Abstract
Thirty human EDTA plasma samples from male and female subjects ranging in age from 24 to 74 years were collected on ice, processed ice cold and stored frozen at −80 °C, in liquid nitrogen (LN2), or freeze dried and stored at room temperature in a desiccator (FDRT) or freeze dried and stored at −20 °C for 1 year (FD-20). In a separate experiment, EDTA plasma samples were collected onto ice, processed ice cold and maintained on ice ± protease inhibitors versus incubated at room temperature for up to 96 h. Random and independent sampling by liquid chromatography and tandem mass spectrometry (LC–ESI–MS/MS), as correlated by the MASCOT, OMSSA, X!TANDEM and SEQUEST algorithms, showed that tryptic peptides from complement component 4B (C4B) were rapidly released in plasma at room temperature. Random sampling by LC–ESI–MS/MS showed that peptides from C4B were undetectable on ice, but peptides were cleaved from the mature C4B protein including NGFKSHALQLNNR within as little as 1 h at room temperature. The frequency and intensity of precursors within ± 3 m/z of the C4B peptide NGFKSHALQLNNR was confirmed by automated targeted analysis where the precursors from MS/MS spectra that correlated to the target sequence were analyzed in SQL/R. The C4B preproprotein was processed at the N terminus to release the mature chain that was cleaved on the carboxyl side of the isoprene C2 domain within a polar C terminal sequence of the mature C4B protein, to reveal the thioester reaction site, consistent with LC–ESI–MS/MS and Western blot. Random sampling showed that proteolytic peptides from complement component C4B were rarely observed with long term storage at − 80 °C in a freezer or in liquid nitrogen (LN2), freeze drying with storage at − 20 °C (FD-20 °C) or freeze drying and storage at room temperature (FDRT). Plasma samples maintained at room temperature (RT) showed at least 10-fold to 100-fold greater frequency of peptide correlation to C4B and measured peptide intensity compared to samples on ice for up to 72 h or stored at − 80 °C, LN2, FDRT or FD-20 °C for up to a year.
Similar content being viewed by others
Background
The proteins and endogenous peptides of human plasma samples may be purified by partition chromatography with identification and quantification by liquid chromatography, electrospray ionization and tandem mass spectrometry (LC–ESI–MS/MS) [1, 2]. Peptides from blood proteins might facilitate the diagnosis of diseases and the evaluation of the efficacy of therapeutic treatments for individual patients [3,4,5,6]. Plasma expresses a weak tryptic protease activity that may slowly degrade the sample proteins over time thus releasing endogenous peptides that may be unrelated to the disease process [7]. The proteolytic activation of the complement system is an important mediator of the acute inflammatory response and humoral immunity [8, 9]. C4B is cleaved to expose a thioester group that permits covalent modification of target macromolecules [10]. It has been shown that levels of C4B peptides may be associated with sampling conditions [2, 11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29]. The steady state balance of ex vivo endo-proteinase and exopeptidase activity may change over time and result in a large variation in the blood peptides observed [2, 30]. Pre-analytical variation in the time the serum or plasma sample remains at room temperature before aliquoting and freezing may be a source of bias in subsequent mass spectrometric measurements [11,12,13,14,15,16,17,18,19,20,21,22,23,24]. The evidence to date indicates that variation in handling immediately after sample collection and prior to centrifugation is the largest source of variation in blood samples [2, 20, 25,26,27]. To prevent degradation, the sample should be kept on ice during sample handling [31]. Adding serine centered endo-peptidase inhibitors like PMSF or AEBSF [32, 33] to blood fluids in order to preserve the proteins will result in alterations of endogenous peptides [2, 25, 30, 32, 34, 35]. Alternatively, it may be possible to quench ex vivo reactions and store blood samples by freeze drying [3], or rapid drying on filter paper [36] or PVDF [37]. Sensitive and reproducible methods to isolate the cleaved peptides from human plasma have been compared and showed C18 solid phase extraction was a reliable method [38, 39]. C18 solid phase extraction of peptides was used to establish that peptides from C4B are released into plasma at room temperature. Here release of the C4B-peptide (NGFKSHALQLNNRQIR) in human plasma was compared over storage and incubation conditions by random and independent sampling with LC–ESI–MS/MS.
The experiments showed that a plasma sample remaining at room temperature undergoes polypeptide degradation compared to ice cold, frozen samples or freeze dried samples stored at room temperature. Proteolytic degradation of plasma at room temperature resulted in the production of peptides containing the C4B-peptide sequence. Here release of the C4B-peptide (NGFKSHALQLNNRQIR) in human plasma was compared over storage and incubation conditions by random and independent sampling with LC–ESI–MS/MS as confirmed by automated targeted analysis. The C4B peptides showed a low frequency of detection and low ion intensity values in samples collected on ice ± protease inhibitors, but showed a sharp increase at room temperature and remained strongly detectable for days on the laboratory bench. Plasma samples temporarily stored on ice ± protease inhibitors, stored at − 80 °C in or liquid nitrogen, or freeze dried and stored at − 20 °C or at room temperature, all show low levels of C4B-peptide, compared to plasma samples incubated at room temperature for a few hours or days. Here, three methods, random and independent sampling, automatic-targeted quantification, and Western blot all showed that processing of C4B reflects the proteolytic degradation of plasma at room temperature. An additional aim of the experiment was to determine if blood cells are required for the processing of plasma proteins.
Methods
Materials
The freeze dryer was from Labconco (Kansas City, MO, USA) and the 30 L pump was from Edwards (Sanborn, NY, USA). The Agilent 1100 HPLC (Santa Clara, CA, USA) for LC–ESI–MS/MS was coupled to an LTQ XL linear ion trap mass spectrometer from the Thermo Electron Corporation (Waltham, MA, USA). The HPLC grade water and acetonitrile were obtained from Caledon Laboratories (Georgetown, Ontario, Canada). The C4B antibody (PA1-9534, Lot No. QB1980891) and the Pierce EZ Link NHS biotinyation kit was obtained from Thermo Fisher Scientific (Waltham, MA, USA). The streptavidin-HRP conjugate was obtained from Jackson ImmunoResearch (West Grove, PA, USA).
Sample collection
Human plasma was collected under a protocol approved by the Comité National d’Ethique de Recherche (CNER) Protocol #201107 “Biospecimen Research” at the Centre Hospitalier de Luxembourg. EDTA blood samples from 30 male and female healthy subjects, ranging in age from 24 to 74 were collected immediately onto ice, centrifuged at 2000×g for 20 min at 4 °C and plasma was aliquoted in 225 μl volumes on ice, and briefly held at − 80 °C prior to randomly assigning to short-term or long-term experimental storage treatments. Short term storage experiments were ice (ICE) or ice plus protease inhibitors (ICE-INH) or room temperature (RT) for up to 96 h as indicated. Long term storage conditions included − 80 °C or liquid nitrogen, (LN2), or freeze drying (FD) followed by storage at − 20 °C (FD-20 °C) or room temperature (FDRT). Freeze dried samples were first frozen to − 80 °C and then rapidly placed in a Labconco rotary sample speedvac with the condenser maintained at − 90 °C with a 30 L per minute pump (a vacuum strong enough to rapidly freeze water) for 24 h before rapidly resealing the vials. There was no heat applied, and no organic solvents or salts were added, and the samples remained frozen under strong vacuum during the drying by means of a − 90 °C condenser and thus were lyophilized, i.e. freeze dried.
Short term storage experiment
Plasma aliquots (225 µl) from the thirty donors were thawed on ice and then randomly assigned to incubation at room temperature or on ice for varying times. The room temperature (RT) and control samples on ice ± protease inhibitors (ICE and ICE-INH) were incubated for various times, up to 72 h. The protease inhibitors AEBSF, PMSF, benzamidine HCl, and caproic acid were used at 2 mM each with the Sigma Eukaryotic protease inhibitor cocktail at 1/100 (v/v). The Sigma Mammalian Protease inhibitor cocktail contains at least: AEBSF, 104 mM, Aprotinin, 80 μM, Bestatin, 4 mM, E-64, 1.4 mM, Leupeptin, 2 mM, Pepstatin A, 1.5 mM used at 1/100 (v/v). Plasma samples (225 µl) from at least 10 different donors were tested at each time point and over the time course of degradation up to 72 h. At the end of each time period, the samples were frozen, freeze dried and stored dried at − 80 °C until analysis.
Long term storage experiment
Plasma aliquots from the 30 donors (225 µl) were randomly assigned to a − 80 °C freezer (− 80 °C), liquid nitrogen (LN2), freeze dried and stored in a desiccator at room temperature (FDRT) or freeze dried and stored at −20 °C (FD-20 °C) until analysis.
Random and independent sampling by LC–ESI–MS/MS
Plasma samples of 25 μl were dissolved in 225 μl of ice cold 5% formic acid prior to collection of the peptides over a preparative, ZipTip, C18 column [39]. The ~ 2 µl elution volume was aspirated and ejected across the C18 resin bed carefully 5 times to avoid permitting air bubbles into the resin bed. Collected peptides were eluted off the ZipTip in 2 µl of acidified 65% acetonitrile and immediately diluted with 18 µl of 5% formic acid and injected for analytical HPLC separation over a 15 cm × 300 µm ID column coupled to an electrospray source for the LTQ XL linear ion trap mass spectrometer (Thermo Electron Corporation). A federated library of human proteins was assembled from NCBI, Ensembl and Swiss-Prot and made non-redundant using Structured Query Language (SQL) [40, 41]. The experimental MS and MS/MS spectra of peptides recorded were correlated to predicted spectra from the federated library at a charge state of 2+ and 3+ to identify fully tryptic peptides using the X!TANDEM [42], OMSSA [43], MASCOT [44] and SEQUEST algorithms [45, 46] set within ± 3 m/z in the precursor mass and within ± 0.5 Da in the fragment mass with up to three missed cleavages [47, 48], as proteins may be only partially digested by proteases. Only the best fit peptide to each MS/MS spectra in terms of charge state or amino acid sequence was accepted.
Automated targeted LC–ESI–MS/MS
The LC–ESI–MS/MS was repeated with targeted analysis to monitor the C4B-peptide (NGFKSHALQLNNR) released from complement component 4B. The LC–ESI–MS/MS analysis by micro electrospray was repeated but instead of random and independent sampling, the LTQ ion trap was set to monitor NGFKSHALQLNNR [with a monoisotopic mass 1497.78 and so at + 2 charge [M + 2H]2+ then (1497.78 + 2)/2 = 749.5 m/z and at + 3 charge [M + 3H]3+ then (1497.78 + 3)/3 = 500.26 m/z] with ± 3 m/z. The MS/MS spectra from the targeted LC–MS/MS of the C4B-peptide was confirmed by searching the results against a protein library that only contained the target sequence NGFKSHALQLNNR from C4B using SEQUEST as described above, but with no cut off at 1000 counts, in order to increase sensitivity. The frequency and intensity of precursors within ± 3 m/z of the C4B peptide NGFKSHALQLNNR were confirmed by automated targeted analysis where only the MS/MS spectra that correlated to the target sequence was stored in SQL for Statistical Analysis in R.
Western blot
A total of 2 µl of EDTA plasma was dissolved in 25 µl of 2 × SDS-PAGE sample-buffer and boiled for 10 min. The EDTA plasma samples were separated over 9% acrylamide discontinuous tris gels with a pH 6.8 stacking gel and a pH 8.8 separating gel in tricine tank buffer at 100 V [49]. The samples were transferred to PVDF (that was pre-wetted in methanol) at 100 V in Towbin buffer [50]. The transfer was confirmed by staining with CBBR in 50% methanol, the position of the molecular weight markers was marked in pencil and the blots erased in pure methanol prior to equilibrating in PBST and Western blot with 1/500 (v/v) biotinylated primary antibody. The binding of the primary antibody was detected with 1/10,000 (v/v) streptavidin HRP conjugate using enhanced chemiluminescence [51].
Statistical analysis
The precursor ion intensity values that were correlated to C4B tryptic peptides by SEQUEST, together with the parent and fragment m/z and intensity values were automatically parsed into an SQL database [40]. The peptide intensity and frequency values were statistically analyzed using classical statistical approaches such as Chi Square and ANOVA for each protein [2, 52,53,54,55,56]. The peptide intensity values were log10 transformed to approximate a normal distribution and then analyzed by ANOVA prior to the Tukey–Kramer Honestly Significant Difference (HSD) and plotted using the R Statistical Analysis System [52, 53, 57]. The Chi Square analysis of the frequency versus intensity histogram for the various treatments was performed and graphed using the generic open-source R statistical system.
Results
Random and independent sampling of all peptides from all proteins
The tryptic peptides in human EDTA plasma were analyzed by random and independent sampling of the 5 highest intensity peptides eluting at any moment from the HPLC–ESI–MS/MS system. The MS/MS spectra were correlated to the tryptic peptides of the human proteome in a federated protein library with up to three missed cleavages that revealed the processing of complement component 4B (C4B) (Table 1, Fig. 1). Random and independent sampling of C4B from ice cold or room temperature EDTA plasma by LC–ESI–MS/MS with correlation by MASCOT, OMSSA, X!TANDEM and SEQUEST showed p values for individual MS/MS spectra that ranged from p < 0.1 (E-1) to p < E − 150 (Fig. 1). Plasma samples that were maintained on ice over time showed very little cleavage of C4B in contrast to samples incubated at room temperature that showed the cleavage of the C4B peptides (Table 2, Fig. 2). The C4B peptides NGFKSHALQLNNR and GLEEELQFSLGSK were among the most commonly observed in agreement with previous results [29]. The release of peptides from complement C4B was readily detected in room temperature samples by unbiased LC–ESI–MS/MS (Table 2, Fig. 2). The C4B peptides were almost undetectable at time zero (ICE), but were clearly detectable in as little as 1 h or more at room temperature. After incubation at room temperature for 4–8 h the most characteristic peptide from C4B (NGFKSHALQLNNR) was observed with greater frequency that was apparently significant by the Chi Square test (Fig. 3) that indicated a low probability (< 0.0001) that the room temperature samples were the same as the control. The intensity distribution of the C4B-peptide approached normality as assessed by quantile plots (Fig. 4a). The C4B-peptide intensity values were typically less than 1000 intensity counts at time zero (ICE). The C4B-peptide showed detectable intensity values after 1 h with a mean of just above 1000 intensity counts, that increased by almost an order of magnitude over 4–8 h at room temperature (RT).
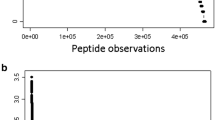
The distribution of p values (or Xcorr values) versus the peptide expected [M + H]+ of C4B from the random and independent sampling degraded plasma. The log10 expectation values (p values) from MASCOT, OMSSA, X!TANDEM and (or Xcorr values) SEQUEST algorithms are plotted against peptide [M + H]+ values. a MASCOT a heuristic probability-based MOWSE score algorithm; b OMSSA a heuristic probability algorithm; c X!TANDEM a goodness of fit algorithm; d SEQUEST a modified Pearson cross correlation algorithm (Xcorr). C4B was the most commonly observed protein by many independent peptides in room temperature samples
Typical examples of the MS/MS spectra of unique peptides from complement C4B. a MS/MS spectra that correlated to the fully tryptic amino acid sequence GLEEELQFSLGSK with [M + H]+ 1437.33 and a z value of 2+; b MS/MS spectra that correlated to the fully tryptic amino acid sequence NGFKSHALQLNNR with [M + H]+ 1500.23 and a z value of 3+. The MS/MS fragment ions are labelled exactly as provided by the Proteome Discoverer system that used the SEQUEST algorithm provided by Thermo Fisher. The MS/MS spectra were obtained from random and independent sampling of all precursor ions during unbiased LC–ESI–MS/MS
The histogram of log10 precursor intensity of the C4B-peptide (NGFKSHALQLNNR) by random and independent sampling over incubation of EDTA plasma on ice versus room temperature over time. The precursor ion intensity filter of 1000 counts was imposed before examining the precursor with the fitting algorithms. Random and independent sampling showed a 10-fold to 100-fold increase in C4B-peptide detection with incubation at room temperature
The quantification of C4B-peptide (NGFKSHALQLNNR) by random and independent sampling by LC–ESI–MS/MS. Panels: a, the quantile plot of the randomly sampled C4B peptides; b, the intensity values of the randomly sampled C4B-peptide log10 intensity value (arbitrary counts) over time. The probability that the time points were the same by ANOVA was p ≤ 0.001. The time point 0 refers to samples maintained on ice
Automatic-targeted LC–ESI–MS/MS analysis
Random and independent sampling showed a large increase in C4B detection frequency and intensity in plasma with time at room temperature but has greater sampling error compared to targeted measurement of C4B peptides. The MS/MS spectra from peptides with precursors within ± 3 m/z of the predicted 2+ or 3+ m/z values were automatically searched against the C4B-peptide NGFKSHALQLNNR using the SEQUEST algorithm. The correlated peptide count and precursor intensity values were automatically collected in SQL Server for the automatic transformation, computation of means, normality, intensity differences by ANOVA and frequency differences by Chi Square using the R statistical analysis system (Fig. 5). The fully automated targeted analysis of the C4B peptide NGFKSHALQLNNR using SQL and R showed a log10 intensity distribution of the peptide that approached Gaussian normality (Fig. 5a). Comparing baseline plasma samples versus those incubated for 1 or 72 h at RT confirmed a sharp increase in average peptide intensity values from time 0 to 1 or 72 h at room temperature by ANOVA followed by the Tukey–Kramer test (Fig. 5b). The NGFKSHALQLNNR peptide counts from all precursors increased dramatically from time 0 (on ice) to thousands or even tens of thousands of counts by as little as 1 h and up to 72 h at room temperature (Fig. 5c).
The automatic-targeted analysis of the complement C4B-peptide (NGFKSHALQLNNR) at time 0, 1 and 72 h incubation at room temperature. a, The normality of the log10 transformed intensity data as assessed by quantile plot; b the mean and standard deviation of the targeted precursor intensity value (different letters indicate a significant difference by the Tukey–Kramer Honestly Significant Difference Test); c the histogram showing the frequency of observing peptides in each log10 intensity bin. Automatic targeted sampling showed a 100-fold increase in frequency and about a 10-fold to 100-fold increase in C4B-peptide intensity with incubation at room temperature
Primary structural analysis of C4B
Peptides consistent with the cleavage of the C4B preproprotein to yield the mature chain were observed (Fig. 6). The most commonly detected cleavage site on the mature C4B chain was on the carboxyl side of the isoprene C2 domain shared with alpha 2 macroglobulin as reflected by LC–ESI–MS/MS. A frequent C4B cleavage site was within the polar C terminal sequence 1337RNGFKSHALQLNNRQIRGLEEELQFSLGSKINVK1370 (NP_001002029.3) that contained the most commonly observed peptides NGFKSHALQLNNR and GLEEELQFSLGSK (Fig. 6).
The location of the most common peptide sequences that were cleaved within the complement C4B protein from the protein accession NP_001002029.3 The peptide sequence NGFKSHALQLNNRQIR shows no significant relationship to any other protein and contained 31% of all observed peptides from C4B. The peptide sequence GLEEELQFSLGSKINVK accounted for 11% of all peptides. The arrows show the locations of the main peptide cleavage sites on the carboxyl side of the isoprene C2 domain at a site within a sequence of basic, acid and polar amino acids. The bracket shows a section of the preproprotein that is also cleaved upon warming to room temperature producing the mature chain detectable by Western blot. The C4B amino acid sequence 1337RNGFKSHALQLNNRQIRGLEEELQFSLGSKINVK1370 on the carboxyl side of the isoprene C2-like superfamily domain was the most frequent site of cleavage (see arrows). The sub-sequence 1137NGFKSHALQLNNR1352 is located just to the carboxyl terminal side of a local hydrophilic maximum and flanks the evolutionarily conserved isoprene C2 sequence shared by the innate defense proteins complement 4A/B and alpha 2 macroglobulin. The cleavage sites of the peptide(s) NGFKSHALQLNNRQIR are flanked on the amino side by short stretches of hydrophilic amino acids, the released peptide includes asparagine and glutamine, and there is a stretch of three glutamic acid residues and glutamine adjacent on the carboxyl side of the cleavage site
Western blot against the mature C4B protein
Samples of EDTA plasma were resolved by tricine SDS-PAGE and blotted to PVDF supports for Western analysis. The Western blot showed that the antibody recognizes the processed form of mature C4B as expected. Western analysis strongly detected the parent C4B protein in close agreement with the predicted relative mass of 84,163 Da and its major cleavage product at 71,539 kD based on the annotation found at NCBI NP_001002029.3 (Fig. 7). The cleavage of the C-terminal portion of the C4B chain should yield a protein of about 63 kDa as observed (Fig. 7).
Western blot against the mature C4B protein chain in human EDTA plasma over the time course of degradation at room temperature. The monoclonal antibody against C4B was biotinylated and detected with Streptavidin-HRP. The arrow indicates the expected C4B peptide after proteolysis. Lanes: MW, Molecular weight marker as indicated; A, Streptavidin HRP alone; 1, time 0; 2, 1 h; 3, 1 h; 4, 4 h; 5, 8 h; 6, 24 h; 7, 36 h; 8, 48 h; 9, 72, hr; 10, 96 h. Molecular weight markers for a 9% Tricine SDS-PAGE gel are shown in kilodaltons (kDa). The Western blot analysis showed the increased formation of mature C4B with time and the development of an additional band with time at room temperature confirming the proteolytic processing of C4B. Complement C4 is expressed as a high molecular mass preproprotein that reacts poorly with the antibody raised against the mature protein that exits in blood as a 79 kDa mature chain in close agreement with the most intense band in the Western blot that appears roughly in line with the 75 kDa molecular mass marker. Proteolytic cleavage of the C-terminal domain from C4B starting at NGFKSHALQLNNR to the carboxyl terminus of the C4B protein should yield a 63 kDa protein (see arrow). Control blots stained with Coomassie Blue confirmed equal loading of all lanes (see Additional file 1: Fig. S1). These data confirm the finding shown in Table 2 and Fig. 2. The manufacturer (THERMO) indicates the antibody recognizes an 88, 75 and 33 kDa form of C4B in good agreement
Plasma sample long-term storage by freezing and freeze-drying
Plasma was protected from degradation, as measured by the release of C4B-peptide, by freeze drying followed by 1 year storage at room temperature (FDRT), freeze drying followed by 1 year storage at − 20 °C (FD-20 °C), 1 year freezing at − 80 °C or 1 year freezing in liquid nitrogen (LN2) (Fig. 8). Furthermore, there were no signs of proteolytic degradation after short-term storage of plasma samples on ice ± protease inhibitors, for up to 3 days (Fig. 8). The Chi Square test showed a low probability (< 0.0001) that the degraded samples were the same as the control.
The histogram showing a comparison of the preservation methods of ice ± protease inhibitors, freezing versus freeze drying alongside a positive control for sample degradation as illustrated by C4B-peptide (NGFKSHALQLNNR) by random and independent sampling by LC–ESI–MS/MS. Storage treatments: freeze drying and storage at room temperature (FDRT); freeze drying and storage at −20 (FD-20); freezing storage at −80 °C (−80); or liquid nitrogen (LN2); versus plasma degraded for 96 h at room temperature (96 h RT). The histogram shows the frequency of peptides in each log10 intensity bin. The precursor ion intensity filter of 1000 counts was imposed before examining the data in R
Discussion
This study regarded the cleavage of the C4B protein at room temperature compared to ice cold, frozen, or freeze dried plasma samples by random and independent sampling, or automatic targeted quantification by LC–ESI–MS/MS, and SDS-PAGE with Western blot. All three analytical methods show a similar trend of little detectable C4B-peptide cleavage in ice cold, frozen or freeze dried plasma samples. Cleavage of C4B rapidly became apparent after incubation for 1–4 h at room temperature using sensitive LC–ESI–MS/MS.
Random and independent sampling
Without any pre-conceived notion of what proteins or peptides might be the best for measuring sample degradation, random and independent sampling was used to compare all possible human tryptic peptides across all sample storage conditions. The counting of peptides to proteins across treatments with SQL/R was a simple means to identify the proteins in human plasma that degrade with incubation at room temperature. The C4B-peptide levels may serve as a marker of sample degradation based on the ≥ tenfold increase in the frequency of detection or peptide intensity over time at room temperature. We conclude that the use of random and independent sampling of peptide frequency and/or intensity values may serve as a means to quantify the cleavage of C4B. In randomly and independently sampled experiments the intensity values at time zero for the C4B-peptide NGFKSHALQLNNR were apparently below the noise cut off used in this statistical experiment but the peptide was detectable by 1 h at room temperature and showed a marked increase in both frequency and intensity by 8 h. The large differences in the peptides frequency and intensity over time indicated that the cleavage of the peptide NGFKSHALQLNNR from complement C4B may serve as a measure of the degradation of the sample due to endopeptidase activity at room temperature after sample collection [1, 2]. Random and independent sampling using unbiased LC–ESI–MS/MS is a costly and time consuming approach that is necessary to make unbiased discoveries of candidate markers but is not an efficient means to assay an individual protein.
Automatic-targeted analysis of Complement 4B NGFKSHALQLNNR
Automatic targeted analysis showed that the C4B-peptide (NGFKSHALQLNNR) was readily detected in the ice control and degraded plasma samples and increased by about ≥ tenfold by 1 h at room temperature. After log10 transformation, the C4B-peptide intensity was Gaussian and could be used to provide relative quantification and statistical analysis using ANOVA. The automatic-targeted quantification of the precursor peptide ion of the C4B-peptide is a promising step towards creating a quantitative assay for the quality control of EDTA plasma samples. In this approach a low resolution, yet robust and sensitive, ion trap is set to monitor the precursor window where automatic computations based on the fit of the MS/MS spectra to the target peptide sequence resolves the intended precursor from any ions with similar m/z values.
Complementary sampling strategy
Random and independent sampling from a totally random experimental design is the statistical gold standard for avoiding false discovery. Pooling representative samples, or taking ratios of isotopic or isobaric tags may reduce or eliminate measured biological and sampling error leading to false discovery. Moreover, making a ratio of isotopic or isobaric peptides, such as ICAT ratios, lacks independence and tends to multiply the error in the two samples (denominator error × numerator error) leading to reduced statistical power. Hence the random and independent sampling of separate biological samples is an appropriate strategy to discover real differences between populations that provides a realistic assessment of population variance. In a complementary approach, the automatic targeted method captured biological variation but avoided sampling error by a high frequency sampling of the targeted analyte that shows that C4B intensity levels increased within 1 h at room temperature and remained elevated and increased for at least 72 h that showed little sampling error. Only the intensity and frequency values of precursors within ± 3 m/z that yielded MS/MS spectra automatically correlated to the target peptide by the SEQUEST algorithm were accepted in SQL. Thus the fit of the MS/MS spectra ensured the validity of the precursors accepted into the dataset for automated statistical analysis in R. The combination of random and independent sampling, together with automatic targeted confirmation, as demonstrated for the first time here, is a sensitive and practical approach to biomarker discovery in human plasma that avoids false discovery and yet automatically provides confirmatory measurements with low technical error.
Room temperature versus ice, frozen or freeze-dried samples
Storing samples on ice was an effective means to prevent proteolytic degradation for up to 3 days. The use of C4B-peptide counting and precursor intensity versus frequency analysis clearly indicated that freeze drying and storage at room temperature, or freeze drying and storage at − 20 °C were just as effective in preventing sample degradation as freezing at − 80 °C or liquid nitrogen for up to 1 year. However degradation of complement C4B commences very rapidly upon thawing and does not reflect the stability of most plasma proteins and so unless the samples are collected on ice, freeze dried and reconstituted on ice ± protease inhibitors some degradation of C4B is likely unavoidable.
Conclusion
Three different methods, random and independent sampling, automatic-targeted analysis, and Western blot all agreed that the cleavage of C4B gives a good indication of the sample incubation at RT. The time that a clinical plasma sample remains at room temperature was a key factor in the cleavage of C4B. The simplest explanation of the observations here is that a tryptic protease activity in human plasma acts ex vivo at room temperature to cleave a proportion of the total pool of complement C4B and this processing may be directly detectable by sensitive LC–ESI–MS/MS or immunological methods. We conclude that the complement proteins, that are part of a proteolytic cascade, are especially sensitive to ex vivo degradation of EDTA plasma during sample warming. The peptide NGFKSHALQLNNR from complement 4B showed a rapid increase in the frequency and intensity of detection after warming to room temperature and increased with hours of incubation and then remained elevated over the subsequent days. Thus, after all blood cells are removed from plasma on ice, C4B peptide levels remained low, but yet subsequently increased with time at room temperature entirely in the absence of white blood cells. All of the results are consistent with the expression of endogenous complement fragments that represents a balance of endoproteinase activity versus exopeptidase activity. The C4B peptide assays indicates that the highly soluble proteins in human plasma may be preserved by freeze drying and entirely reconstituted upon the addition of water and so freeze drying is an attractive option for sample preservation that would also permit robust and reliable shipment of samples at low cost.
Abbreviations
- AEBSF:
-
4-benzenesulfonyl fluoride hydrochloride
- ACN:
-
acetonitrile
- C4B:
-
complement component 4B
- C4B-peptide:
-
NGFKSHALQLNNR
- ICE:
-
fresh EDTA plasma stored on wet ice
- ICE_INHIB:
-
fresh EDTA plasma stored on wet ice with protease inhibitors
- RT:
-
fresh EDTA plasma stored at room temperature
- LN2 :
-
fresh EDTA plasma stored in liquid nitrogen
- FD:
-
freeze dried EDTA plasma
- FD-20 °C:
-
freeze dried EDTA plasma stored in an electric freezer at − 20 °C
- FDRT:
-
EDTA plasma freeze dried and stored in a desiccator at room temperature
- PMSF:
-
phenylmethane sulfonyl fluoride
- PVDF:
-
polyvinylidene fluoride
References
Marshall J, Jankowski A, Furesz S, Kireeva I, Barker L, Dombrovsky M, Zhu W, Jacks K, Ingratta L, Bruin J, Kristensen E, Zhang R, Stanton E, Takahashi M, Jackowski G. Human serum proteins preseparated by electrophoresis or chromatography followed by tandem mass spectrometry. J Proteome Res. 2004;3:364–82.
Marshall J, Kupchak P, Zhu W, Yantha J, Vrees T, Furesz S, Jacks K, Smith C, Kireeva I, Zhang R, Takahashi M, Stanton E, Jackowski G. Processing of serum proteins underlies the mass spectral fingerprinting of myocardial infarction. J Proteome Res. 2003;2:361–72.
Zhu P, Bowden P, Zhang D, Marshall JG. Mass spectrometry of peptides and proteins from human blood. Mass Spectrom Rev. 2011;30:685–732.
Wiita AP, Hsu GW, Lu CM, Esensten JH, Wells JA. Circulating proteolytic signatures of chemotherapy-induced cell death in humans discovered by N-terminal labeling. Proc Natl Acad Sci USA. 2014;111:7594–9.
Qin S, Zhou Y, Gray L, Kusebauch U, McEvoy L, Antoine DJ, Hampson L, Park BK, Campbell DS, Caballero J, Glusman G, Yan X, Kim TK, Yuan Y, Wang K, Rowen L, Moritz RL, Omenn GS, Pirmohamed M, Hood L. Identification of blood protein biomarkers of acute liver injury by targeted quantitative proteomics in acetaminophen and carbon tetrachloride treated mouse models and acetaminophen overdose patients. J Proteome Res. 2016;15:3724–40.
Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, Reissfelder C, Pilarsky C, Fraga MF, Piwnica-Worms D, Kalluri R. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–82.
Sardesai VM, Provido HS. A fluorometric method for determining the tame esterase (tryptic) activity of plasma. J Lab Clin Med. 1965;65:1023–9.
Weisman HF, Bartow T, Leppo MK, Marsh HC Jr, Carson GR, Concino MF, Boyle MP, Roux KH, Weisfeldt ML, Fearon DT. Soluble human complement receptor type 1: in vivo inhibitor of complement suppressing post-ischemic myocardial inflammation and necrosis. Science. 1990;249:146–51.
Volanakis JE. Participation of C3 and its ligands in complement activation. Curr Top Microbiol Immunol. 1990;153:1–21.
Fung KW, Wright DW, Gor J, Swann MJ, Perkins SJ. Domain structure of human complement C4b extends with increasing NaCl concentration: implications for its regulatory mechanism. Biochem J. 2016;473:4473–91.
Banks RE. Preanalytical influences in clinical proteomic studies: raising awareness of fundamental issues in sample banking. Clin Chem. 2008;54:6–7.
Tuck MK, Chan DW, Chia D, Godwin AK, Grizzle WE, Krueger KE, Rom W, Sanda M, Sorbara L, Stass S, Wang W, Brenner DE. Standard operating procedures for serum and plasma collection: early detection research network consensus statement standard operating procedure integration working group. J Proteome Res. 2009;8:113–7.
Karsan A, Eigl BJ, Flibotte S, Gelmon K, Switzer P, Hassell P, Harrison D, Law J, Hayes M, Stillwell M, Xiao Z, Conrads TP, Veenstra T. Analytical and preanalytical biases in serum proteomic pattern analysis for breast cancer diagnosis. Clin Chem. 2005;51:1525–8.
Semmes OJ, Feng Z, Adam BL, Banez LL, Bigbee WL, Campos D, Cazares LH, Chan DW, Grizzle WE, Izbicka E, Kagan J, Malik G, McLerran D, Moul JW, Partin A, Prasanna P, Rosenzweig J, Sokoll LJ, Srivastava S, Thompson I, Welsh MJ, White N, Winget M, Yasui Y, Zhang Z, Zhu L. Evaluation of serum protein profiling by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry for the detection of prostate cancer: I. Assessment of platform reproducibility. Clin Chem. 2005;51:102–12.
de Noo ME, Tollenaar RA, Ozalp A, Kuppen PJ, Bladergroen MR, Eilers PH, Deelder AM. Reliability of human serum protein profiles generated with C8 magnetic beads assisted MALDI-TOF mass spectrometry. Anal Chem. 2005;77:7232–41.
Li J, Orlandi R, White CN, Rosenzweig J, Zhao J, Seregni E, Morelli D, Yu Y, Meng XY, Zhang Z, Davidson NE, Fung ET, Chan DW. Independent validation of candidate breast cancer serum biomarkers identified by mass spectrometry. Clin Chem. 2005;51:2229–35.
Pilny R, Bouchal P, Borilova S, Ceskova P, Zaloudik J, Vyzula R, Vojtesek B, Valik D. Surface-enhanced laser desorption ionization/time-of-flight mass spectrometry reveals significant artifacts in serum obtained from clot activator-containing collection devices. Clin Chem. 2006;52:2115–6.
Albrethsen J, Bogebo R, Olsen J, Raskov H, Gammeltoft S. Preanalytical and analytical variation of surface-enhanced laser desorption-ionization time-of-flight mass spectrometry of human serum. Clin Chem Lab Med. 2006;44:1243–52.
Koomen JM, Li D, Xiao LC, Liu TC, Coombes KR, Abbruzzese J, Kobayashi R. Direct tandem mass spectrometry reveals limitations in protein profiling experiments for plasma biomarker discovery. J Proteome Res. 2005;4:972–81.
Rai AJ, Gelfand CA, Haywood BC, Warunek DJ, Yi J, Schuchard MD, Mehigh RJ, Cockrill SL, Scott GB, Tammen H, Schulz-Knappe P, Speicher DW, Vitzthum F, Haab BB, Siest G, Chan DW. HUPO Plasma Proteome Project specimen collection and handling: towards the standardization of parameters for plasma proteome samples. Proteomics. 2005;5:3262–77.
Callesen AK, Vach W, Jorgensen PE, Cold S, Mogensen O, Kruse TA, Jensen ON, Madsen JS. Reproducibility of mass spectrometry based protein profiles for diagnosis of breast cancer across clinical studies: a systematic review. J Proteome Res. 2008;7:1395–402.
van Winden AW, Gast MC, Beijnen JH, Rutgers EJ, Grobbee DE, Peeters PH, van Gils CH. Validation of previously identified serum biomarkers for breast cancer with SELDI-TOF MS: a case control study. BMC Med Genomics. 2009;2:4.
Spencer K. The influence of different sample collection types on the levels of markers used for Down’s syndrome screening as measured by the Kryptor immunosassay system. Ann Clin Biochem. 2003;40:166–8.
Govorukhina NI, de Vries M, Reijmers TH, Horvatovich P, van der Zee AG, Bischoff R. Influence of clotting time on the protein composition of serum samples based on LC-MS data. J Chromatogr B Anal Technol Biomed Life Sci. 2008;877:1281–91.
Gast MC, van Gils CH, Wessels LF, Harris N, Bonfrer JM, Rutgers EJ, Schellens JH, Beijnen JH. Influence of sample storage duration on serum protein profiles assessed by surface-enhanced laser desorption/ionisation time-of-flight mass spectrometry (SELDI-TOF MS). Clin Chem Lab Med. 2009;47:694–705.
West-Nielsen M, Hogdall EV, Marchiori E, Hogdall CK, Schou C, Heegaard NH. Sample handling for mass spectrometric proteomic investigations of human sera. Anal Chem. 2005;77:5114–23.
West-Norager M, Kelstrup CD, Schou C, Hogdall EV, Hogdall CK, Heegaard NH. Unravelling in vitro variables of major importance for the outcome of mass spectrometry-based serum proteomics. J Chromatogr B Anal Technol Biomed Life Sci. 2007;847:30–7.
Jin Y, Manabe T. Differences in protein distribution between human plasma preparations, EDTA-plasma and heparin-plasma, analyzed by non-denaturing micro-2-DE and MALDI-MS PMF. Electrophoresis. 2009;30:931–8.
Kaisar M, van Dullemen LF, Thezenas ML, Zeeshan Akhtar M, Huang H, Rendel S, Charles PD, Fischer R, Ploeg RJ, Kessler BM. Plasma degradome affected by variable storage of human blood. Clin Proteomics. 2016;13:26.
Williams D, Ackloo S, Zhu P, Bowden P, Evans KR, Addison CL, Lock C, Marshall JG. Precipitation and selective extraction of human serum endogenous peptides with analysis by quadrupole time-of-flight mass spectrometry reveals posttranslational modifications and low-abundance peptides. Anal Bioanal Chem. 2010;396:1223–47.
van den Broek I, Sparidans RW, Schellens JH, Beijnen JH. Liquid chromatography/tandem mass spectrometric method for the quantification of eight proteolytic fragments of ITIH4 with biomarker potential in human plasma and serum. Rapid Commun Mass Spectrom. 2008;22:2915–28.
Yi J, Kim C, Gelfand CA. Inhibition of intrinsic proteolytic activities moderates preanalytical variability and instability of human plasma. J Proteome Res. 2007;6:1768–81.
Yi J, Liu Z, Craft D, O’Mullan P, Ju G, Gelfand CA. Intrinsic peptidase activity causes a sequential multi-step reaction (SMSR) in digestion of human plasma peptides. J Proteome Res. 2008;7:5112–8.
Zheng X, Baker H, Hancock WS, Fawaz F, McCaman M, Pungor E Jr. Proteomic analysis for the assessment of different lots of fetal bovine serum as a raw material for cell culture. Part IV. Application of proteomics to the manufacture of biological drugs. Biotechnol Prog. 2006;22:1294–300.
Yang X, Clifton J, Huang F, Kovac S, Hixson DC, Josic D. Proteomic analysis for process development and control of therapeutic protein separation from human plasma. Electrophoresis. 2009;30:1185–93.
Kulik W, van Lenthe H, Stet FS, Houtkooper RH, Kemp H, Stone JE, Steward CG, Wanders RJ, Vaz FM. Bloodspot assay using HPLC-tandem mass spectrometry for detection of Barth syndrome. Clin Chem. 2008;54:371–8.
Gharahdaghi F, Kirchner M, Fernandez J, Mische SM. Peptide-mass profiles of polyvinylidene difluoride-bound proteins by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in the presence of nonionic detergents. Anal Biochem. 1996;233:94–9.
Tucholska M, Florentinus A, Williams D, Marshall JG. The endogenous peptides of normal human serum extracted from the acetonitrile-insoluble precipitate using modified aqueous buffer with analysis by LC-ESI-Paul ion trap and Qq-TOF. J Proteomics. 2010;73:1254–69.
Tucholska M, Scozzaro S, Williams D, Ackloo S, Lock C, Siu KWM, Evans KR, Marshall JG. Endogenous peptides from biophysical and biochemical fractionation of serum analyzed by matrix-assisted laser desorption/ionization and electrospray ionization hybrid quadrupole time-of-flight. Anal Biochem. 2007;370:228–45.
Bowden P, Beavis R, Marshall J. Tandem mass spectrometry of human tryptic blood peptides calculated by a statistical algorithm and captured by a relational database with exploration by a general statistical analysis system. J. Proteomics. 2009;73:103–11.
Bowden P, Pendrak V, Zhu P, Marshall JG. Meta sequence analysis of human blood peptides and their parent proteins. J Proteomics. 2010;73:1163–75.
Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–7.
Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open mass spectrometry search algorithm. J Proteome Res. 2004;3:958–64.
Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–67.
Yates JR III, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem. 1995;67:1426–36.
Yates JR III. Database searching using mass spectrometry data. Electrophoresis. 1998;19:893–900.
Zhu P, Bowden P, Tucholska M, Marshall JG. Chi square comparison of tryptic peptide-to-protein distributions of tandem mass spectrometry from blood with those of random expectation. Anal Biochem. 2011;409:189–94.
Zhu P, Bowden P, Tucholska M, Zhang D, Marshall JG. Peptide-to-protein distribution versus a competition for significance to estimate error rate in blood protein identification. Anal Biochem. 2011;411:241–53.
Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–79.
Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4.
Haan C, Behrmann I. A cost effective non-commercial ECL-solution for Western blot detections yielding strong signals and low background. J Immunol Methods. 2007;318:11–9.
Florentinus AK, Bowden P, Sardana G, Diamandis EP, Marshall JG. Identification and quantification of peptides and proteins secreted from prostate epithelial cells by unbiased liquid chromatography tandem mass spectrometry using goodness of fit and analysis of variance. J Proteomics. 2012;75:1303–17.
Florentinus AK, Jankowski A, Petrenko V, Bowden P, Marshall JG. The Fc receptor–cytoskeleton complex from human neutrophils. J Proteomics. 2011;75:450–68.
Eckel-Passow JE, Oberg AL, Therneau TM, Bergen HR III. An insight into high-resolution mass-spectrometry data. Biostatistics. 2009;10:481–500.
Baggerly KA, Morris JS, Wang J, Gold D, Xiao LC, Coombes KR. A comprehensive approach to the analysis of matrix-assisted laser desorption/ionization-time of flight proteomics spectra from serum samples. Proteomics. 2003;3:1667–72.
Sorace JM, Zhan M. A data review and re-assessment of ovarian cancer serum proteomic profiling. BMC Bioinformatics. 2003;4:24.
Bowden P, Thavarajah T, Zhu P, McDonell M, Thiele H, Marshall JG. Quantitative statistical analysis of standard and human blood proteins from liquid chromatography, electrospray ionization, and tandem mass spectrometry. J Proteome Res. 2012;11:2032–47.
Authors’ contributions
JD calibrated, tuned and quality controlled the instruments and performed LC–ESI–MS/MS analysis. TH performed LC–ESI–MS/MS analysis. AF-M performed the sample aliquoting and performed LC–ESI–MS/MS analysis and edited the manuscript. JA performed LC–ESI–MS/MS analysis and helped to edit the manuscript. AF performed protein assays and SDS-PAGE. PB collected the data into an SQL database for statistical analysis in R. JM planned the experiment, and performed the clinical plasma collection and sample treatments and storage experiments.
Acknowledgements
This research was paid for by the Fonds National de la Recherche, Mobility of Researcher award through Luxembourg Institute of Health (formerly CRP Sante) and a direct grant from the Integrated Biobank of Luxembourg to JGM. We gratefully acknowledge the review and critique of this manuscript by Dr. Fay Betsou and Dr. R.A. Phillips of IBBL.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The raw data is provided in the supplemental materials or companion publications.
Consent for publication
No material from any other publication was used in this publication.
Ethics approval and consent to participate
Ethical approval and informed consent was obtained through the Comité National d’Ethique de Recherche (CNER) Protocol #201107 “Biospecimen Research” at the Centre Hospitalier de Luxembourg.
Funding
This research was paid for by the Fonds National de la Recherche, Mobility of Researcher award through Luxembourg Institute of Health (formerly CRP Sante) and the Integrated Biobank of Luxembourg to JGM.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional file
12014_2017_9170_MOESM1_ESM.docx
Additional file 1: Fig. S1. A replicate CBBR stained gel to confirm the equal loading of the Western blot shown in Figure 8 of the main paper. (see legend of Figure 8 for details).
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Dufresne, J., Hoang, T., Ajambo, J. et al. Freeze-dried plasma proteins are stable at room temperature for at least 1 year. Clin Proteom 14, 35 (2017). https://doi.org/10.1186/s12014-017-9170-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12014-017-9170-0