Abstract
Genome-wide association analyses on large patient cohorts are generating large sets of candidate disease genes. This is coupled with the availability of ever-increasing genomic databases and a rapidly expanding repository of biomedical literature. Computational approaches to disease-gene association attempt to harness these data sources to identify the most likely disease gene candidates for further empirical analysis by translational researchers, resulting in efficient identification of genes of diagnostic, prognostic and therapeutic value. Existing computational methods analyze gene structure and sequence, functional annotation of candidate genes, characteristics of known disease genes, gene regulatory networks, protein-protein interactions, data from animal models and disease phenotype. To date, a few studies have successfully applied computational analysis of clinical phenotype data for specific diseases and shown genetic associations. In the near future, computational strategies will be facilitated by improved integration of clinical and computational research, and by increased availability of clinical phenotype data in a format accessible to computational approaches.
Similar content being viewed by others
Historically, disease phenotype has informed the selection of candidate disease genes through observations of the effects of perturbations in these candidates in vitro, in tissue cultures and in animal models. This hypothesis-driven approach is increasingly being superseded by genome-wide analyses that assume no prior knowledge of the underlying genotype, and hypotheses about the associated genes are inferred from large-scale genetic studies of samples with the disease phenotype. These studies include genome-wide linkage and association studies in affected and healthy patient populations to identify chromosomal regions most likely to contain etiological genes [1–3], and the detailed analysis of genome-wide changes in the disease state by high-throughput techniques, such as single nucleotide polymorphism (SNP) [4] and microarray expression analysis [5], serial analysis of gene expression (SAGE) [6] and cap analysis of gene expression (CAGE) [7]. Current approaches include next generation sequencing of linked regions, high-density SNP analysis and the study of copy number variation [8].
Typically, genome-wide approaches generate large sets of potential genetic associations for further analysis; for example, multifactorial disease loci identified by linkage analysis can be approximately 30 Mb in size and contain several hundred genes [9]. This synergizes with ongoing research on many complex diseases, in which multiple gene variations, rather than single dysfunctional genes, are believed to underlie the disease phenotype [10]. Genome-wide analyses have therefore massively increased the number of candidate genes to be investigated for a given phenotype.
Concurrently, available genetic information has increased as a result of more sophisticated experimental methods and centralization of genetic information in public genome databases (such as Ensembl [11], NCBI [12] and UCSC [13]), gene expression databases (such as GEO [14]) and human variation databases (such as HapMap [15]). Additional data on gene regulatory networks and pathways are becoming increasingly accessible (for example, KEGG [16] and Reactome [17]). In addition, biomedical literature has become too massive a resource to be assimilated by individuals (for example, 17.8 million abstracts are listed by PubMed in May 2009, of which 10 million deal with human data).
The subsequent challenge is to use this wide variety of data sources to identify relevant disease gene candidates within the lists of genes generated from genome-wide analyses for further empirical research, an overwhelming task to undertake manually. Computational analysis can facilitate efficient and accurate utilization of all such sources of information, and the resulting early prioritization allows streamlined empirical research and quicker and cheaper identification of disease-causing genes.
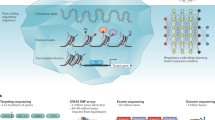
To date, many computational methods have focused on the prediction of candidates by analysis of inherent sequence characteristics of genes, sequence similarity to known disease genes, and functional annotation of candidate genes [18]. These approaches are briefly reviewed here. The computational analysis of phenotypes for the prioritization of disease candidates is less utilized, and is explored later in this article (Figure 1).
Miners in Germany (1952). As with mining of minerals, data mining of associations between genes and diseases can be dirty and disheartening, but the potential for reward is great. Photo: Günther Paalzow. Reproduced from Bundesarchiv, Bild 183-13175-0020 under the Creative Commons Attribution ShareAlike 3.0 license (Germany).
Approaches for the identification of candidate disease genes
Using intrinsic gene properties
By analyzing the intrinsic properties of genes already associated with an inherited disease regardless of its phenotype, differences can be found between disease genes and all human genes. The pattern of differences can be used to predict novel disease genes. Properties associated with disease genes include gene structure, such as longer size of the gene and its associated proteins, and longer regulatory regions such as the mRNA 3' un-translated region (UTR); phylogenetic information, including lower mutation rates, broader phylogenetic breadth and fewer paralogs (that is, fewer highly similar genes giving less opportunity for functional redundancy); and genomic properties such as a higher proportion of CpG islands in promoters and longer intergenic distances.
The first method to apply this type of approach was DGP [19], followed by PROSPECTR [20], which included additional gene properties (for an extensive review of these approaches see [21]). Such analyses rely on the definition of genes as 'disease genes' and 'non-disease genes' and, although suited to analysis of monogenic (Mendelian) diseases, such approaches may preclude the selection of genes that do not produce an obvious phenotype but rather contribute to disease susceptibility or the severity of the effect of a simultaneous mutation in another gene. The efficacy of such approaches thus becomes limited in the study of complex phenotypes, in which the association between the gene and disease may not be one of direct or exclusive causation.
Similarity to genes previously associated with disease
Several methods of associating genes with diseases rely on the functional annotation of the gene and, under the hypothesis that similar diseases may have associated genes with similar functions, propose associations on the basis of genes already known to be associated with a disease. This approach is supported by multiple lines of evidence and is a logical way to initiate a search for candidate genes. For example, genes related to the detection or synthesis of neurotransmitters are likely to be good candidates for association with neural disorders, or immune-related genes with asthma and allergy phenotypes.
This is a logical inference, but when there is a growth in the number of gene candidates it becomes difficult to get all the information on known diseases and related literature manually, and computational approaches are helpful. Computational analyses take advantage of both controlled vocabularies describing disease features (such as MeSH terms - an ontology developed at the National Library of Medicine covering different subject categories, including disease phenotype [22]) and similarity between gene functions measured by using their annotations with controlled vocabularies, such as the Gene Ontology [23]. Methods such as G2D [9], POCUS [24], ENDEAVOUR [25] and TOM [26] use this approach.
A limitation of methods relying on the functional annotations of genes is that just a small percentage of genes in the databases have an experimentally verified function (6% have links to non-genomic literature [27]). Most annotation (for approximately 71% of genes [27]) is based on functions assumed to associate with predicted protein domains from manually curated databases (such as the Gene Ontology Annotation (GOA) project [28]).
Implication of genes in regulatory networks or in protein-protein interaction networks
Information from interactions between genes can be used to find disease-related genes. These data are available from multiple public resources and may describe protein-protein interactions (such as STRING [29] and UniHI [30]), proteins regulating gene expression (such as TRANSFAC [31]), and metabolic pathways (such as KEGG [16]). Some of these categories can overlap to some extent with functional annotations (for example, several genes encoding proteins from the same pathway or protein complex may be described by the same functional annotation comprising a common Gene Ontology term).
The assumption made is that if two genes work together, the known association of one with a disease suggests that the other may also be associated with the same disease. For example, mutations in different subunits of the sarcoglycan complex can result in muscular dystrophy [32]. For genes in a regulatory cascade, if the mutation of a gene produces a given phenotype, then mutations in genes further upstream, such as a transcription factor for the downstream gene or a protein kinase that phosphorylates it, could result in the same phenotype. Methods such as ENDEAVOUR [25] and recent versions of G2D [33] exploit this hypothesis.
Gene expression information
The methods described earlier can be complemented using gene expression data. This can be done in relation to the particular disease under analysis (for example selecting genes that are expressed in an affected tissue, such as neural tissue in the case of a neurodegenerative disease); or gene co-expression can be used as another measure of gene similarity to find associations between genes. The second approach is based on the premise that genes acting together will be expressed together, as seen for subunits of protein complexes (such as is described in [21, 25, 34]).
Disease phenotypes
Clinical knowledge is fundamental to defining disease phenotype, and some existing methods aim to make use of this knowledge directly. For example, GeneSeeker [35] is a web tool that uses phenotype search terms input directly by the researcher and filters positional candidate disease genes based on expression and phenotypic data. The Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER) centralizes clinician-sourced phenotype data and their relationship to copy number variation [36] and is ground-breaking in its aim to make accessible a global wealth of phenotype descriptions submitted directly by clinicians.
Animal models allow in-depth studies of phenotypic variation associated with genes, which are impossible in human subjects (for example, genetic manipulations such as gene knockouts). The data thus generated can be used to associate human genes with phenotypes according to the properties of orthologous genes in such model organisms. In some cases, phenotype association may be in the form of quantitative trait loci (QTLs) involving a number of genes. These methods have to overcome the challenge of identifying the appropriate orthology relations between human and animal genes, which becomes harder with increasing evolutionary distance between the species under study and humans. This approach is used by methods such as GeneSeeker [35] and ToppGene [37] with mouse phenotype data, and Fraser and Plotkin have used a similar approach with yeast data [38].
Some available methods define the disease phenotype in a formalized way that involves the use of existing or customized ontologies. As ontologies are formal representations of a set of concepts within a domain of knowledge and the relationships between those concepts, they are preferable to a definition of the problem phenotype by means of a mere set of keywords provided by the user. Ontologies can facilitate optimal use of available knowledge because many pieces of information can be linked through queries in the databases in which they are used. For example, most of the articles in MEDLINE are annotated with MeSH terms. In this way, they can directly link the phenotype described by a MeSH term to the information contained in the article annotated with it. Phenotype ontologies are used to mine textual databases, such as MEDLINE abstracts in PubMed and/or Online Mendelian Inheritance in Man (OMIM) records, and relate them to gene features and lists of candidate genes. eVOC, a controlled vocabulary for unifying gene expression data, is a purposely developed anatomical ontology that can integrate text mining of biomedical literature and data mining of available human gene expression data [39]. The GFINDer method uses an ontology developed from OMIM entries [40]. G2D uses disease MeSH terms linked in the OMIM record associated with the phenotype of interest to link phenotypes and Gene Ontology terms [9]. PhenoGO, another ontology that assigns phenotypic context to Gene Ontology annotations, also mines the literature to associate phenotypes to Gene Ontology terms [41]. A Human Phenotype Ontology has been developed and used to annotate OMIM entries [42], and the broader Mammalian Phenotype Ontology [43] is used in both the Mouse and Rat Genome Databases [44, 45].
Despite the obvious limitations of transferring information between animal models and humans, the broad range of phenotypic measures that can be obtained from animals is impossible to collect from humans. In the context of complex phenotypes, mice are being predominantly used to study the (usually small) quantitative phenotypic differences associated with a genetic variation (QTLs) [46]. Large-scale projects are underway to induce knockouts in mice and analyze the corresponding genotypes by high-throughput techniques [47, 48]. These projects will need extensive and more sophisticated annotation systems such as Phenotype and Trait Ontology (PATO [49]), which combines existing phenotype ontologies with phenotype qualities; for example 'insect eye', from the fly anatomy ontology, can bear the quality 'red', giving the combined 'red eye' phenotype.
Application of computational approaches to specific diseases
Only a few studies so far have used phenotype-based computational approaches to identify disease genes and followed this through in patient samples. The GeneSeeker tool was used to prioritize candidates for skeletal dysplasia, and the contribution of a selected candidate to the disease phenotype was demonstrated [50]. In this study, linkage analysis identified 77 candidate genes in a 17.1 cM interval. GeneSeeker identified the disease-causing gene as RMRP (an untranslated RNA gene), and its etiological role was confirmed in patients with the disease. Mutations in this gene have been identified previously as disease-causing in milder types of autosomal recessive skeletal dysplasias with differing phenotypes; identification of this additional disease phenotype associated with the gene, however, has furthered understanding of gene function and suggested its involvement in other related disease phenotypes.
The G2D method was used to prioritize candidate genes for asthma and atopy (a type of allergic hypersensitivity) at two previously linked loci in a French Canadian population [51]. Ten genes were selected by G2D for a subsequent association study, and SNPs within the candidate genes were genotyped and analyzed using a family-based association test. The results suggested a protective association with allergic asthma for the protein tyrosine phosphatase gene PTPRE in this French Canadian population, although the association could not be replicated in a different cohort [51].
Other than these translational studies, computational analyses have generally been applied to specific diseases and where there are known etiological genes for the disease in question, the accuracy of the results has been assessed by the ranking of these known genes. The candidates that have been flagged as most likely and warranting further empirical research are published for use by the research community, as in our previous studies on candidate genes for metabolic syndrome [52] and type 2 diabetes (T2D) [53]. In these, we used multiple computational techniques, including those based on phenotype [35, 39], for disease gene prioritization, using sets of genes defined by multiple linkage analysis data available through the biomedical literature as starting point.
In the case of metabolic syndrome [52], we initially selected candidates for discrete phenotypes that are associated with the disorder from a starting set of 13,882 genes, and identified candidate genes showing commonality across multiple phenotypes. The phenotype-specific candidates were then weighted according to the prevalence of each phenotype in patient populations, with 19 candidates prioritized as the most likely etiological genes. For T2D, we used multiple computational approaches to identify obesity- and T2D-specific candidates from a starting set of 9,556 positional candidates. This allowed us to generate a final list of nine primary T2D candidates, two of which were also primary candidates for obesity. A SNP in the lipoprotein lipase gene LPL, which was one of the proposed two top candidates, has since been associated with T2D in Korean patients [54].
Similar approaches have been used for other diseases, such as the use of multiple existing computational methods for prediction of genes associated with osteoporosis [55], and the use of extensive phenotype data to select candidate etiological genes for fetal alcohol syndrome[56]. ENDEAVOUR has also been used to prioritize candidate genes for a variety of phenotypes, reviewed in [25].
Future directions for computational prioritization of candidate disease genes
With increased understanding and availability of human genome and transcriptome data, additional resources can refine the computational prioritization of candidate disease genes. These include data on copy number variation, which has already been used to identify candidate genes for autism [57], and on RNA editing in candidate genes [58]. Phenotype can be affected by perturbations in additional elements such as long non-coding genes [59]; long range non-coding RNAs, as identified for short stature phenotypes [60] and cancer [61]; natural antisense transcripts [62]; promoter elements, such as those associated with degenerative heart disease [63]; and microRNAs [64]. Epidemiological data for disease occurrence used in conjunction with genome-wide data on population variation [65] can facilitate associations between disease phenotypes prevalent in particular populations and their underlying genotypes. Finally, collation and standardization of phenotype data (as undertaken in the DECIPHER project [36]) and the further development of phenotype ontologies that have an appropriate degree of granularity and are accessible to scientists are essential for the compilation of clinical phenotype data in a format that allows the computational analysis of associations between disease phenotype and genotype.
Conclusions
Understanding underlying disease genetics is crucial for the development of appropriate disease-specific diagnostic, prognostic and therapeutic approaches, and increasing the efficiency of this process can result in substantial progress in the clinical management of disease. Computational approaches for the identification of disease genes have contributed significantly to our understanding of gene and protein characteristics. These include the tendency of enzymes and transporters to underlie recessive diseases, while transcription regulators and structural molecules often underlie dominant inheritance [19]. More generally, they have shown evidence that disease gene function and expression patterns correlate with the type of disease they cause [66]. The computational analysis of disease phenotypes has revealed the tendency for similar disease phenotypes to be caused by functionally related genes [67]. Such analyses have also shown that the phenotypic similarity between syndromes correlates with the sequence similarity of their associated genes [18, 68]. We believe, however, that there is great scope to better harness clinical phenotypic data to improve computational disease gene prioritization. In an ideal scenario, extensive clinical phenotypic data would be available to computational scientists to use in conjunction with genome-wide empirical data, allowing for effective prediction of most likely disease gene candidates and leading to rapid and economical empirical identification of etiological genes.
For this to become a reality, several objectives need first to be realized. Most importantly, clinical phenotype data need to be routinely standardized in an accessible, patient-anonymous format for computational use. To this end, prospective studies on patient populations could include a computational component at the design stage, so that standardized clinical/phenotypic data can be collected throughout the study. To ensure that this becomes standard practice, however, clinicians need to be convinced of the utility of computational approaches in determining candidate disease genes, and computational studies for specific diseases need to reach the appropriate clinical audience. Collaborative studies between clinical researchers and computational scientists are invaluable in bridging this gap and promoting recognition of computational applications in clinical research, but these are not yet standard practice.
In addition, many computational scientists focus on the development of novel generic approaches for candidate gene prediction for diseases in general. This should not, however, preclude the application of existing methods to specific diseases. Such disease-specific studies, presented in a format accessible to non-computational researchers, would facilitate translation of computational research into the clinical environment and promote recognition of the role of computational studies in disease gene identification.
The ability to investigate the genetics underlying disease phenotypes at a genome-wide level will result in more rapid disease gene identification, as genome-wide analyses can return multiple potential candidate genes simultaneously, rather than verifying or refuting the implication of individual genes in a sequential way. This transition is serendipitous, given that the field of disease genetics is moving on from Mendelian diseases and focusing on complex diseases in which multiple etiological genes are believed to act in concert [69]. As increasingly sophisticated techniques uncover the stronger and more frequent gene-disease associations, research techniques will shift towards defining our understanding of more subtle or indirect effects of genes on disease phenotype, in parallel with our increased understanding of the subtleties and complexities of the biological mechanisms of the human cell.
The challenge now lies in finding relevant candidates within the lists of potential disease genes generated from genome-wide approaches. Computational methods are well suited to the systematic analysis of these large gene lists to generate encompassing hypotheses about disease genotype, predict the most likely disease gene candidates from large datasets, and rapidly disseminate the results to clinical researchers performing translational research. Computational disease gene prediction can thus contribute substantially to faster and more cost-efficient empirical identification of disease-causing genes.
Abbreviations
- MeSH:
-
Medical Subject Heading
- OMIM:
-
Online Mendelian Inheritance in Man
- QTL:
-
quantitative trait locus
- SNP:
-
single nucleotide polymorphism
- T2D:
-
type 2 diabetes.
References
Zeggini E, Ioannidis JP: Meta-analysis in genome-wide association studies. Pharmacogenomics. 2009, 10: 191-201. 10.2217/14622416.10.2.191
Iles MM: What can genome-wide association studies tell us about the genetics of common disease?. PLoS Genet. 2008, 4: e33- 10.1371/journal.pgen.0040033
Altshuler D, Daly M: Guilt beyond a reasonable doubt. Nat Genet. 2007, 39: 813-815. 10.1038/ng0707-813
Marchini J, Howie B, Myers S, McVean G, Donnelly P: A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007, 39: 906-913. 10.1038/ng2088
Farber CR, Lusis AJ: Integrating global gene expression analysis and genetics. Adv Genet. 2008, 60: 571-601. full_text
Hene L, Sreenu VB, Vuong MT, Abidi SH, Sutton JK, Rowland-Jones SL, Davis SJ, Evans EJ: Deep analysis of cellular transcriptomes - LongSAGE versus classic MPSS. BMC Genomics. 2007, 8: 333- 10.1186/1471-2164-8-333
de Hoon M, Hayashizaki Y: Deep cap analysis gene expression (CAGE): genome-wide identification of promoters, quantification of their expression, and network inference. Biotechniques. 2008, 44: 627-628. 630, 632. 10.2144/000112802
Mardis ER: The impact of next-generation sequencing technology on genetics. Trends Genet. 2008, 24: 133-141.
Perez-Iratxeta C, Bork P, Andrade MA: Association of genes to genetically inherited diseases using data mining. Nat Genet. 2002, 31: 316-319.
Risch NJ: Searching for genetic determinants in the new millennium. Nature. 2000, 405: 847-856. 10.1038/35015718
Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, Down T, Dyer SC, Eyre T, Fitzgerald S, Fernandez-Banet J, Gräf S, Haider S, Hammond M, Holland R, Howe KL, Howe K, Johnson N, Jenkinson A, Kähäri A, Keefe D, Kokocinski F, Kulesha E, Lawson D, Longden I, Megy K, et al.: Ensembl 2008. Nucleic Acids Res. 2008, 36: D707-D714. 10.1093/nar/gkm988
Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, Feolo M, Geer LY, Helmberg W, Kapustin Y, Khovayko O, Landsman D, Lipman DJ, Madden TL, Maglott DR, Miller V, Ostell J, Pruitt KD, Schuler GD, Shumway M, Sequeira E, Sherry ST, Sirotkin K, Souvorov A, Starchenko G, et al.: Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008, 36: D13-D21. 10.1093/nar/gkm1000
Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, Meyer L, Hsu F, Hinrichs AS, Harte RA, Giardine B, Fujita P, Diekhans M, Dreszer T, Clawson H, Barber GP, Haussler D, Kent WJ: The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009, 37: D755-D761. 10.1093/nar/gkn875
Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Muertter RN, Edgar R: NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009, 37: D885-D890. 10.1093/nar/gkn764
, Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, et al.: A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007, 449: 851-861. 10.1038/nature06258
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y: KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36: D480-D484. 10.1093/nar/gkm882
Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, Kanapin A, Lewis S, Mahajan S, May B, Schmidt E, Vastrik I, Wu G, Birney E, Stein L, D'Eustachio P: Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009, 37: D619-D622. 10.1093/nar/gkn863
Oti M, Brunner HG: The modular nature of genetic diseases. Clin Genet. 2007, 71: 1-11. 10.1111/j.1399-0004.2006.00708.x
Lopez-Bigas N, Ouzounis CA: Genome-wide identification of genes likely to be involved in human genetic disease. Nucleic Acids Res. 2004, 32: 3108-3114. 10.1093/nar/gkh605
Adie EA, Adams RR, Evans KL, Porteous DJ, Pickard BS: Speeding disease gene discovery by sequence based candidate prioritization. BMC Bioinformatics. 2005, 6: 55- 10.1186/1471-2105-6-55
Oti M, Snel B, Huynen MA, Brunner HG: Predicting disease genes using protein-protein interactions. J Med Genet. 2006, 43: 691-698. 10.1136/jmg.2006.041376
United States National Library of Medicine, National Institutes of Health, Medical Subject Headings. http://www.nlm.nih.gov/mesh/meshhome.html
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G: Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000, 25: 25-29. 10.1038/75556
Turner FS, Clutterbuck DR, Semple CA: POCUS: mining genomic sequence annotation to predict disease genes. Genome Biol. 2003, 4: R75- 10.1186/gb-2003-4-11-r75
Tranchevent LC, Barriot R, Yu S, Van Vooren S, Van Loo P, Coessens B, De Moor B, Aerts S, Moreau Y: ENDEAVOUR update: a web resource for gene prioritization in multiple species. Nucleic Acids Res. 2008, 36: W377-W384. 10.1093/nar/gkn325
Masotti D, Nardini C, Rossi S, Bonora E, Romeo G, Volinia S, Benini L: TOM: enhancement and extension of a tool suite for in silico approaches to multigenic hereditary disorders. Bioinformatics. 2008, 24: 428-429. 10.1093/bioinformatics/btm588
Perez-Iratxeta C, Palidwor G, Andrade-Navarro MA: Towards completion of the Earth's proteome. EMBO Rep. 2007, 8: 1135-1141. 10.1038/sj.embor.7401117
Camon E, Magrane M, Barrell D, Lee V, Dimmer E, Maslen J, Binns D, Harte N, Lopez R, Apweiler R: The Gene Ontology Annotation (GOA) Database: sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 2004, 32: D262-D266. 10.1093/nar/gkh021
Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C: STRING 8 - a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009, 37: D412-D416. 10.1093/nar/gkn760
Chaurasia G, Malhotra S, Russ J, Schnoegl S, Hanig C, Wanker EE, Futschik ME: UniHI 4: new tools for query, analysis and visualization of the human protein-protein interactome. Nucleic Acids Res. 2009, 37: D657-D660. 10.1093/nar/gkn841
Wingender E: The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief Bioinform. 2008, 9: 326-332. 10.1093/bib/bbn016
Duggan DJ, Hoffman EP: Autosomal recessive muscular dystrophy and mutations of the sarcoglycan complex. Neuromuscul Disord. 1996, 6: 475-482. 10.1016/S0960-8966(96)00388-4
Perez-Iratxeta C, Bork P, Andrade-Navarro MA: Update of the G2D tool for prioritization of gene candidates to inherited diseases. Nucleic Acids Res. 2007, 35: W212-W216. 10.1093/nar/gkm223
Lage K, Karlberg EO, Storling ZM, Olason PI, Pedersen AG, Rigina O, Hinsby AM, Tumer Z, Pociot F, Tommerup N, Moreau Y, Brunak S: A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007, 25: 309-316. 10.1038/nbt1295
van Driel MA, Cuelenaere K, Kemmeren PP, Leunissen JA, Brunner HG, Vriend G: GeneSeeker: extraction and integration of human disease-related information from web-based genetic databases. Nucleic Acids Res. 2005, 33: W758-W761. 10.1093/nar/gki435
Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, Van Vooren S, Moreau Y, Pettett RM, Carter NP: DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet. 2009, 84: 524-533. 10.1016/j.ajhg.2009.03.010
Chen J, Xu H, Aronow BJ, Jegga AG: Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinformatics. 2007, 8: 392- 10.1186/1471-2105-8-392
Fraser HB, Plotkin JB: Using protein complexes to predict phenotypic effects of gene mutation. Genome Biol. 2007, 8: R252- 10.1186/gb-2007-8-11-r252
Tiffin N, Kelso JF, Powell AR, Pan H, Bajic VB, Hide WA: Integration of text- and data-mining using ontologies successfully selects disease gene candidates. Nucleic Acids Res. 2005, 33: 1544-1552. 10.1093/nar/gki296
Masseroli M, Galati O, Pinciroli F: GFINDer: genetic disease and phenotype location statistical analysis and mining of dynamically annotated gene lists. Nucleic Acids Res. 2005, 33: W717-W723. 10.1093/nar/gki454
Sam LT, Mendonca EA, Li J, Blake J, Friedman C, Lussier YA: PhenoGO: an integrated resource for the multiscale mining of clinical and biological data. BMC Bioinformatics. 2009, 10 (Suppl 2): S8- 10.1186/1471-2105-10-S2-S8
Robinson PN, Kohler S, Bauer S, Seelow D, Horn D, Mundlos S: The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am J Hum Genet. 2008, 83: 610-615. 10.1016/j.ajhg.2008.09.017
Smith CL, Goldsmith CA, Eppig JT: The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol. 2005, 6: R7- 10.1186/gb-2004-6-1-r7
Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA: The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008, 36: D724-D728. 10.1093/nar/gkm961
Twigger SN, Shimoyama M, Bromberg S, Kwitek AE, Jacob HJ: The Rat Genome Database, update 2007 - easing the path from disease to data and back again. Nucleic Acids Res. 2007, 35: D658-D662. 10.1093/nar/gkl988
Valdar W, Solberg LC, Gauguier D, Burnett S, Klenerman P, Cookson WO, Taylor MS, Rawlins JN, Mott R, Flint J: Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat Genet. 2006, 38: 879-887. 10.1038/ng1840
Auwerx J, Avner P, Baldock R, Ballabio A, Balling R, Barbacid M, Berns A, Bradley A, Brown S, Carmeliet P, Chambon P, Cox R, Davidson D, Davies K, Duboule D, Forejt J, Granucci F, Hastie N, de Angelis MH, Jackson I, Kioussis D, Kollias G, Lathrop M, Lendahl U, Malumbres M, von Melchner H, Müller W, Partanen J, Ricciardi-Castagnoli P, Rigby P, et al.: The European dimension for the mouse genome mutagenesis program. Nat Genet. 2004, 36: 925-927. 10.1038/ng0904-925
Collins FS, Rossant J, Wurst W: A mouse for all reasons. Cell. 2007, 128: 9-13. 10.1016/j.cell.2006.12.018
Phenotypic Quality Ontology, PATO. http://obofoundry.org/wiki/index.php/PATO:Main_Page
Thiel CT, Horn D, Zabel B, Ekici AB, Salinas K, Gebhart E, Rüschendorf F, Sticht H, Spranger J, Müller D, Zweier C, Schmitt ME, Reis A, Rauch A: Severely incapacitating mutations in patients with extreme short stature identify RNA-processing endoribonuclease RMRP as an essential cell growth regulator. Am J Hum Genet. 2005, 77: 795-806. 10.1086/497708
Tremblay K, Lemire M, Potvin C, Tremblay A, Hunninghake GM, Raby BA, Hudson TJ, Perez-Iratxeta C, Andrade-Navarro MA, Laprise C: Genes to diseases (G2D) computational method to identify asthma candidate genes. PLoS One. 2008, 3: e2907- 10.1371/journal.pone.0002907
Tiffin N, Okpechi I, Perez-Iratxeta C, Andrade-Navarro MA, Ramesar R: Prioritization of candidate disease genes for metabolic syndrome by computational analysis of its defining phenotypes. Physiol Genomics. 2008, 35: 55-64. 10.1152/physiolgenomics.90247.2008
Tiffin N, Adie E, Turner F, Brunner HG, van Driel MA, Oti M, Lopez-Bigas N, Ouzounis C, Perez-Iratxeta C, Andrade-Navarro MA, Adeyemo A, Patti ME, Semple CA, Hide W: Computational disease gene identification: a concert of methods prioritizes type 2 diabetes and obesity candidate genes. Nucleic Acids Res. 2006, 34: 3067-3081. 10.1093/nar/gkl381
Cho YS, Go MJ, Han HR, Cha SH, Kim HT, Min H, Shin HD, Park C, Han BG, Cho NH, Shin C, Kimm K, Oh B: Association of lipoprotein lipase (LPL) single nucleotide polymorphisms with type 2 diabetes mellitus. Exp Mol Med. 2008, 40: 523-532. 10.3858/emm.2008.40.5.523
Huang QY, Li GH, Cheung WM, Song YQ, Kung AW: Prediction of osteoporosis candidate genes by computational disease-gene identification strategy. J Hum Genet. 2008, 53: 644-655. 10.1007/s10038-008-0295-x
Lombard Z, Tiffin N, Hofmann O, Bajic VB, Hide W, Ramsay M: Computational selection and prioritization of candidate genes for fetal alcohol syndrome. BMC Genomics. 2007, 8: 389- 10.1186/1471-2164-8-389
Zwaag van der B, Franke L, Poot M, Hochstenbach R, Spierenburg HA, Vorstman JA, van Daalen E, de Jonge MV, Verbeek NE, Brilstra EH, van 't Slot R, Ophoff RA, van Es MA, Blauw HM, Veldink JH, Buizer-Voskamp JE, Beemer FA, Berg van den LH, Wijmenga C, van Amstel HK, van Engeland H, Burbach JP, Staal WG: Gene-network analysis identifies susceptibility genes related to glycobiology in autism. PLoS ONE. 2009, 4: e5324- 10.1371/journal.pone.0005324
Li JB, Levanon EY, Yoon JK, Aach J, Xie B, Leproust E, Zhang K, Gao Y, Church GM: Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009, 324: 1210-1213. 10.1126/science.1170995
Mercer TR, Dinger ME, Mattick JS: Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009, 10: 155-159. 10.1038/nrg2521
Sabherwal N, Bangs F, Roth R, Weiss B, Jantz K, Tiecke E, Hinkel GK, Spaich C, Hauffa BP, Kamp van der H, Kapeller J, Tickle C, Rappold G: Long-range conserved non-coding SHOX sequences regulate expression in developing chicken limb and are associated with short stature phenotypes in human patients. Hum Mol Genet. 2007, 16: 210-222. 10.1093/hmg/ddl470
Clark SJ: Action at a distance: epigenetic silencing of large chromosomal regions in carcinogenesis. Hum Mol Genet. 2007, 16 (Spec 1): R88-R95. 10.1093/hmg/ddm051
Lavorgna G, Dahary D, Lehner B, Sorek R, Sanderson CM, Casari G: In search of antisense. Trends Biochem Sci. 2004, 29: 88-94. 10.1016/j.tibs.2003.12.002
Danko CG, Pertsov AM: Identification of gene co-regulatory modules and associated cis-elements involved in degenerative heart disease. BMC Med Genomics. 2009, 2: 31- 10.1186/1755-8794-2-31
Nam S, Li M, Choi K, Balch C, Kim S, Nephew KP: MicroRNA and mRNA integrated analysis (MMIA): a web tool for examining biological functions of microRNA expression. Nucleic Acids Res. 2009, 37: W356-W362. 10.1093/nar/gkp294
Adeyemo A, Rotimi C: Genetic variants associated with complex human diseases show wide variation across multiple populations. Public Health Genomics. 2009, doi:10.1159/000218711.
Lopez-Bigas N, Blencowe BJ, Ouzounis CA: Highly consistent patterns for inherited human diseases at the molecular level. Bioinformatics. 2006, 22: 269-277. 10.1093/bioinformatics/bti781
Freudenberg J, Propping P: A similarity-based method for genome-wide prediction of disease-relevant human genes. Bioinformatics. 2002, 18 (Suppl 2): S110-S115.
van Driel MA, Bruggeman J, Vriend G, Brunner HG, Leunissen JA: A text-mining analysis of the human phenome. Eur J Hum Genet. 2006, 14: 535-542. 10.1038/sj.ejhg.5201585
Yang Q, Khoury MJ, Botto L, Friedman JM, Flanders WD: Improving the prediction of complex diseases by testing for multiple disease-susceptibility genes. Am J Hum Genet. 2003, 72: 636-649. 10.1086/367923
Acknowledgements
MAA-N acknowledges funding support from the Helmholtz Alliance on Systems Biology. NT is supported by the South African Medical Research Council.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
NT drafted the initial manuscript and coordinated further work on it. All authors wrote the manuscript and have read and approved the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
About this article
Cite this article
Tiffin, N., Andrade-Navarro, M.A. & Perez-Iratxeta, C. Linking genes to diseases: it's all in the data. Genome Med 1, 77 (2009). https://doi.org/10.1186/gm77
Published:
DOI: https://doi.org/10.1186/gm77