Abstract
Background
This study aims to investigate the effect of miR-21-5p on process of colon adenocarcinoma (COAD) cells and its connection with neural cell adhesion molecule L1 (CHL1).
Methods
Different expressions of mRNAs and miRNAs were calculated with microarray analysis. QRT-PCR and western blot were performed to quantify miR-21-5p and CHL1 expression. Flow Cytometry, MTT assay, colony formation assay, transwell assay and ELISA were performed to evaluate propagation and invasiveness of COAD cells. Dual luciferase reporter assay was employed to scrutinize the relationship between miR-21-5P and CHL1. We performed in vivo experiment to detect the impact of miR-21-5p and CHL1 on COAD tumor growth.
Results
Expression level of miR-21-5p increased in both COAD tissues and cells. MTT and Cell cycle assay showed that overexpression of miR-21-5p accelerated proliferation of COAD cells. Transwell assay indicated that miR-21-5p promoted cell invasion. The result of dual luciferase reporter assay indicated that miR-21-5p targeted CHL1 directly and inhibited its expression. The result of in vivo experiments showed that down-regulation of miR-21-5p decreased the volume and weight of tumor, while knockdown of CHLI stimulated tumor growth.
Conclusions
The overexpression of miR-21-5p can promote propagation and invasiveness of COAD cells through inhibiting the expression of CHL1.
Similar content being viewed by others
Impact statement
We aimed to investigate the effect of miR-21-5p on process of colon adenocarcinoma (COAD) cells and its connection with CHL1.The result showed that the over-expression of miR-21-5p promoted proliferation and invasion of COAD cells through targeting CHL1. The findings of our study may assist researchers in discovering the mechanism of progression of COAD and may even help in developing antitumor treatments.
Background
Colon adenocarcinoma (COAD), a type of human colorectal cancer, is the most common cancers leading to mortality (Phang et al., 2017). Treatments for COAD are classified into four methods, surgery, radiation therapy, chemotherapy and targeted therapy. Optimisation of surgery for patients with localised disease has an important impact on prognosis. Once COAD spreads widely, patients are often given drugs directed towards relieving symptoms to improve quality of life (Cunningham et al., 2010). There are three continually used screening modalities of COAD, including flexible sigmoidoscopy, total colonoscopy and fecal occult blood testing, which have assisted in lowering the mortality caused by COAD. Unexpectedly, cases of COAD in Asian countries have experienced a dramatic uprush during the past decade, while the rate of rectal adenocarcinoma (READ) descends at the speed of 3% every year in several western countries (Kostic et al., 2013). The situation seemed even worse for the latent causes of the origin and the trend of this regional malignancy have not been completely elaborated yet.
To control COAD, therapeutic and worthy-of-promotion targets should be causally related to the disease and obey the designed interventions of the corresponding therapy. Besides, desired biomarkers should be easy to measure, detect and related to clinical results. MicroRNAs (miRNAs) could meet both standards described above. MiRNAs are small RNAs with the length of 21 to 25 nucleotides, which often take part in the regulation of progression of cell cycle, differentiation, apoptosis as well as tumorigenesis (Bartel, 2004). MiRNA levels are altered in most types of tumor (Volinia et al., 2006). It has been shown that experimental operations of specific miRNAs regulate development of tumor in mouse-model systems (Georgantas 3rd et al., 2007; Wang et al., 2006; Dews et al., 2006). Previous study revealed that overexpression of miR-141 resulted in proliferation of COAD cells (Ding et al., 2017). It’s also reported that miR-1297 inhibited COAD cell migration, invasion and proliferation in vivo (Chen et al., 2014). If abnormal miRNA expression is a causal factor to carcinogenesis, regulating miRNA expression may be a new strategy in cancer treatments.
MicroRNA-21 (miR-21), located in 17q23.1, is an oncogenic microRNA that regulates some cancer-related gene expressions such as TPM1, PTEN, PDCD4, TIMP3, RHOB and PDCD and has been confirmed to be highly expressed in COAD (Bovell et al., 2013; Leone et al., 2013; Martin del Campo et al., 2015; Xu et al., 2014). MiR-21 also plays a role in several signaling pathways such as, Wnt/β-catenin, PTEN/PI3K/AKT andRAS/MEK/ERK (Xiong et al., 2013; Lan et al., 2015). Several studies confirmed that increasing expression of miR-21 was related to the prognosis of COAD in stage I (Kjaer-Frifeldt et al., 2012; Oue et al., 2014). Most researches uncovered an elevated expression of miR-21 in colorectal cancerous tissues in comparison to paracancerous tissues (Oue et al., 2014; Tokarz & Blasiak, 2012; Nielsen et al., 2011; Chen et al., 2013), while there are few researches on a specific type of cancer cells in colorectal cancer and the mechanism of how miR-21 influences progression of COAD still remains unclear. Therefore, it is essential to carry out deeper analyses of miR-21 and COAD.
Neural cell adhesion molecule L1 (CHL1) is a neural adhesion molecule, which may function in signal transduction pathways and also participate in normal tissues and various of human cancers (He et al., 2013). It was reported that CHL1 had a close relationship with mental retardation in 3p-syndrome (Wei et al., 1998), which was associated with schizophrenia in Chinese population (Chen et al., 2005). Alteration of CHL1 has been implicated in the development of different cancers, including colon cancer (Gavert et al., 2005). CHL1 is verified to inhibit invasive growth and able to suppress further metastatic spread in COAD (Senchenko et al., 2011). Moreover, some researches verified that miR-21 targeting CHL1 to inhibit the neuroblastoma cell growth (Li et al., 2016a). However, few studies have been carried out to research the molecular mechanism between CHL1 and miR-21in COAD at present.
Our study explored dysregulation of miR-21-5p in both COAD tissues and cells. MTT assay, flow cytometry assay and transwell assay were employed to show the effect of miR-21-5p on proliferation, cell cycle and invasion. Furthermore, we investigated the relationship between miR-21-5p and CHL1 through dual-luciferase report and western blot. Moreover, we detected the impact of miR-21-5p/CHL1 axis on proliferation by in vivo experiment. The findings of our study may assist researchers in discovering the mechanism of progression of COAD and may even help in developing antitumor treatments.
Methods
Samples collection
43 paired of cancerous and adjacent tissues were provided by COAD patients. Normal colonic epithelial cells and COAD cell lines CW-2, T84, SW1116, LoVo were offered by BeNa Culture Collection (BNCC, Beijing, China). The Second Hospital of Shandong University approved the study and all informed consent had been signed and gathered beforehand.
Cell culture
Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) together with additional 100 U/ml penicillin/streptomycin and 10% Fetal Bovine Serum (FBS, Invitrogen, Gaithersburg, MD, USA). The cells were incubated in a humidified incubator with 5% CO2 at 37 °C.
Microarray analysis
The microarray data of differentially expressed miRNAs and mRNAs in COAD tissues was analyzed from the Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/). Differentially expressed miRNAs and mRNAs were screened with a t-test (P < 0.05) combined with fold change (FC) (log2 (FC) > 2 for up-regulated, and log2 (FC) < − 2 for down-regulated).The target genes and the binding sites of miRNAs were predicted by TargetScan 7.1 database (www.targetscan.org).
Quantitative real time polymerase chain reaction (qRT-PCR)
Total RNA was extracted by Trizol Reagent (Invitrogen). RNAs were reversely transcribed into cDNA using Takara Reverse Transcription System (Takara, Dalian, China) before qPCR was carried out. The reaction program was set initially at 95 °C for 30 s, followed by other 40 cycles composed of 95 °C for 10 s and 60 °C for 30 s, with a final dissociation stage of 95 °C for 15 s, 60 °C for 1 min and 95 °C for 15 s. GAPDH and U6 acted as internal control respectively for CHL1 and miR-21 measurement. The quantitative results were yielded by the relative quantification approach (\( {2}^{-\Delta \Delta {C}_t} \)). Primer sequences were designed by Sangon Biotech (Shanghai, China) and listed in Table 1.
Cell transfection
MiR-21-5p mimics, mimics negative control (mimics NC), miR-21-5p inhibitor, inhibitor negative control (inhibitor NC), CHL1 siRNA, pcDNA3.1 CHL1, siRNA negative control(siRNA NC)and miR-21-5p inhibitor + CHL1 siRNA were manufactured by Sangon Biotech. Cells were plated and cultured in complete medium at 37 °C for 24 h. Next, cells were transferred into serum-free medium, mixed with plasmid and liposome transfection reagent. Cells were prepared by rinsing with serum-free medium. The transfection was conducted by Lipidosome™2000 (Invitrogen). Transfection efficiency was detected after 48 h.
Western blot
Total protein in cells was extracted by radio immunoprecipitation buffer (RIPA, Sigma-Aldrich, St. Louis, MO, USA) and quantified by bicinchoninic acid (BCA, Pierce, Rock ford, IL, USA). After loading the equivalent amount of protein to SDS-PAGE (Bio-Rad, Hercules, CA, USA), proteins were then transferred to polyvinylidene fluoride (PVDF, Invitrogen) membranes. Followed blocked by 5% skim milk for 1 h, primary rabbit monoclonal antibody CHL1 solution (Epitomics, Burlingame, CA, USA) was placed into the membranes. After 2 h incubation, the membranes were incubated with second antibody: horseradish peroxidase (HRP) conjugated goat anti-rabbit (1:1000, Proteintech Group, Rosemont, IL, USA) at room temperature for 1 h. After washed by Tris Buffered Saline Tween (TBST), proteins were observed with an enhanced chemiluminescent (ECL, Thermo Scientific, Waltham, MA, USA) followed by exposure to X-ray films. The murine monoclonal antibody actin (1:2000, Proteintech Group) was used as internal reference.
MTT assay
2 × 103 cells were sowed into 96-well plates (100 μL/well) with 20 μL/well of MTT (5 mg/mL, Sigma-Aldrich) and incubated for 4 h. After washed by PBS, cells were dissolved and crystallized by 150 μL dimethyl sulfoxide (DMSO, Sigma-Aldrich). The optical density (OD) was detected by a microplate reader (Molecular Devices, Sunnyvale, CA, USA) at wavelength of 570 nm.
Flow cytometry assay
Cells were re-suspended in 100 μL PBS and fixed with 400 μL anhydrous ethanol for 1 h. After incubated at 37 °C for 1 h, 500 μL PBS was added and ethanol was removed. After incubated with 100 μg/ml RNase A (Sigma) in the dark at 37 °C for 30 min, 5 μL of Annexin V-fluorescein isothiocyanate (Annexin V-FITC) and propidium iodide (PI) (Becton Dickinson, NJ, USA) were added into the suspension for 15 min at room temperature in the dark. Each tube was supplemented with 400 μL binding buffer and cells were calculated by a flow cytometry (Beckman FC 500 MCL/MPL, Beckman Coulter, Miami, FL, USA).
Transwell assay
The upper surface of transwell chambers were covered with 40 μL Matrigel (BD Biosciences, San Jose, CA, USA) diluted with DMEM in proportion 1:3 for 24 h at 37 °C. After lysed with trypsin, cells were centrifuged, collected and re-suspended in 200 μL serum-free medium. Then the upper chambers were added with the cell suspension and 600 μL medium containing 20% FBS (Invitrogen) was added onto the substratum of chambers. After 24 h cultivation, cells were then fixed by methyl alcohol for 20 min. In the end, we used 0.1% crystal violet to stain the cells, and counted the cells under a microscope (Nikon, Japan).
Colony formation assay
Cells were collected when they were in logarithmic phase. After complete medium was removed, cells were washed by PBS before stained with 0.1% crystal violet for 30 min. Methanol was utilized to fix the cells. The colony cells were counted under an optical microscope (Nikon).
Enzyme-linked immunosorbent assay (ELISA)
Human MMP-2 Quantikine ELISA kit (R&D Systems, USA) and human MMP-9 ELISA kit (Abnova, Taiwan) were applied to test the expression of MMP-2 and MMP-9 respectively. After transfection, cells were incubated with fresh DMEM for 24 h, after which the culture medium was collected and centrifuged at 2000 rpm. Clarified medium was analyzed with ELISA kit (Institut Pourquier).
Dual luciferase reporter assay
CHL1 wt and mut were both synthesized by Sangon Biotech, which were transfected in LoVo cells. After post-transfection for 48 h, luciferase activities were analyzed by dual-luciferase reporter assay system (Promega, Madison, WI, USA) as the protocol suggested. The ratio of firefly to renilla luciferase signal was used to normalize efficiency of firefly activity in intra experimental transfection.
Mouse xenograft model
We divided the 4-weeks-old BALB/C athymic female nude mice into 5 groups (4 mice each group), and injected them with cells transfected by miR-21-5p inhibitor, miR-21-5p inhibitor negative control, CHL1 siRNA, siRNA negative control, miR-21-5p inhibitor together with CHL1 siRNA respectively. 1 × 107 treated LoVo cells were subcutaneously injected into the back of each subject. Then tumor volume measurement of nude mice in all groups was performed with calipers every week using the following method: volume = (length × width2) /2, with each parameter standing for the maximum and the minimum diameter respectively. The weight of tumor was measured after 4 weeks.
Statistical analysis
Statistical results were assessed by SPSS18.0 and all data were displayed in the form as mean ± standard deviation. Student t-test was employed to evaluate the difference between two groups, while one-way ANOVA analysis was used to compare multiple groups. P < 0.05 indicated statistically significance.
Results
Clinicopathological features of COAD patients
43 samples of COAD patients were used in this study. There were 32 samples with high expression of miR-21-5p and 11 samples with low expression of miR-21-5p. As shown in Table 2, miR-21-5p had close relationship with clinical stage, lymph node metastasis and distant metastasis of the patient samples (all P < 0.05). However, it seems that there was no significant correlation between the expression of miR-21-5p and gender or age (all P > 0.05).
MiR-21-5p highly expressed in COAD tissues and cells
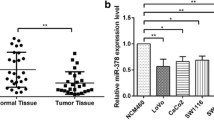
Microarray analysis was used to identify differentially expressed miRNAs between COAD tissues and normal tissues. The results showed that miR-21-5p expression in the COAD samples was up-regulated by about 6.22-fold compared to the normal adjacent tissues (P < 0.001, Fig. 1a). The result of qRT-PCR indicated that the expression level of miR-21-5p in colorectal cancer tissues was higher than that in adjacent normal tissues, which was more active in the higher pathological grade (both P < 0.001, Fig. 1b-c). The expression level of miR-21-5p was also remarkably up-regulated in 4 kinds of COAD cells, especially in LoVo cells compared with normal colonic epithelium cells NCM-460 (P < 0.001, Fig. 1d). Thus, LoVo cell line was chosen to be employed in the subsequent experiments.
MiR-21-5p highly expressed in COAD tissues and cells. a The heat map showed that miR-21-5p was high expressed in COAD tissues compared with normal tissues. b The relative expression of miR-21-5p in cancer tissues was much higher than that in adjacent tissues detected by qRT-PCR. ***P < 0.001, compared with adjacent tissue. c The expression of miR-21-5p in stage III-IV was notably higher than that in stage I-II detected by qRT-PCR. ***P < 0.001, compared with stage I-II. d The expression of miR-21-5p in COAD cell lines (CW-2, T84, SW116, LoVo) was higher than that in normal colonic epithelial cell line (NCM-460) detected by qRT-PCR, which reached the highest level in LoVo cell line. **P < 0.01, ***P < 0.001, compared with NCM-460
MiR-21-5p promoted the propagation and invasiveness of COAD cells
MiR-21-5p expression was up-regulated followed transfected with miR-21-5p mimics (P < 0.001, Fig. 2a), while it was down-regulated after transfected with miR-21-5p inhibitor (P < 0.001, Fig. 2b). According to the results of colony formation and MTT assay, with the passage of time, overexpression of miR-21-5p facilitated the propagation of COAD cells (P < 0.05, Fig. 3a & c), while inhibition of miR-21-5p suppressed COAD cells proliferation (P < 0.05, Fig. 3b & d). Flow cytometry result showed that miR-21-5p mimics promoted the process of CODA cell cycle in S phase (P < 0.01), while miR-21-5p inhibitor restrained the process of COAD cell cycle at G0/G1 phase (P < 0.05, Fig. 4). The results illustrated that overexpression of miR-21-5p can accelerate proliferation of COAD cells. Transwell assay indicated that overexpression of miR-21-5p promoted the invasion of COAD cells (P < 0.01, Fig. 5a), while inhibition of miR-21-5p suppressed it (P < 0.01, Fig. 5b). The result of ELISA verified that miR-21-5p stimulated the expression of invasiveness-related factor MMP-2 (P < 0.01, Fig. 5c) as well as MMP-9 (P < 0.01, Fig. 5d), which were suppressed by miR-21-5p inhibitor. Therefore, miR-21-5p can promote the proliferation and invasion of COAD cells.
The transfection of miR-21-5p mimics and miR-21-5p inhibitor. a The result of qRT-PCR showed that miR-21-5p expression in miR-21-5p mimics was much higher than that in NC mimics. ***P < 0.001, compared with NC mimics. b MiR-21-5p expression in miR-21-5p inhibitor was significantly lower than that in NC inhibitor. ***P < 0.001, compared with NC inhibitor
MiR-21-5p promoted COAD cell proliferation. a, b According to the results of colony formation assay and MTT assay, colony cells in miR-21-5p mimics group were more than that in NC mimics group since 48 h transfection. *P < 0.05, compared with NC mimics. c, d Colony cells in miR-21-5p inhibitor group were less than that in NC inhibitor group. *P < 0.05, compared with NC inhibitor group
MiR-21-5p promoted the cell cycle of COAD cells. a, b Compared with NC mimics, more cells in miR-21-5p mimics group were arrested in S phase and less cells appeared in G0/G1 phase. *P < 0.05, **P < 0.01, compared with NC mimics group. c, d Compared with NC inhibitor, more cells appeared in G0/G1 phase and less cells were arrested in S phase. *P < 0.05, **P < 0.01, compared with NC inhibitor group
MiR-21-5p promoted the invasion of COAD cells. a, b Cell number in miR-21-5p mimics group was more than that in NC mimics group, while that in miR-21-5p inhibitor group was less than that in NC inhibitor group detected by transwell assay. The scale bar was 50 μm. c, d The expressions of invasiveness-related factor MMP-2 and MMP-9 in miR-21-5p mimics group were higher than that in NC mimics group, while those in miR-21-5p inhibitor group were lower than that in NC inhibitor group detected by ELISA. **P < 0.01, compared with NC mimics group, ##P < 0.01, compared with NC inhibitor group
MiR-21-5p inhibited the expression of CHL1
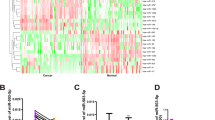
TargetScan was used to predict the target mRNA of miR-21-5p and the outcome showed that a binding site of miR-21-5p existed in the 3’UTR region of CHL1 (Fig. 6b). We screened the top 10 differentially expressed mRNAs by microarray analysis. The results of microarray analysis indicated that the expression of CHL1 was down-regulated by 2.57-fold compared with normal tissues (P < 0.001, Fig. 6a). Since CHL1 has been reported as a tumor suppressor (Martin-Sanchez et al., 2017), we supposed that miR-21-5p may promote tumor development via inhibiting CHL1 expression. Dual luciferase reporter assay was conducted to reconfirm the predicted targeting relationship between miR-21-5p and CHL1 (P < 0.01, Fig. 6b). Overexpression of miR-21-5p significantly inhibited CHL1 expression (P < 0.001), while inhibition of miR-21-5p up-regulated CHL1 expression (P < 0.01, Fig. 6c-d). Hence, miR-21-5p inhibited CHL1 expression by binding to 3’UTR of CHL1.
MiR-21-5p inhibited the expression of CHL1. a The heat map showed that the expression of CHL1 in COAD tissues was lower than that in normal tissues. b TargetScan predicted that there was a binding site for miR-21-5p in the 3’UTR of CHL1. The result of dual-luciferase reporter assay showed that the luciferase activity in miR-21-5p mimics and CHL1 3’UTR-wt co-combination group was lower than that in NC mimics and CHL1 3’UTR-wt co-combination group. In the meantime, there was no significant difference between miR-21-5p mimics and CHL1 3’UTR-mut co-combination group NC mimics and CHL1 3’UTR-mut co-combination group. c, d CHL1 expression in miR-21-5p mimics group was much lower than that in NC mimics group, while that in miR-21-5p inhibitor group was higher than that in NC inhibitor group. **P < 0.01, ***P < 0.001, compared with NC mimics group, &&P < 0.01, compared with NC inhibitor group
CHL1 inhibited progression of COAD cells
After transfection, colony formation and MTT assay was used to evaluate cell propagation. The results indicated that cell viability was inhibited by pcDNA3.1 CHL1 (P < 0.05), while it was enhanced by deletion of CHL1 (P < 0.05). In addition, the effect of CHL1 siRNA was rescued by miR-21-5p inhibitor (P < 0.05, Fig. 7a-b). The result of flow cytometry assay revealed that the cells were arrested in G0/G1 phase in pcDNA3.1 CHL1 group (P < 0.05), while the number of cells in S phase increased in CHL1 siRNA group compared with NC group (P < 0.01) and the cells in S phase decreased after co-transfected with CHL1 siRNA and miR-21-5p inhibitor (P < 0.05, Fig. 7c-d). Transwell assay proved that the ability of invasion of COAD cells was attenuated in pcDNA3.1 CHL1 group (P < 0.01). However, in CHL1 siRNA group, cell invasion ability was enhanced (P < 0.01), and it was suppressed by miR-21-5p inhibitor (P < 0.05, Fig. 7e-f). As a result, CHL1 suppressed COAD growth as well as invasion, and blocked cell cycle at G0/G1 phase.
CHL1 inhibited COAD cells development. a Colony formation assay indicated that CHL1 suppressed COAD cells growth. b MTT assay revealed that overexpression of CHL1 inhibited cell viability, while knockdown of CHL1 enhanced the proliferation ability of COAD cells. c, d The result of flow cytometry suggested that CHL1 arrested cell cycle in G0/G1 phase, while deletion of CHL1 increased cells in S phase. e, f Transwell assay was performed to detect invasion ability. The cell number in pcDNA3.1 CHL1 group was increased, while that in CHL1 siRNA group was decreased compared with control group. The scale bar was 50 μm. *P < 0.05, **P < 0.01, compared with NC group. #P < 0.05, ##P < 0.01, compared with CHL1 siRNA group
MiR-21-5p regulated COAD tumor growth in vivo through inhibiting CHL1 expression
The result of in vivo experiment indicated that inhibition of miR-21-5p suppressed COAD tumor growth and knockdown of CHL1 promoted the growth of COAD tumors (Fig. 8a-c). The expression of miR-21-5p and CHL1 in tumor was accessed by western blot and qRT-PCR. The result of western blot showed that the expression of CHL1 was up-regulated by inhibiting miR-21-5p (P < 0.01, Fig. 8d-e). The expression of miR-21-5p was also down-regulated by miR-21-5p inhibitor in vivo (P < 0.01, Fig. 8f).Therefore, miR-21-5p promoted the tumor growth by inhibiting CHL1 expression in vivo as well.
MiR-21-5p promoted COAD tumor growth in vivo through inhibiting CHL1 expression. a-c The volume and weight of tumors in miR-21-5p inhibitor group were much lower than that in NC inhibitor group, while that in CHL1 siRNA group was higher than that in siRNA NC group. **P < 0.01, compared with NC inhibitor group, &&P < 0.01, compared with siRNA NC group. d, e CHL1 expression in miR-21-5p inhibitor group was much higher than that in NC inhibitor group, while that in CHL1 siRNA group was lower than that in siRNA group. Meanwhile, there was no significant difference between NC inhibitor group, siRNA NC group and miR-21-5p inhibitor+CHL1 siRNA co-combination group. **P < 0.01, compared with NC inhibitor group, ##P < 0.01, compared with siRNA NC group. f The expression of miR-21-5p was down-regulated by miR-21-5p inhibitor in vivo evaluated by qRT-PCR. **P < 0.01, compared with NC inhibitor group, ##P < 0.01, compared with siRNA NC group
Discussion
In this research, we explored the correlation between miR-21-5p and clinicopathological features of COAD patients as well as miR-21-5p expression in COAD cells. In our experiments, the results illustrated that miR-21-5p stimulated the propagation and invasiveness of COAD cells. Moreover, we detected that miR-21-5p inhibited CHL1 expression. In the meantime, we explored in vivo experiment which confirmed that miR-21-5p promoted the tumor growth by inhibiting CHL1 expression.
COAD is a common type of colorectal cancer, and it is the fourth most common cause of cancer-related death around the world. Although chemotherapy is generally effective in weakening growth of tumor cells and impeding metastasis, it often loses virtue in advanced stages of COAD due to development of chemoresistance (Germani et al., 2014), which leads to recurrence of cancer or even death of patients. MiRNAs are often dysregulation in various types of tumor, which play crucial roles in tumorigenesis (Lambert & Becker, 1989; Hur, 2015). Our findings suggested that miR-21-5p highly expressed in COAD cells. Consistent with our results, a previous study indicates that miR-21 expression increased in COAD tissues as well as COAD cell lines (Liu et al., 2013). A recent study finds that miRNA-21 promotes invasion, migration and proliferation of COAD (Li et al., 2016b), which indicates that miR-21 plays a vital role in COAD development. To further explore the impact of miR-21-5p on cell activity of COAD, we did MTT assay and transwell assay, which showed that miR-21-5p promoted the propagation and invasiveness of COAD cells. These results corresponded with the previous reports reveal that miR-21 induces invasion and proliferation of COAD cells (Asangani et al., 2008). Moreover, we found that miR-21-5p over-expression enhanced the development of tumor in vivo, which coincided with a previous report that over-expression of miR-21 in COAD cells fortified their tumorigenic potential (Xu et al., 2014).
CHL1, highly related to the L1 protein, is an adhesion molecule of cells. A great number of studies have shown that miRNAs were able to target several transcription factors to regulate the metabolism and progression of cells (Li et al., 2017; Cao et al., 2017; Zhuo et al., 2017; Qiu & Dou, 2017). In order to ascertain the regulatory relationship between miR-21-5p and CHL1, we employed TargetScan as well as dual luciferase reporter assay and found that CHL1 3’UTR has a binding site with miR-21-5p. Moreover, the expression level of CHL1 was up-regulated by inhibiting miR-21-5p. Previous study verified that CHL1 gene was one of the putative tumor suppressor genes localized on human chromosome 3 (Qin et al., 2008). In our experiments, knockdown of CHL1 promoted the tumor growth. Previous report also revealed that miR-21 promotes the propagation and invasiveness of neuroblastoma cells by suppressing CHL1 (Li et al., 2016a).
Even though we have observed some meaningful results in our study, there were also some clear limitations. For example, our study only looked into the functions of miR-21-5p in promoting proliferation and invasion of COAD cells, whereas the underlying detailed mechanism of miR-21-5p in COAD cells remained blurry which was quite essential to completely understand the pathogenesis of COAD. In addition, miR-21-5p was controlled by one gene called miR-21 and in previous studies, miR-21 has been proved to be capable of targeting many tumor suppressors such as PTEN (Meng et al., 2007), RECK (Gabriely et al., 2008), BTG2 (Liu et al., 2009) and TP63 (Papagiannakopoulos et al., 2008). The correlation of miR-21 and its other target genes in COAD cells need to be further studied.
Conclusions
In conclusion, our experiments demonstrated that over-expression of miR-21-5p enhanced proliferation and invasion of COAD cells through targeting CHL1. Nevertheless, further research is necessarily required to deeply and comprehensively understand the mechanism of miR-21 on COAD.
Abbreviations
- Annexin V-FITC:
-
Annexin V-fluorescein isothiocyanate
- BCA:
-
Bicinchoninic acid
- BNCC:
-
BeNa Culture Collection
- CHL1 :
-
Cell adhesion molecule L1
- COAD:
-
Colon adenocarcinoma
- DMEM:
-
Dulbecco’s Modified Eagle Medium
- DMSO:
-
Dimethyl sulfoxide
- ECL:
-
Enhanced chemiluminescent
- FBS:
-
Fetal Bovine Serum
- FC:
-
Fold change
- inhibitor NC:
-
Inhibitor negative control
- mimics NC:
-
mimics negative control
- miR-21:
-
MicroRNA-21
- miRNAs:
-
MicroRNAs
- OD:
-
Optical density
- PI:
-
Propidium iodide
- PVDF:
-
Polyvinylidene fluoride
- READ:
-
Rectal adenocarcinoma
- RIPA:
-
Radio immunoprecipitation buffer
- RP:
-
Horseradish peroxidase
- siRNA NC:
-
siRNA negative control
- TBST:
-
Tris Buffered Saline Tween
- TCGA:
-
Cancer Genome Atlas
References
Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–36.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism. and function Cell. 2004;116:281–97.
Bovell LC, Shanmugam C, Putcha BD, Katkoori VR, Zhang B, Bae S, et al. The prognostic value of microRNAs varies with patient race/ethnicity and stage of colorectal cancer. Clin Cancer Res. 2013;19:3955–65.
Cao H, Yu H, Feng Y, Chen L, Liang F. Curcumin inhibits prostate cancer by targeting PGK1 in the FOXD3/miR-143 axis. Cancer Chemother Pharmacol. 2017;79:985–94.
Chen P, Wang BL, Pan BS, Guo W. MiR-1297 regulates the growth, migration and invasion of colorectal cancer cells by targeting cyclo-oxygenase-2. Asian Pac J Cancer Prev. 2014;15:9185–90.
Chen QY, Chen Q, Feng GY, Lindpaintner K, Chen Y, Sun X, et al. Case-control association study of the close homologue of L1 (CHL1) gene and schizophrenia in the Chinese population. Schizophr Res. 2005;73:269–74.
Chen TH, Chang SW, Huang CC, Wang KL, Yeh KT, Liu CN, et al. The prognostic significance of APC gene mutation and miR-21 expression in advanced-stage colorectal cancer. Color Dis. 2013;15:1367–74.
Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, et al. Colorectal cancer. Lancet. 2010;375:1030–47.
Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–5.
Ding L, Yu LL, Han N, Zhang BT. miR-141 promotes colon cancer cell proliferation by inhibiting MAP2K4. Oncol Lett. 2017;13:1665–71.
Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–80.
Gavert N, Conacci-Sorrell M, Gast D, Schneider A, Altevogt P, Brabletz T, et al. L1, a novel target of beta-catenin signaling, transforms cells and is expressed at the invasive front of colon cancers. J Cell Biol. 2005;168:633–42.
Georgantas RW 3rd, Hildreth R, Morisot S, Alder J, Liu CG, Heimfeld S, et al. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci U S A. 2007;104:2750–5.
Germani A, Matrone A, Grossi V, Peserico A, Sanese P, Liuzzi M, et al. Targeted therapy against chemoresistant colorectal cancers: inhibition of p38alpha modulates the effect of cisplatin in vitro and in vivo through the tumor suppressor FoxO3A. Cancer Lett. 2014;344:110–8.
He LH, Ma Q, Shi YH, Ge J, Zhao HM, Li SF, et al. CHL1 is involved in human breast tumorigenesis and progression. Biochem Biophys Res Commun 2013;438: 433–8.
Hur K. MicroRNAs: promising biomarkers for diagnosis and therapeutic targets in human colorectal cancer metastasis. BMB Rep. 2015;48:217–22.
Kjaer-Frifeldt S, Hansen TF, Nielsen BS, Joergensen S, Lindebjerg J, Soerensen FB, et al. The prognostic importance of miR-21 in stage II colon cancer: a population-based study. Br J Cancer. 2012;107:1169–74.
Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–15.
Lambert RG, Becker EJ. Diffuse skeletal hyperostosis in idiopathic hypoparathyroidism. Clin Radiol. 1989;40:212–5.
Lan F, Pan Q, Yu H, Yue X. Sulforaphane enhances temozolomide-induced apoptosis because of down-regulation of miR-21 via Wnt/beta-catenin signaling in glioblastoma. J Neurochem. 2015;134:811–8.
Leone E, Morelli E, Di Martino MT, Amodio N, Foresta U, Gulla A, et al. Targeting miR-21 inhibits in vitro and in vivo multiple myeloma cell growth. Clin Cancer Res. 2013;19:2096–106.
Li C, Zhao L, Chen Y, He T, Chen X, Mao J, et al. MicroRNA-21 promotes proliferation, migration, and invasion of colorectal cancer, and tumor growth associated with down-regulation of sec23a expression. BMC Cancer. 2016b;16:605.
Li Y, Shang YM, Wang QW. MicroRNA-21 promotes the proliferation and invasion of neuroblastoma cells through targeting CHL1. Minerva Med. 2016a;107:287–93.
Li Y, Su J, Li F, Chen X, Zhang G. MiR-150 regulates human keratinocyte proliferation in hypoxic conditions through targeting HIF-1alpha and VEGFA: implications for psoriasis treatment. PLoS One. 2017;12:e0175459.
Liu GH, Zhou ZG, Chen R, Wang MJ, Zhou B, Li Y, et al. Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumour Biol. 2013;34:2175–81.
Liu M, Wu H, Liu T, Li Y, Wang F, Wan H, et al. Regulation of the cell cycle gene, BTG2, by miR-21 in human laryngeal carcinoma. Cell Res. 2009;19:828–37.
Martin del Campo SE, Latchana N, Levine KM, Grignol VP, Fairchild ET, Jaime-Ramirez AC, et al. MiR-21 enhances melanoma invasiveness via inhibition of tissue inhibitor of metalloproteinases 3 expression: in vivo effects of MiR-21 inhibitor. PLoS One. 2015;10:e0115919.
Martin-Sanchez E, Mendaza S, Ulazia-Garmendia A, Monreal-Santesteban I, Blanco-Luquin I, Cordoba A, et al. CHL1 hypermethylation as a potential biomarker of poor prognosis in breast cancer. Oncotarget. 2017;8:15789–801.
Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–58.
Nielsen BS, Jorgensen S, Fog JU, Sokilde R, Christensen IJ, Hansen U, et al. High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin Exp Metastasis. 2011;28:27–38.
Oue N, Anami K, Schetter AJ, Moehler M, Okayama H, Khan MA, et al. High miR-21 expression from FFPE tissues is associated with poor survival and response to adjuvant chemotherapy in colon cancer. Int J Cancer. 2014;134:1926–34.
Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–72.
Phang CW, Karsani SA, Abd Malek SN. Induction of apoptosis and cell cycle arrest by Flavokawain C on HT-29 human Colon adenocarcinoma via enhancement of reactive oxygen species generation, Upregulation of p21, p27, and GADD153, and inactivation of inhibitor of apoptosis proteins. Pharmacogn Mag. 2017;13:S321–S8.
Qin YR, Fu L, Sham PC, Kwong DL, Zhu CL, Chu KK, et al. Single-nucleotide polymorphism-mass array reveals commonly deleted regions at 3p22 and 3p14.2 associate with poor clinical outcome in esophageal squamous cell carcinoma. Int J Cancer. 2008;123:826–30.
Qiu X, Dou Y. miR-1307 promotes the proliferation of prostate cancer by targeting FOXO3A. Biomed Pharmacother. 2017;88:430–5.
Senchenko VN, Krasnov GS, Dmitriev AA, Kudryavtseva AV, Anedchenko EA, Braga EA, et al. Differential expression of CHL1 gene during development of major human cancers. PLoS One. 2011;6:e15612.
Tokarz P, Blasiak J. The role of microRNA in metastatic colorectal cancer and its significance in cancer prognosis and treatment. Acta Biochim Pol. 2012;59:467–74.
Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61.
Wang CL, Wang BB, Bartha G, Li L, Channa N, Klinger M, et al. Activation of an oncogenic microRNA cistron by provirus integration. Proc Natl Acad Sci U S A. 2006;103:18680–4.
Wei MH, Karavanova I, Ivanov SV, Popescu NC, Keck CL, Pack S, et al. In silico-initiated cloning and molecular characterization of a novel human member of the L1 gene family of neural cell adhesion molecules. Hum Genet. 1998;103:355–64.
Xiong B, Cheng Y, Ma L, Zhang C. MiR-21 regulates biological behavior through the PTEN/PI-3 K/Akt signaling pathway in human colorectal cancer cells. Int J Oncol. 2013;42:219–28.
Xu LF, Wu ZP, Chen Y, Zhu QS, Hamidi S, Navab R. MicroRNA-21 (miR-21) regulates cellular proliferation, invasion, migration, and apoptosis by targeting PTEN, RECK and Bcl-2 in lung squamous carcinoma, Gejiu City, China. PLoS One. 2014;9:e103698.
Zhuo C, Xue R, Luo L, Ji F, Tian H, Qu H, et al. Efficacy of antidepressive medication for depression in Parkinson disease: a network meta-analysis. Medicine (Baltimore). 2017;96:e6698.
Availability of data and materials
The datasets generated and analysed during the current study are available in the Cancer Genome Atlas (TCGA) repository (https://cancergenome.nih.gov/) and the TargetScan 7.1 database (www.targetscan.org).
Author information
Authors and Affiliations
Contributions
WY and KZ made substantial contributions to conception and design. YW made acquisition of data. HY and JG made analysis and interpretation of data. WY has been involved in drafting the manuscript. KZ, YW, HY and JG have been involved in revising it critically for important intellectual content. All authors have given final approval of the version to be published and agreed to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study involved human participants or animals and was approved by the Second Hospital of Shandong University. All the patients and participants have signed the informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yu, W., Zhu, K., Wang, Y. et al. Overexpression of miR-21-5p promotes proliferation and invasion of colon adenocarcinoma cells through targeting CHL1. Mol Med 24, 36 (2018). https://doi.org/10.1186/s10020-018-0034-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s10020-018-0034-5