loss-of-function of a rice gibberellin biosynthetic gene, GA20 oxidase (GA20ox-2), led to the rice ‘green revolution.’ Breed Sci 52:143–150
Biswas S, Tian J, Li R, Chen X, Luo Z, Chen M, Zhao X, Zhang D, Persson S, Yuan Z, Shi J (2020) Investigation of CRISPR/Cas9-induced SD1 rice mutants highlights the importance of molecular characterization in plant molecular breeding. J Genet Genom 47:273–280
Brewer PB, Koltai H, Beveridge CA (2013) Diverse roles of strigolactones in plant development. Mol Plant 6(1):18–28
Chandler RF (1968) Dwarf rice-a giant in tropical Asia. USDA Yearbook, 252–255
Chao J, Li Z, Sun Y, Aluko OO, Wu X, Wang Q, Liu G (2021) MG2C: a user-friendly online tool for drawing genetic maps. Mol Hortic 1(1):1–4
Chen X, Xu P, Zhou J, Tao D, Yu D (2018) Mapping and breeding value evaluation of a semi-dominant semi-dwarf gene in upland rice. Plant Divers 40(5):238–244
Chen K, Wang Y, Zhang R, Zhang H, Gao C (2019) CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol 70:667–697
Dong T, Park Y, Hwang I (2015) Abscisic acid: biosynthesis, inactivation, homoeostasis and signalling. Essays Biochem 58:29–48
Eckardt NA (2007) GA perception and signal transduction: molecular interactions of the GA receptor GID1 with GA and the DELLA Protein SLR1 in rice. Plant Cell 19(7):2095–2097
Evans LT (1993) Crop evolution, adaptation and yield. Cambridge University Press, Cambridge
Evans LT (1998) Feeding the ten billion. Plant and population growth. Cambridge Uniersity Press, Cambridge
Fàbregas N, Caño-Delgado AI (2014) Turning on the microscope turret: a new view for the study of brassinosteroid signaling in plant development. Physiol Plant 151(2):172–183
Fan S, Yao X, Liu J, Dong X, Mao T, Wang J (2016) Characterization and fine mapping of osh15(t), a novel dwarf mutant gene in rice (Oryza sativa L.). Genes Genom 38(9):849–856
Farooq M, Siddique KHM, Rehman H, Aziz T, Lee D-J, Wahid A (2011) Rice direct seeding: experiences, challenges and opportunities. Soil Tillage Res 111:87–98
Ferrero-Serrano Á, Cantos C, Assmann SM (2019) The role of dwarfing traits in historical and modern agriculture with a focus on rice. Cold Spring Harb Perspect Biol 11:a034645
Fleet CM, Sun T-p (2005) A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr Opin Plant Biol 8(1):77–85
Food, Nations Agriculture Organization of the United (2009) Global agriculture towards 2050. High Level Expert Forum - How to Feed the World in 2050. Rome Oct. 12–13
Frankel OH (1970) Genetic conservation in perspective. Genetic conservation in perspective
Fukagawa NK, Ziska LH (2019) Rice: importance for global nutrition. J Nutr Sci Vitaminol (tokyo) 65(Supplement):S2–S3
Futsuhara Y, Kikuchi F (1997) Inheritance of morphological characters. 2. Inheritance of dwarfism. Sci Rice Plant 3:300–308
Gao Z, Qian Q, Liu X, Yan M, Feng Q, Dong G, Liu J, Han B (2009) Dwarf 88, a novel putative esterase gene affecting architecture of rice plant. Plant Mol Biol 71(3):265–276
Gaur VS, Channappa G, Chakraborti M, Sharma TR, Mondal TK (2020) ‘Green revolution’ dwarf gene sd1 of rice has gigantic impact. Brief Funct Genom 19:390–409
Godfray HC, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C (2010) Food security: the challenge of feeding 9 billion people. Science 327(5967):812–818
Gomi K, Sasaki A, Itoh H, Ueguchi-Tanaka M, Ashikari M, Kitano H, Matsuoka M (2004) GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J 37(4):626–634
Guo T, Huo X, De Rao H, Liu YZ, Zhang JG, Chen ZQ, Wang H (2011) Identification and fine mapping of semidwarf gene iga 1 in rice. Acta Agron Sin 37(6):955–964
Hafeez-ur-Rehman, Nawaz A, Awan MI, Ijaz M, Hussain M, Ahmad S, Farooq M (2019) Direct seeding in rice: problems and prospects. In: Hasanuzzaman M (ed) Agronomic crops: Volume 1: Production technologies, Springer, Singapore Singapore, pp 199–222
Hamiaux C, Drummond RS, Janssen BJ, Ledger SE, Cooney JM, Newcomb RD, Snowden KC (2012) DAD2 Is an α/β hydrolase likely to be involved in the perception of the plant branching hormone. Strigolactone Curr Biol 22(21):2032–2036
Harlan JR (1975) Our vanishing genetic resources. Science 188(4188):618–621
Hedden P (2003) Constructing dwarf rice. Nat Biotechnol 21(8):873–874
Hedden P (2017) Gibberellins. Thomas B, Murray BG, Murphy DJ (eds) Encyclopedia of applied plant sciences, 2nd Edn. Academic Press, Oxford, 411–420
Hirano K, Asano K, Tsuji H, Kawamura M, Mori H, Kitano H, Ueguchi-Tanaka M, Matsuoka M (2010) Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice. Plant Cell 22(8):2680–2696
Hirose N, Makita N, Kojima M, Kamada-Nobusada T, Sakakibara H (2007) Overexpression of a type-A response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol 48:523–539
Hong Z, Ueguchi-Tanaka M, Shimizu-Sato S, Inukai Y, Fujioka S, Shimada Y, Takatsuto S, Agetsuma M, Yoshida S, Watanabe Y, Uozu S (2002) Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J 32(4):495–508
Hong Z, Ueguchi-Tanaka M, Umemura K, Uozu S, Fujioka S, Takatsuto S, Yoshida S, Ashikari M, Kitano H, Matsuoka M (2003) A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15(12):2900–2910
Hong Z, Ueguchi-Tanaka M, Fujioka S, Takatsuto S, Yoshida S, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M (2005) The Rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell 17(8):2243–2254
Hu X, Qian Q, Xu T, Zhang YE, Dong G, Gao T, Xie Q, Xue Y (2013) The U-Box E3 ubiquitin ligase TUD1 functions with a heterotrimeric G α subunit to regulate brassinosteroid-mediated growth in rice. PLoS Genet 9:e1003391
Hu Q, He Y, Wang L, Liu S, Meng X, Liu G, Jing Y, Chen M, Song X, Jiang L, Yu H, Wang B, Li J (2017) DWARF14, a receptor covalently linked with the active form of strigolactones, undergoes strigolactone-dependent degradation in rice. Front Plant Sci 8:1935–1945
Hu X, Cui Y, Dong G, Feng A, Wang D, Zhao C, Zhang Y, Hu J, Zeng D, Guo L, Qian Q (2019) Using CRISPR-Cas9 to generate semi-dwarf rice lines in elite landraces. Sci Rep 9:19096
Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J (2001) slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13(5):999–1010
Institue International Rice Research (1967) Annual Report for 1966. 59–82
Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J (2005) Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol 46(1):79–86
Jiang S-Y, Ramachandran S (2010) Natural and artificial mutants as valuable resources for functional genomics and molecular breeding. Int J Biol Sci 6(3):228–251
Jiang L, Liu X, Xiong G, Liu H, Chen F, Wang L, Meng X, Liu G, Yu H, Yuan Y, Yi W, Zhao L, Ma H, He Y, Wu Z, Melcher K, Qian Q, Xu HE, Wang Y, Li J (2013) DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504(7480):401–405
Jung YJ, Kim JH, Lee HJ, Kim DH, Yu J, Bae S, Cho YG, Kang KK (2020) Generation and transcriptome profiling of Slr1-d7 and Slr1-d8 mutant lines with a new semi-dominant dwarf allele of SLR1 using the CRISPR/Cas9 system in rice. Int J Mol Sci 21(15):5492
Khoury CK, Brush S, Costich DE, Curry HA, de Haan S, Engels JM, Guarino L, Hoban S, Mercer KL, Miller AJ, Nabhan GP, Perales HR, Richards C, Riggins C, Thormann I (2022) Crop genetic erosion: understanding and responding to loss of crop diversity. New Phytol 233:84–11
Kinoshita T (1984) Current linkage maps. RGN 1:16–27
Koh H-J, Heu M-H (1993) A new dominant dwarfing gene in rice. Rice Genet Newsl 10:77–79
Li J (2013) Brassinosteroids. Maloy S, Hughes K (eds) Brenner's Encyclopedia of Genetics, 2nd Edn. Academic Press, San Diego, pp 377–380
Li Y, Yang L, Pathak M, Li D, He X, Weng Y (2011) Fine genetic mapping of cp: a recessive gene for compact (dwarf) plant architecture in cucumber, Cucumis sativus L. Theor Appl Genet 123(6):973
Li S, Tian Y, Wu K, Ye Y, Yu J, Zhang J, Liu Q, Hu M, Li H, Tong Y, Harberd NP, Fu X (2018) Modulating plant growth–metabolism coordination for sustainable agriculture. Nature 560:595–600
Li S, Huang Y-Z, Liu X-Y, Fu X-D (2021) Genetic improvement of nitrogen use efficiency in crops. Hereditas 43(7):629–641
Liang F, Xin X, Hu Z, Xu J, Wei G, Qian X, Yang J, He H, Luo X (2011) Genetic analysis and fine mapping of a novel semidominant dwarfing gene LB4D in rice. J Integr Plant Biol 53(4):312–323
Lin H, Wang R, Qian Q, Yan M, Meng X, Fu Z, Yan C, Jiang B, Su Z, Li J, Wang Y (2009) DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21(5):1512–1525
Liu XM, Yang YZ, Chen CY, Tang PL, Liu B, Fu CJ (2002) Breeding of dwarfing variants with the technique of somaclonal variation for photo-(thermo-) sensitive genic male sterile line Zhu 1S. Chin J Rice Sci 16(4):321–325
Liu BM, Wu YJ, Fu XD, Qian Q (2008a) Characterizations and molecular mapping of a novel dominant semi-dwarf gene Sdd(t) in rice (Oryza sativa). Plant Breed 127(2):125–130
Liu B, Wu Y, Xu X, Song M, Zhao M, Fu XD (2008b) Plant height revertants of dominant semidwarf mutant rice created by low-energy ion irradiation. Nucl Instrum Methods Phys Res Sect B Beam Interact Mater Atoms 266(7):1099–1104
Liu W, Wu C, Fu Y, Hu G, Si H, Zhu L, Luan W, He Z, Sun Z (2009) Identification and characterization of HTD2: a novel gene negatively regulating tiller bud outgrowth in rice. Planta 230(4):649–658
Liu C, Zheng S, Gui J, Fu C, Yu H, Song D, Shen J, Qin P, Liu X, Han B, Yang Y, Li L (2018) Shortened basal internodes encodes a gibberellin 2-oxidase and contributes to lodging resistance in rice. Mol Plant 11(2):288–299
Mashiguchi K, Seto Y, Yamaguchi S (2021) Strigolactone biosynthesis, transport and perception. Plant J 105(2):335–350
Mitsunaga S, Tashiro T, Yamaguchi J (1994) Identification and characterization of gibberellin-insensitive mutants selected from among dwarf mutants of rice. Theor Appl Genet 87(6):705–712
Miura K, Wu J, Sunohara H, Wu X, Matsumoto T, Matsuoka M, Ashikari M, Kitano H (2009) High-resolution mapping revealed a 1.3-Mbp genomic inversion in Ssi1, a dominant semidwarf gene in rice (Oryza sativa). Plant Breed 128(1):63–69
Monna L, Kitazawa N, Yoshino R, Suzuki J, Masuda H, Maehara Y, Tanji M, Sato M, Nasu S, Minobe Y (2002) Positional cloning of rice semidwarfing gene, sd-1: rice “green revolution gene” encodes a mutant enzyme involved in gibberellin synthesis. DNA Res 9(1):11–17
Nagai K, Hirano K, Angeles-Shim RB, Ashikari M (2018) Breeding applications and molecular basis of semi-dwarfism in rice. In: Sasaki T, Ashikari M (eds) Rice genomics, genetics and breeding. Springer Singapore, Singapore, pp 155–176
Nakagawa H, Tanaka A, Mori M (2011) Brassinosteroid signaling in rice. In: Brassinosteroids: a class of plant hormone, Springer, Dordrecht, pp 83–117
Ogawa M, Kusano T, Katsumi M, Sano H (2000) Rice gibberellin-insensitive gene homolog, OsGAI, encodes a nuclear-localized protein capable of gene activation at transcriptional level. Gene 245(1):21–29
Oki K, Inaba N, Kitano H, Takahashi S, Fujisawa Y, Kato H, Iwasaki Y (2009a) Study of novel d1 alleles, defective mutants of the alpha subunit of heterotrimeric G-protein in rice. Genes Genet Syst 84(1):35–42
Oki K, Kitagawa K, Fujisawa Y, Kato H, Iwasaki Y (2009b) Function of alpha subunit of heterotrimeric G protein in brassinosteroid response of rice plants. Plant Signal Behav 4(2):126–128
Peng S, Khush GS, Virk P, Tang Q, Zou Y (2008) Progress in ideotype breeding to increase rice yield potential. Field Crop Res 108(1):32–38
Peng Y, Hu Y, Qian Q, Ren D (2021) Progress and prospect of breeding utilization of green revolution gene SD1 in rice. Agriculture 11(7):611
Piao R, Chu SH, Jiang W, Yu Y, Jin Y, Woo MO, Lee J, Kim S, Koh HJ (2014) Isolation and characterization of a dominant dwarf gene, D-h, in rice. PLoS ONE 9(2):e86210
Qin R, Qiu Y, Cheng Z, Shan X, Guo X, Zhai H, Wan J (2008) Genetic analysis of a novel dominant rice dwarf mutant 986083D. Euphytica 160(3):379–387
Sakai H, Itoh T (2010) Massive gene losses in Asian cultivated rice unveiled by comparative genome analysis. BMC Genomics 11(1):121
Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, Miyao A, Hirochika H, Kitano H, Ashikari M, Matsuoka M (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134(4):1642–1653
Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, Ueguchi-Tanaka M, Mizutani M, Sakata K, Takatsuto S, Yoshida S, Tanaka H, Kitano H, Matsuoka M (2006) Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol 24(1):105–109
Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, Kitano H, Matsuoka M (2002) A mutant gibberellin-synthesis gene in rice. Nature 416:701–702
Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong DH, An G, Kitano H, Ashikari M, Matsuoka M (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299(5614):1896–1898
Sato Y, Sentoku N, Miura Y, Hirochika H, Kitano H, Matsuoka M (1999) Loss-of-function mutations in the rice homeobox gene OSH15 affect the architecture of internodes resulting in dwarf plants. EMBO J 18:992–1002
Schmülling T, Werner T, Riefler M, Krupková E, Bartrina y Manns I (2003) Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. J Plant Res 116(3): 241–252
Seto Y, Kameoka H, Yamaguchi S, Kyozuka J (2012) Recent advances in strigolactone research: chemical and biological aspects. Plant Cell Physiol 53(11):1843–1853
Shi Z, Rao Y, Xu J, Hu S, Fang Y, Yu H, Pan J, Liu R, Ren D, Wang X, Zhu Y, Dong G, Zhang G, Zeng D, Guo L, Hu J, Qian Q (2015) Characterization and cloning of SMALL GRAIN 4, a novel DWARF11 allele that affects brassinosteroid biosynthesis in rice. Sci Bull 60(10):905–915
Siddiq EA, Vemireddy LR (2021) Advances in genetics and breeding of rice: an overview. In: Ali J, Wani SH (eds) Rice improvement: physiological, molecular breeding and genetic perspectives. Springer International Publishing, Cham, pp 1–29
Spielmeyer W, Ellis MH, Chandler PM (2002) Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc Natl Acad Sci 99(13):9043–9048
Sun P, Zhang W, Wang Y, He Q, Shu F, Liu H, Wang J, Wang J, Yuan L, Deng H (2016) OsGRF4 controls grain shape, panicle length and seed shattering in rice. J Integr Plant Biol 58:836–847
Sun T-p, Gubler F (2004) Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol 55:197–223
Sunohara H, Kitano H (2003) New dwarf mutant controlled by a dominant gene, twisted dwarf1. Rice Genet Newsl 20:20–21
Sunohara H, Kawai T, Shimizu-Sato S, Sato Y, Sato K, Kitano H (2009) A dominant mutation of TWISTED DWARF 1 encoding an α-tubulin protein causes severe dwarfism and right helical growth in rice. Genes Genet Syst 84(3):209–218
Sunohara H, Miura K, Wu X, Saeda T, Mizuno S, Ashikari M, Matsuoka M, Kitano H (2006) Effects of Ssi1 gene controlling dm-type internode elongation pattern on lodging resistance and panicle characters in rice. Breed Sci 56(3):261–268
Takeda K (1974) Studies on the character expression and inheritance of dm-type internode distribution pattern in dwarf rice plants (preliminary report). Bull Fac Agric Hirosaki Univ 22:19–30
Takeda K (1977) Internode elongation and dwarfism in some gramineous plants. Gamma Field Symp 16:1–18
Tanabe S, Ashikari M, Fujioka S, Takatsuto S, Yoshida S, Yano M, Yoshimura A, Kitano H, Matsuoka M, Fujisawa Y, Kato H, Iwasaki Y (2005) A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17(3):776–790
Tar’an B, Warkentin T, Somers DJ, Miranda D, Vandenberg A, Blade S, Woods S, Bing D, Xue A, DeKoeyer D, Penner G (2003) Quantitative trait loci for lodging resistance, plant height and partial resistance to mycosphaerella blight in field pea (Pisum sativum L.). Theor Appl Genet 107(8):1482–1491
Tomita M (2009) Introgression of Green Revolution sd1 gene into isogenic genome of rice super cultivar Koshihikari to create novel semidwarf cultivar ‘Hikarishinseiki’ (Koshihikari-sd1). Field Crop Res 114(2):173–181
Tong H, Chu C (2012) Brassinosteroid signaling and application in rice. J Genet Genom 39(1):3–9
Tong J-P, Liu X-J, Zhang S-Y, Li S-Q, Peng X-J, Yang J, Zhu Y-G (2007) Identification, genetic characterization, GA response and molecular mapping of Sdt97: a dominant mutant gene conferring semi-dwarfism in rice (Oryza sativa L.). Genet Res 89(4):221–230
Tong J, Han Z, Han A, Liu X, Zhang S, Fu B, Hu J, Su J, Li S, Wang S, Zhu Y (2016) Sdt97: a point mutation in the 5’ untranslated region confers semidwarfism in rice. G3 (bethesda, Md.) 6(6):1491–1502
Tong J-p, Wu Y-j, Wu J-d, Liu G-f, Zheng L-y, Yu Z-l (2001) Discovery of a dominant semi-dwarf japonica rice mutant and its preliminary study. Zhongguo Shuidao Kexue 15(4):314–316
Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow T-Y, Hsing Yue-ie C, Kitano H, Yamaguchi I, Matsuoka M (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437:693–698
Wang F, Yoshida H, Matsuoka M (2021a) Making the ‘green revolution’truly green: improving crop nitrogen use efficiency. Plant Cell Physiol 62(6):942–947
Wang H, Tong X, Tang L, Wang Y, Zhao J, Li Z, Liu X, Shu Y, Yin M, Adegoke TV, Liu W, Wang S, Xu H, Ying J, Yuan W, Yao J, Zhang J (2021b) RLB (Rice Lateral Branch) recruits PRC2-mediated H3K27 tri-methylation on OsCKX4 to regulate lateral branching. Plant Physiol 188(1):460–476
Wang Y, Shang L, Yu H, Zeng L, Hu J, Ni S, Rao Y, Li S, Chu J, Meng X, Wang L, Hu P, Yan J, Kang S, Qu M, Lin H, Wang T, Wang Q, Hu X, Chen H, Wang B, Gao Z, Guo L, Zeng D, Zhu X, Xiong G, Li J, Qian Q (2020) A Strigolactone biosynthesis gene contributed to the green revolution in rice. Mol Plant 13(6):923–932
Wei L-R, Xu J-C, Li X-B, Qian Q, Zhu L-H (2006) Genetic analysis and mapping of the dominant dwarfing gene D-53 in rice. J Integr Plant Biol 48(4):447–452
Wu X (1997) Dominant gene, Ssil (t) located on the chromosome 1 shows the dm-type internode elongation pattern accompanied by a semidwarfness. Rice Genet Newslett 14:45–47
Wu Y, Tong J, Wu J, Liu G, Zheng L (2002) A dominant semi-dwarf mutant in rice. Chin Rice Res Newsl 10:7–8
Wu Z, Tang D, Liu K, Miao C, Zhuo X, Li Y, Tan X, Sun M, Luo Q, Cheng Z (2018) Characterization of a new semi-dominant dwarf allele of SLR1 and its potential application in hybrid rice breeding. J Exp Bot 69(20):4703–4713
Xin M, Qin Z, Wang L, Zhu Y, Wang C, Zhou X (2014) Differential proteomic analysis of dwarf characteristics in cucumber (Cucumis sativus Linn.) stems. Acta Physiol Plant 37(1):1703
Yamaguchi H (1976) Morphological mutants, specially with reference to dm-type of internode elongation pattern, induced by X-ray irradiation to growing rice plants. Jpn J Breed 24(2):31–32
Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M (2000) Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12(9):1591–1605
Zhang C, Bai M-y, Chong K (2014) Brassinosteroid-mediated regulation of agronomic traits in rice. Plant Cell Rep 33(5):683–696
Zhang Y-h, Bian X-f, Zhang S-b, Ling J, Wang Y-j, Wei X-y, Fang X-w (2016) Identification of a novel gain-of-function mutant allele, slr1-d5, of rice DELLA protein. J Integr Agric 15(7):1441–1448
Zhao M, Liu B, Wu K, Ye Y, Huang S, Wang S, Wang Y, Han R, Liu Q, Fu X, Wu Y (2015) Regulation of OsmiR156h through alternative polyadenylation improves grain yield in rice. PLoS ONE 10(5):e0126154
Zhao Z, Zhang C, Liu X, Lin Y, Liu L, Tian Y, Chen L, Liu S, Zhou J, Jiang L, Tao D, Wan J (2018) Genetic analysis and fine mapping of a dominant dwarfness gene from wild rice (Oryza barthii). Plant Breed 137(1):50–59
Zhou F, Lin Q, Zhu L, Ren Y, Zhou K, Shabek N, Wu F, Mao H, Dong W, Gan L, Ma W, Gao H, Chen J, Yang C, Wang D, Tan J, Zhang X, Guo X, Wang J, Jiang L, Liu X, Chen W, Chu J, Yan C, Ueno K, Ito S, Asami T, Cheng Z, Wang J, Lei C, Zhai H, Wu C, Wang H, Zheng N, Wan J (2013) D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504(7480):406–410
Zhu L, Lai G, Teng Y, Zhou A, Wang H (1995) Studies on the inheritance of dominant dwarfism in rice. I. On the genetic nature of dwarfism of a Japonica line KL908. J Nanjing Agric Univ 18(2):1–8
Zou J, Zhang S, Zhang W, Li G, Chen Z, Zhai W, Zhao X, Pan X, Xie Q, Zhu L (2006) The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J 48(5):687–698
Acknowledgements
We thank Dr. Ming-Shun Chen (United States Department of Agriculture-Agricultural Research Service :USDA-ARS) for proof reading and reviewing. We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Funding
This work was supported by Jiangxi “Double Thousand Plan” (2F2522100), Modern Agricultural Research Collaborative Innovation Special Project (JXXTCXBSJJ202205), and the Scientific and Technological Program of Jiangxi Province (20161BBF60129).
Author information
Authors and Affiliations
Contributions
XC and GZ proposed the concept. XC wrote the manuscript. YH, JY, YT and LT prepared the supplemental materials and revised the manuscript. GZ revised and finalized the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical Approval and Consent to participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1
Details on Rice dwarf and semi-dwarf genes from Rice Annotation Project Database (https://rapdb.dna.affrc.go.jp/).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cheng, X., Huang, Y., Tan, Y. et al. Potentially Useful Dwarfing or Semi-dwarfing Genes in Rice Breeding in Addition to the sd1 Gene. Rice 15, 66 (2022). https://doi.org/10.1186/s12284-022-00615-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-022-00615-y