Abstract
Traditional pathogen detection methods in public health infectious disease surveillance rely upon the identification of agents that are already known to be associated with a particular clinical syndrome. The emerging field of metagenomics has the potential to revolutionize pathogen detection in public health laboratories by allowing the simultaneous detection of all microorganisms in a clinical sample, without a priori knowledge of their identities, through the use of next-generation DNA sequencing. A single metagenomics analysis has the potential to detect rare and novel pathogens, and to uncover the role of dysbiotic microbiomes in infectious and chronic human disease. Making use of advances in sequencing platforms and bioinformatics tools, recent studies have shown that metagenomics can even determine the whole-genome sequences of pathogens, allowing inferences about antibiotic resistance, virulence, evolution and transmission to be made. We are entering an era in which more novel infectious diseases will be identified through metagenomics-based methods than through traditional laboratory methods. The impetus is now on public health laboratories to integrate metagenomics techniques into their diagnostic arsenals.
Similar content being viewed by others
How do we detect infectious diseases? An introduction to public health laboratory surveillance
Public health infectious disease surveillance employs two strategies to detect cases and outbreaks of communicable diseases: laboratory-based surveillance and syndromic surveillance, which relies on non-laboratory data. Although syndromic surveillance is sometimes the only viable option for population-level monitoring of certain diseases, laboratory-based surveillance is usually more accurate, as the definitive diagnosis of most infectious diseases requires laboratory confirmation. A range of methods are available in public health laboratories: traditional assays include microscopy and culture-based analyses, as well as immunoassays that detect antigens from the pathogen or immune responses from the host; modern techniques include nucleic acid amplification tests. Nevertheless, many samples entering a public health laboratory remain undiagnosed despite being subjected to a battery of conventional laboratory tests.
Conventional laboratory assays fail to detect a causative agent in approximately 40% of gastroenteritis [1] and as many as 60% of encephalitis cases [2], complicating surveillance of these diseases. Presuming a pathogenic agent is present in the sample, the undetected disease agents in these cases may simply be known species that are not targeted by a laboratory’s testing algorithm or they may be truly novel pathogens. The emergence of novel microorganisms challenges laboratory surveillance efforts, which must constantly evolve to identify new pathogens, such as the Middle East respiratory syndrome coronavirus (MERS-CoV) [3, 4] and H7N9 influenza [5]. Over 60% of these emerging pathogens are zoonotic in origin [6], with their entry into human populations facilitated by both human encroachment into previously uninhabited regions and vector redistribution resulting from habitat loss and climate change [7].
One potential way to improve laboratory surveillance would be to employ molecular methods and analytical algorithms that are pathogen-agnostic. Metagenomics, the culture-independent sequencing and analysis of all nucleic acids recovered from a sample, has the potential to revolutionize the detection of both known and novel microorganisms. Its holistic nature means that instead of performing multiple targeted assays, each looking for a specific pathogen, laboratories can use a single sequencing-based test that is capable of identifying most, if not all, microorganisms in a sample without the need for culture. Furthermore, the use of sequencing technology permits the assembly of the complete, or near-complete, genomes of pathogens from a sample. These sequence data can then be used to predict antibiotic resistance phenotypes, to identify virulence genes and to inform enhanced outbreak investigations [8].
In recent years, metagenomics has proven useful in investigating novel species and strains [9–11], outbreaks [12, 13] and complex diseases [14, 15]. As next-generation sequencing technologies improve and costs continue to drop, we anticipate that metagenomic approaches to infectious disease investigations will become increasingly common in public health laboratories, particularly given recent technical improvements that mean that metagenomics can detect pathogens at very low abundance and be can performed directly from clinical samples [16] or even single cells [17].
This review explores metagenomics approaches from the public health laboratory perspective, beginning with a description of two metagenomics approaches and their utility in pathogen detection. We then discuss the implementation of metagenomics approaches in the public health setting, covering challenges that still need to be addressed, such as diagnostic sensitivities, computational resources, and ascertainment of causation.
Where are we now? Traditional laboratory methods for pathogen detection
The traditional paradigm in diagnostic microbiology relies upon tests tailored to etiological agents that are definitively associated with a specific syndrome. Most reference laboratories currently employ a panel of laboratory assays, including culture, microscopy, serology, and PCR. These tools can be powerful methods for identifying microbes, but only if the respective culture conditions, microscope sensitivity, serologic reagents, and PCR primers are compatible with the microbial target and specimen type.
When conventional tests fail to identify microbial pathogens in a sample, an enhanced molecular approach can be used in which PCR-based analyses designed to capture a wider array of targets are employed. These include single or multiplexed PCR assays for conserved regions within a species or genus [18, 19], differentially labeled nucleic acid probes, and direct sequencing of PCR products [20–22]. Computational analyses are used to identify conserved regions in the genomes of known species or genus members; these regions are chosen as primer or probe targets. In multiplexed assays, regions from multiple targets can be selected to detect the most common pathogens known to be associated with a clinical syndrome. Consensus PCR using degenerate primers has also been used successfully to identify a large variety of bacteria and viruses from various hosts and environments, including the recently emerged MERS-CoV [23].
Where are we going? Metagenomics approaches for pathogen detection
When both conventional and enhanced molecular testing fail to identify a causative agent in a sample, the culture-free, holistic metagenomics approach might provide an answer. As with metagenomics studies in human and environmental microbiology [24–26], public health metagenomics can take one of two forms. The first is a targeted strategy called deep amplicon sequencing (DAS), which employs a pre-sequencing PCR amplification step to amplify selectively a taxonomic marker such as a rRNA gene. The second is a broader strategy known as shotgun metagenomics, in which the total nucleic acid content of a sample is sequenced either directly or after applying an enrichment step, which might be a capture-based approach or subtraction prior to sequencing (Table 1). There are benefits and disadvantages to both methods, with some groups adopting a parallel approach in which both techniques are applied to a sample of interest [27].
Deep amplicon sequencing
Within a given taxonomic group, certain gene families occur in every known member species. The Human Microbiome Project (HMP), along with many environmental microbiome studies investigating various microbial communities, have used PCR primers to target conserved gene families. By designing PCR primers to amplify regions within these genes, researchers generate PCR products called amplicons. The DNA sequences of these amplicons are specific to different microbial species, allowing the identification of the various members of the microbial community. Using next-generation-based ‘deep sequencing’, the many different amplicons in a sample can all be sequenced. The resulting sequences are compared to a reference database of the conserved gene to identify the species and/or genus associated with each sequence. The DAS technique is capable of identifying novel microorganisms, describing the microbiome of a specimen, and quantifying the abundance of various taxa in a sample (Table 1).
Bacterial DAS strategies typically utilize primers that are specific to conserved genes, such as 16S rRNA, chaperonin-60 (cpn-60; also named heat shock protein-90 (hsp-90) or groEL) [41], or the RNA polymerase (rpoB) [42]. Similarly, protozoan [43, 44] and fungal [45] DAS studies often target conserved 18S rRNA gene regions. The extraordinary genomic diversity of viruses precludes the amplification of universally conserved genes and the ability to take a complete viral census of a sample; however, primers that are specific to large phylogenetic groups, such as the picorna-like virus superfamily, have enabled large-scale viral DAS studies of previously uncharacterized viral populations (Table 1) [33].
With respect to pathogen detection, the PCR amplification step inherent in the DAS protocol increases the assay’s sensitivity for the microorganisms being targeted, potentially allowing higher resolution and more confident identification of strains or species. Despite its utility in detecting otherwise unidentifiable organisms, however, potential biases in PCR amplification or variable copy numbers of the targeted genes can cause DAS to generate artificially inflated counts of certain taxa in a sample [46, 47]. Furthermore, the ‘universal’ primers used in DAS might not be truly universal, potentially causing certain species, or even groups of species, to be missed [48]. Thus, DAS can give an inaccurate estimation of the microbial community composition. Given that DAS introduces an inherent bias into pathogen detection and requires some a priori knowledge of the potential pathogenic agent of interest in order to select the appropriate gene for amplification, an unbiased sequence-independent shotgun metagenomics approach is better suited to the task of identifying unknown organisms in a sample of interest.
Metagenomics
In contrast to the approach taken by DAS of leveraging conserved gene families across bacteria, fungi, protists or viruses, shotgun metagenomics can potentially catalogue all of the microbes present in a sample, irrespective of their kingdom of origin, by sequencing all the nucleic acid extracted from a specimen. Extracted material is sequenced on a next-generation sequencing platform, and the resulting reads compared to a reference database. These databases are much larger than those used in DAS, as they must contain all known sequences from all organisms rather than a set of sequences from a single gene family. Although this makes the analytical part of a shotgun study computationally intensive, the advantages over DAS are numerous. Shotgun methods are less biased and generate data that better reflect the sample’s true population structure, as recently shown by the HMP team [39]. Furthermore, only shotgun methods can interrogate the accessory genome, that is, the non-core set of genes that often differentiate pathogenic bacteria within a genus or species from closely related commensal strains. For example, Escherichia coli strains K12 and O157:H7 are identical by 16S rRNA DAS analysis, yet the latter strain is considerably more virulent [13].
Shotgun metagenomics studies, which are sometimes followed by Sanger sequencing to generate complete, finished genomes of novel viruses, have identified several novel pathogens from clinical samples (Table 1) [49–55]. A recent notable discovery is the Bas-Congo virus, a rhabdovirus that was associated with a 2009 hemorrhagic fever outbreak in the African Congo [37]. After metagenome-based detection and subsequent de novo assembly of the full-length virus genome, this novel rhabdovirus was shown to share only 25% amino acid identity with its closest known relative. Other examples of novel pathogens that have been discovered through metagenomics include previously unknown cycloviruses found in the cerebrospinal fluid of patients with paraplegia of unknown etiology [56] and a unique hybrid circo/parvovirus (NIH-CQV) in seronegative hepatitis patients [57].
How does it work? Technical and computational aspects of shotgun metagenomics
Given the advantages of shotgun metagenomics over DAS for pathogen detection, the former is becoming increasingly prominent in the public health laboratory setting. Laboratories must adapt to the new technical challenges presented by this technique, including the preparation of samples and sequencing libraries, sequencing, and bioinformatics analysis.
Sample and library preparation
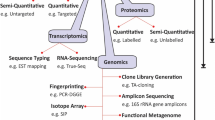
Although some metagenomics studies opt to sequence all of the nucleic acid extracted from a sample regardless of its origin, others adopt a focused strategy in which only a select population of microorganisms (bacteria or viruses) or a specific nucleic acid population (DNA or RNA) is targeted. In these cases, sample preparation pipelines must be modified to target specifically the group of interest. In a virus-specific metagenomics study, cellular material is first removed using filtration or centrifugation to maximize the recovery of virions [58, 59]. Enzymatic treatment of the sample with nucleases can further enrich for viral nucleic acids by removing non-viral nucleic acids while viral nucleic acids remain protected within the nucleocapsid (Figure 1) [60].
Workflow outlining a pipeline of laboratory and bioinformatics methods required for metagenomic pathogen detection. The left side (pale blue) lists each step in the metagenomics workflow and the right side lists the tools used for each stage. Boxes on the right are color-coded to indicate the type of tool used: dark blue, laboratory method; gray, data format; green, computer software; maroon, database. BWA, Burrows-Wheeler Aligner; BLAST, Basic Local Alignment Search Tool; IMG, integrated microbial genomics; MG-RAST, Metagenomics Rapid Annotation Server.
A second technical issue is that the microbial fraction of nucleic acid in a clinical sample can be extremely small, as most of the DNA present is often of human origin. Human DNA can be removed prior to and post-DNA sequencing, using laboratory and bioinformatics methods, respectively. If human DNA is removed using laboratory methods, the small amount of microbial nucleic acid remaining will require newer techniques in sequence library construction to generate usable DNA libraries. Library preparation kits such as Nextera XT (Illumina, San Diego, CA, USA) now require just one nanogram of input DNA. Nextera XT was recently used in a metagenomics characterization of the pathogen in the 2011 German Shiga-Toxigenic E. coli outbreak [12]. To further aid in the detection of microorganisms in samples with very low levels of nucleic acid, or nucleic acid present at very low concentrations, higher sequencing depth might also be necessary to uncover rare sequences [61].
Other laboratory protocols can be implemented to sequence directly from a clinical sample with low nucleic acid concentration, including random PCR as well as multiple displacement amplification (MDA). A recent example by McLean et al.[17] coupled fluorescence-activated cell sorting (FACS) and MDA for single-cell genomic analyses of microbial communities in the biofilm from a hospital sink. The incorporation of MDA also avoids the introduction of mutations in the single cell approach, which may result from culturing the individually sorted bacterial cells [62–64].
DNA sequencing
Following nucleic acid extraction and library preparation (Figure 1), DNA sequencing is performed. Multiple next-generation sequencing platforms are available (Table 2), but the two most commonly used methods in metagenomics studies are Illumina sequencing-by-synthesis and Roche/454 pyrosequencing. The choice of platform is dependent on the sample being sequenced, the questions being asked, and the laboratory’s budget and capacity. An increasing number of public health laboratories are acquiring their own ‘bench-top’ sequencing machines, such as the Illumina MiSeq and Ion Torrent PGM, which are small, affordable and ideally suited for microbial work, but other laboratories outsource samples to a larger sequencing center.
Bioinformatics
Following sequencing, the hundreds of thousands to millions of short reads generated must be computationally transformed into meaningful data reflective of the presence and abundance of the microbes of interest. Bioinformatics analysis is often performed using a staged approach, as outlined in Figure 1. A public health laboratory working in metagenomics must have sufficient computational power and analytic expertise to execute these steps, which may require hiring an experienced bioinformatician to design an in-house data analysis pipeline.
Computational pipelines typically begin with the removal of sequencing library adapters and filtering of low-quality sequences, although this step is sometimes handled by software embedded in the DNA sequencer itself. For clinical samples of human origin, in which human-derived sequence reads comprise the majority of data generated by the sequencer, it is necessary to remove the human reads, often by comparing all of the reads to a human reference genome and discarding those that map to the human genome [68, 69].
Reads that remain after filtering can then be directly compared to microbial reference sequence databases or assembled de novo into larger clusters of contiguous sequence reads (contigs), which are then compared to reference sequence databases. Often, these two approaches are used sequentially (Figure 1). First, individual reads are compared to a reference database in order to assign as many reads as possible to their species, genus, or phylum of origin. The database used for reference-based assembly, as well as the parameters used to call a match, must be chosen carefully because they have a large impact on the assembly generated and on the time taken to generate it. Large databases increase the chance of finding a match but significantly increase the analysis time. Similarly, more permissive parameters might enable the identification of species that are divergent from the reference organism, but may also lead to incorrect taxonomic assignments. Thus, reference-based assembly can also be performed in stages, with increasing database size and decreasing stringency.
Next, remaining reads that did not map to any microbial sequence can be assembled de novo, often using specific algorithms that have been developed for metagenomic assemblies [70]. For taxonomic assignment of the contigs generated, algorithms that are capable of identifying more distant taxonomic matches must be used [71]. Alternatively, a strategy to identify novel sequence reads by using paired-end information to increase iteratively the size of contigs of known classification has recently been developed and successfully implemented to identify two novel arenaviruses in snakes [72].
What can it do? Applications of metagenomics in public health infectious disease surveillance
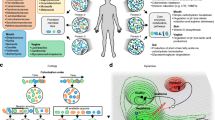
Currently, public health infectious disease surveillance requires a priori knowledge of the pathogen of interest, in that there must be a validated test for the pathogen, and it must be included in the laboratory’s test portfolio. This approach often cannot detect the emergence of completely novel pathogens or pathogens that are not known to be present in a given region. When such an unknown or unusual infectious disease syndrome is encountered, patient specimens will be serially tested against a list of known and suspected pathogens (Figure 2). Nevertheless, conventional laboratory testing might remain negative even after multiple samples are collected and multiple tests are conducted. It is at this point that an investigational pathogen-agnostic method such as metagenomics should be deployed, with the results helping both to uncover unknown etiological agents and to inform the development of new laboratory diagnostic tests or the testing of algorithms to detect future instances of the pathogen in question.
Strategy for novel pathogen detection in public health. Currently, specimens are sent for conventional laboratory tests. If one of these tests is positive (dashed arrows), then an actionable result is generated. If these are all negative, then investigational methods such as metagenomics can be employed afterwards (white arrows). With advances in metagenomics, these methods may be performed earlier in the diagnostic algorithm in the future (black arrows) instead of following multiple traditional laboratory tests.
There are several examples of instances in which a metagenomic approach was able to detect pathogens missed by traditional techniques (Table 3). These include scenarios in which the pathogen was present at very low levels in the sample [73], where the suspected pathogen was not the true cause and was not detected by the tests used [12, 53, 73], and where the causal agent was either a distantly related variant of the suspect pathogen or an entirely new species [9–11]. For example, metagenomics was able to detect pathogens that were missed because of each of the above reasons in a study of condyloma samples that were negative for human papillomavirus (HPV) by PCR. Metagenomics detected both HPV type 6 and putative novel HPV types, as well as the molluscum contagiosum virus (MCV), all of which would have gone undetected using conventional techniques [73].
Beyond pathogen detection, many shotgun metagenomics studies have successfully generated complete or nearly complete pathogen genome assemblies from a sample [16, 17, 40], allowing a comprehensive characterization of the microbe in question at multiple levels. Such assemblies might allow prediction of the microbial phenotype, as indicated by the presence or absence of antimicrobial resistance or virulence genes. They can also be used to calculate the microbial genotype by using in silico techniques in concert with epidemiological information to identify person-to-person transmission events and outbreak or epidemic dynamics [74, 79]. In the case of functional profiling, metagenomics offers the significant advantage of replacing multiple tests with a single analysis (Figure 2), although it must be noted that it is not always possible to detect which species within a metagenome a particular gene comes from [78].
Rather than a single etiological agent, a particular combination of species within an individual can sometimes cause a disease. Through a mixture of DAS and shotgun metagenomics, the HMP has characterized the healthy microbiomes of 242 individuals, collecting microbial nucleic acids from 18 body habitats from five sites (oral, nasal, skin, gut, and urogenital) [81]. By comparing the microbial diversity of these sites across individuals, the HMP demonstrated that the healthy human microbiome differs significantly in taxonomic composition between individuals and between body sites, while the microbial metabolic pathways at each site remain stable [28, 82]. Through a public-health lens, metagenomics thus provides the opportunity to compare taxonomic and functional differences between the microbes present in healthy individuals and those with a range of conditions, from acute infections to chronic diseases of both known and unknown etiology.
Metagenomic studies employing a case-control association discovery approach have identified associations between inflammatory bowel disease and Enterobacteriaceae [83], colorectal carcinoma and Fusobacterium[35], and type 2 diabetes and butyrate-producing bacteria [14, 84]. However, this study design requires careful matching of characteristics, including age, gender, ethnicity and underlying co-morbidities across cases and controls, and any associations identified must be verified in additional samples. Two recent studies investigating metagenomic associations with type 2 diabetes highlight such difficulties. The studies used two populations, one of mixed gender from China and the other of elderly European females. Although both populations demonstrated an association between butyrate-producing bacteria and type 2 diabetes, other discriminatory characteristics differed between the two groups; when the classification generated from one sample set was used on the other, its predictive power was much reduced [14, 84, 85].
Despite these difficulties, association studies provide valuable information about the nature of dysbiotic microbiomes, that is, the disruption in the membership or functional capacity of the healthy microbiome [80]. This altered state could result from a pathogenic species changing the abundance and distribution of other microbial community members, or could be caused by iatrogenic interventions such as antibiotic treatment. Understanding dysbiosis and its role in disease opens the door to the development of alternative forms of treatment, such as probiotics and stool transplants [82], which have been effective in treating Clostridium difficile infections [86].
Beyond profiling of bacterial and viral microbiomes, the fungal component of the human microbiome, the ‘mycobiome’, is an emerging field. Akin to early bacteriological DAS studies, only culturable fungal species associated with illnesses have been studied in detail. Recently, targeted DAS of 18S rRNA revealed that the species diversity of the endogenous fungal community is richer than previously assumed [87]. Furthermore, the mycobiome is not unique to humans: diverse fungal populations have also been recovered from a variety of mammals [45]. The impact that the mycobiome has on human health and disease is currently unknown, and additional investigations using metagenomics approaches are required to further characterize the mycobiome and its potential public health impacts.
In addition to identifying unknown or novel pathogens and characterization of normal and disease-associated microbial communities, metagenomics can also be advantageous in the characterization of the microbiomes of environments that are relevant to public health, such as hospitals and healthcare facilities. Previous non-metagenomic studies have looked for the presence of specific pathogens in healthcare environments [88, 89], but metagenomic profiling of these environments allows the simultaneous detection of multiple pathogens. For example, a single metagenomics study was able to identify 78 candidate species from a biofilm in a hospital sink [17], including the identification of a new bacterial phylum [90]. Metagenomic investigation of healthcare environments promises to provide important insight into the microbial ecology and dynamics of settings such as hospitals or clinics. This is the focus of the recent Hospital Microbiome Project [91], which aims to investigate interactions between the microbiomes of patients and their surrounding hospital environment.
What stands in our way? Challenges facing metagenomics in public health
Despite the successes described above, the application of metagenomics to laboratory-based diagnostics is still in its infancy (Table 4). A recent study using metagenomics to investigate diarrhea samples that were positive for Shiga-Toxigenic E. coli showed a sensitivity of only 67% compared to culture [12, 78], implying that further advances are necessary if metagenomics is to replace traditional culture-based and molecular diagnostics. However, the same study demonstrated metagenomics’ utility in identifying ‘unknown unknowns’, with the authors able to identify co-infections that were not detected by conventional testing.
At the moment, metagenomics has proven most useful in the detection of novel microorganisms. Discovering a novel pathogen or an unusual collection of microorganisms within a clinical sample is, however, merely the first step in the process of determining its role in a disease. The identification of a microbial species through its genome alone does not establish causation. In fact, many of the pathogens that have been discovered through this approach fail to meet Koch’s postulates for causality as it is sometimes not possible to culture the pathogen or to identify a suitable animal model for further studies [99]. Confronted with these challenges, several groups have suggested alternative Koch’s postulates. After the introduction of PCR- and DNA-based identification methods, Fredericks and Relman [99] suggested modified postulates, but even these might not be adequate for recognizing complex diseases in which a combination of multiple microorganisms and/or environmental factors are required to cause disease. More recently, a set of postulates that are applicable to metagenomics has been suggested [96]; but even these require inoculation into a host, which may not be possible for all pathogens.
Other evidence, such as serological and epidemiological analyses, or the ability to stop the disease with microorganism-specific drugs or antibodies, have also been used to address difficulties in meeting Koch’s postulates [11, 75, 100]. For ubiquitous viruses (for example, Epstein-Barr virus, human herpes virus 6 and torque teno virus) or for diseases for which additional variables such as host genetics and environmental factors play a significant role, however, proof of causality can be exceedingly difficult [14, 15]. In such circumstances, care must be taken not to create spurious links between infectious agents and disease, since such false associations could lead to potentially dangerous treatments and might be harder to disprove than to generate initially [75]. For other complex diseases with a polymicrobial etiology, metagenomics can provide a foundation for more targeted quantitative analyses on larger cohorts in order to differentiate between the microorganism(s) driving the disease and non-pathogenic commensals [101].
When interpreting results from metagenomic studies, it is also important to balance scientific plausibility with the possibility of identifying a truly novel association. Research findings are more likely to be true when the prior probability of the finding is high [102]; thus, for unusual metagenomics results, additional lines of evidence are required for confirmation. For example, in a study of nasopharyngeal swabs taken from individuals in the 2009 H1N1 pandemic, one sample contained a pair of reads that mapped with 97% nucleotide identity to Ebola virus, but after further investigation, this finding was concluded to be contamination [13]. Since shotgun metagenomics is a relatively new field, all the possible causes of contamination are not yet known, but they can include experimental reagents, DNA extraction columns [103, 104], cross-contamination during sample processing, and carry-over between sequencing runs [97]. Despite these caveats, all new discoveries must initially arise from novel and unexpected findings, but they must be followed up with the appropriate control samples and experiments.
Conclusions and future perspectives
Although metagenomics pre-dates next-generation sequencing, current sequencing technology has transformed this emerging field, enabling the comprehensive characterization of all of the microbes in a sample. As metagenomic approaches mature and the methods are clinically validated, metagenomics-based approaches might become front-line diagnostic tests for infectious diseases in the public health setting. When faced with an unknown or complex infectious disease, multiple conventional diagnostic tests are often used, potentially leading to unnecessary costs and delays in diagnosis. Instead, metagenomics might be used as a single comprehensive screening test for potential pathogens, both known and novel, as well as to assess the state of an individual’s microbiome (Figure 2). Additional targeted diagnostic tests could then be used to further understand the clinical disease and determine management options.
As sequencing becomes cheaper and faster, it will become possible to serially characterize human microbiomes to investigate changes over time. This could lead to personalized medicine for infectious diseases that accounts for the host genome and microbiome, and to personalized treatments such as the use of narrow-spectrum antibiotics to reduce disruption of the microbiome or specific probiotics to restore an individual’s microbiome to a healthy state [82]. Similar procedures could also be applied to environmental microbiomes in healthcare settings; for example, urinary catheters could be treated with prebiotics to reduce the risk of colonization by harmful bacteria [105]. In fact, it has been suggested that metagenomic investigations of the microbiome could become so standard that DNA sequencers could be used in household toilets to monitor changes in stool microbiome content, which could then be used to guide interventions to maintain health [106].
When a pathogen of interest is known, current metagenomic approaches have limited sensitivity compared to traditional techniques for pathogen detection. Thus, although metagenomics might one day be used for screening clinical samples, it is currently best positioned as a complementary technique to be used alongside culture and other traditional methods. The greatest value of metagenomics is in clinical cases where conventional techniques fail to find a microbial cause. Even then, metagenomics requires skilled scientists to perform the experiments and to analyze the data, and thus, to date it has been exercised primarily in the realm of academic research rather than at the frontlines of public health. To be considered a bona fide clinical test for pathogen detection in a public health laboratory, standard metagenomic protocols are necessary both for testing and analyzing samples and for inter-laboratory comparison of results. As whole-genome sequencing technologies decrease in price and increase in speed and simplicity, however, it is expected that metagenomics approaches will be applied more often in public health emergencies, and routine pipelines are likely to evolve from ongoing collaborations between researchers and clinicians. Such forward steps will be crucial for increasing our arsenal of tools in public health, thus allowing us to rapidly detect and to manage novel and emerging infectious diseases.
Abbreviations
- cpn-60:
-
chaperonin-60
- DAS:
-
deep amplicon sequencing
- HMP:
-
Human Microbiome Project
- HPV:
-
human papillomavirus
- MDA:
-
multiple displacement amplification
- MERS-CoV:
-
Middle East respiratory syndrome coronavirus
- rpoB:
-
RNA polymerase.
References
Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D: Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 2008, 4: e1000011-10.1371/journal.ppat.1000011.
Ambrose HE, Granerod J, Clewley JP, Davies NW, Keir G, Cunningham R, Zuckerman M, Mutton KJ, Ward KN, Ijaz S, Crowcroft NS, Brown DW, UK Aetiology of Encephalitis Study Group: Diagnostic strategy used to establish etiologies of encephalitis in a prospective cohort of patients in England. J Clin Microbiol. 2011, 49: 3576-3583. 10.1128/JCM.00862-11.
Kindler E, Jonsdottir HR, Muth D, Hamming OJ, Hartmann R, Rodriguez R, Geffers R, Fouchier RA, Drosten C, Muller MA, Dijkman R, Thiel V: Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential. mBio. 2013, 4: e00611-e00612.
van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Osterhaus AD, Haagmans BL, Gorbalenya AE, Snijder EJ, Fouchier RA: Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio. 2012, 3: e00473-12.
Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, et al: Human infection with a novel avian-origin influenza A (H7N9) virus. New Engl J Med. 2013, 368: 1888-1897. 10.1056/NEJMoa1304459.
Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P: Global trends in emerging infectious diseases. Nature. 2008, 451: 990-993. 10.1038/nature06536.
Brodie EL, DeSantis TZ, Parker JP, Zubietta IX, Piceno YM, Andersen GL: Urban aerosols harbor diverse and dynamic bacterial populations. Proc Natl Acad Sci USA. 2007, 104: 299-304. 10.1073/pnas.0608255104.
Robinson ER, Walker TM, Pallen MJ: Genomics and outbreak investigation: from sequence to consequence. Genome Med. 2013, 5: 36-
Wan XF, Barnett JL, Cunningham F, Chen S, Yang G, Nash S, Long LP, Ford L, Blackmon S, Zhang Y, Hanson L, He Q: Detection of African swine fever virus-like sequences in ponds in the Mississippi Delta through metagenomic sequencing. Virus Genes. 2013, 46: 441-446. 10.1007/s11262-013-0878-2.
Mokili JL, Dutilh BE, Lim YW, Schneider BS, Taylor T, Haynes MR, Metzgar D, Myers CA, Blair PJ, Nosrat B, Wolfe ND, Rohwer F: Identification of a novel human papillomavirus by metagenomic analysis of samples from patients with febrile respiratory illness. PLoS One. 2013, 8: e58404-10.1371/journal.pone.0058404.
Xu B, Liu L, Huang X, Ma H, Zhang Y, Du Y, Wang P, Tang X, Wang H, Kang K, Zhang S, Zhao G, Wu W, Yang Y, Chen H, Mu F, Chen W: Metagenomic analysis of fever, thrombocytopenia and leukopenia syndrome (FTLS) in Henan Province, China: discovery of a new bunyavirus. PLoS pathogens. 2011, 7: e1002369-10.1371/journal.ppat.1002369.
Loman NJ, Constantinidou C, Christner M, Rohde H, Chan JZ, Quick J, Weir JC, Quince C, Smith GP, Betley JR, Aepfelbacher M, Pallen MJ: A culture-independent sequence-based metagenomics approach to the investigation of an outbreak of Shiga-toxigenic Escherichia coli O104:H4. JAMA. 2013, 309: 1502-1510. 10.1001/jama.2013.3231.
Greninger AL, Chen EC, Sittler T, Scheinerman A, Roubinian N, Yu G, Kim E, Pillai DR, Guyard C, Mazzulli T, Isa P, Arias CF, Hackett J, Schochetman G, Miller S, Tang P, Chiu CY: A metagenomic analysis of pandemic influenza A (2009 H1N1) infection in patients from North America. PLoS One. 2010, 5: e13381-10.1371/journal.pone.0013381.
Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, et al: A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012, 490: 55-60. 10.1038/nature11450.
Cho I, Blaser MJ: The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012, 13: 260-270.
Seth-Smith HM, Harris SR, Skilton RJ, Radebe FM, Golparian D, Shipitsyna E, Duy PT, Scott P, Cutcliffe LT, O’Neill C, Parmar S, Pitt R, Baker S, Ison CA, Marsh P, Jalal H, Lewis DA, Unemo M, Clarke IN, Parkhill J, Thomson NR: Whole-genome sequences of Chlamydia trachomatis directly from clinical samples without culture. Genome Res. 2013, 23: 855-866. 10.1101/gr.150037.112.
McLean JS, Lombardo MJ, Ziegler MG, Novotny M, Yee-Greenbaum J, Badger JH, Tesler G, Nurk S, Lesin V, Brami D, Hall AP, Edlund A, Allen LZ, Durkin S, Reed S, Torriani F, Nealson KH, Pevzner PA, Friedman R, Venter JC, Lasken RS: Genome of the pathogen Porphyromonas gingivalis recovered from a biofilm in a hospital sink using a high-throughput single-cell genomics platform. Genome Res. 2013, 23: 867-877. 10.1101/gr.150433.112.
Mahony JB, Hatchette T, Ojkic D, Drews SJ, Gubbay J, Low DE, Petric M, Tang P, Chong S, Luinstra K, Petrich A, Smieja M: Multiplex PCR tests sentinel the appearance of pandemic influenza viruses including H1N1 swine influenza. J Clin Virol. 2009, 45: 200-202. 10.1016/j.jcv.2009.05.031.
Rohayem J, Berger S, Juretzek T, Herchenroder O, Mogel M, Poppe M, Henker J, Rethwilm A: A simple and rapid single-step multiplex RT-PCR to detect Norovirus, Astrovirus and Adenovirus in clinical stool samples. J Virol Methods. 2004, 118: 49-59. 10.1016/j.jviromet.2004.01.016.
Molenkamp R, van der Ham A, Schinkel J, Beld M: Simultaneous detection of five different DNA targets by real-time Taqman PCR using the Roche LightCycler480: Application in viral molecular diagnostics. J Virol Methods. 2007, 141: 205-211. 10.1016/j.jviromet.2006.12.007.
Raymond F, Carbonneau J, Boucher N, Robitaille L, Boisvert S, Wu WK, De Serres G, Boivin G, Corbeil J: Comparison of automated microarray detection with real-time PCR assays for detection of respiratory viruses in specimens obtained from children. J Clin Microbiol. 2009, 47: 743-750. 10.1128/JCM.01297-08.
Roh KH, Kim J, Nam MH, Yoon S, Lee CK, Lee K, Yoo Y, Kim MJ, Cho Y: Comparison of the Seeplex reverse transcription PCR assay with the R-mix viral culture and immunofluorescence techniques for detection of eight respiratory viruses. Ann Clin Lab Sci. 2008, 38: 41-46.
Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA: Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New Engl J Med. 2012, 367: 1814-1820. 10.1056/NEJMoa1211721.
Bibby K: Metagenomic identification of viral pathogens. Trends Biotechnol. 2013, 31: 275-279. 10.1016/j.tibtech.2013.01.016.
Thomas T, Gilbert J, Meyer F: Metagenomics - a guide from sampling to data analysis. Microb InformExp. 2012, 2: 3-10.1186/2042-5783-2-3.
Temperton B, Giovannoni SJ: Metagenomics: microbial diversity through a scratched lens. Curr Opin Microbiol. 2012, 15: 605-612. 10.1016/j.mib.2012.07.001.
Lozupone C, Cota-Gomez A, Palmer BE, Linderman DJ, Charlson ES, Sodergren E, Mitreva M, Abubucker S, Martin J, Yao G, Campbell TB, Flores SC, Ackerman G, Stombaugh J, Ursell L, Beck JM, Curtis JL, Young VB, Lynch SV, Huang L, Weinstock GM, Knox KS, Twigg H, Morris A, Ghedin E, Bushman FD, Collman RG, Knight R, Fontenot AP, Lung HIV Microbiome Project: Widespread colonization of the lung by Tropheryma whipplei in HIV Infection. Am J Respir Crit Care Med. 2013, 187: 1110-1117. 10.1164/rccm.201211-2145OC.
Human Microbiome Project Consortium: Structure, function and diversity of the healthy human microbiome. Nature. 2012, 486: 207-214. 10.1038/nature11234.
Tito RY, Knights D, Metcalf J, Obregon-Tito AJ, Cleeland L, Najar F, Roe B, Reinhard K, Sobolik K, Belknap S, Foster M, Spicer P, Knight R, Lewis CM: Insights from characterizing extinct human gut microbiomes. PLoS One. 2012, 7: e51146-10.1371/journal.pone.0051146.
Paramel Jayaprakash T, Schellenberg JJ, Hill JE: Resolution and characterization of distinct cpn60-based subgroups of Gardnerella vaginalis in the vaginal microbiota. PLoS One. 2012, 7: e43009-10.1371/journal.pone.0043009.
Khamis A, Raoult D, La Scola B: Comparison between rpoB and 16S rRNA gene sequencing for molecular identification of 168 clinical isolates of Corynebacterium. J Clin Microbiol. 2005, 43: 1934-1936. 10.1128/JCM.43.4.1934-1936.2005.
Schellenberg J, Links MG, Hill JE, Dumonceaux TJ, Peters GA, Tyler S, Ball TB, Severini A, Plummer FA: Pyrosequencing of the chaperonin-60 universal target as a tool for determining microbial community composition. Appl Environ Microbiol. 2009, 75: 2889-2898. 10.1128/AEM.01640-08.
Culley AI, Lang AS, Suttle CA: High diversity of unknown picorna-like viruses in the sea. Nature. 2003, 424: 1054-1057. 10.1038/nature01886.
Wurtzel O, Sesto N, Mellin JR, Karunker I, Edelheit S, Becavin C, Archambaud C, Cossart P, Sorek R: Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol Syst Biol. 2012, 8: 583-
Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA: Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012, 22: 299-306. 10.1101/gr.126516.111.
Rosario K, Breitbart M: Exploring the viral world through metagenomics. Curr Opin Virol. 2011, 1: 289-297. 10.1016/j.coviro.2011.06.004.
Grard G, Fair JN, Lee D, Slikas E, Steffen I, Muyembe JJ, Sittler T, Veeraraghavan N, Ruby JG, Wang C, Makuwa M, Mulembakani P, Tesh RB, Mazet J, Rimoin AW, Taylor T, Schneider BS, Simmons G, Delwart E, Wolfe ND, Chiu CY, Leroy EM: A novel rhabdovirus associated with acute hemorrhagic fever in central Africa. PLoS Pathog. 2012, 8: e1002924-10.1371/journal.ppat.1002924.
Chan JZ, Sergeant MJ, Lee OY, Minnikin DE, Besra GS, Pap I, Spigelman M, Donoghue HD, Pallen MJ: Metagenomic analysis of tuberculosis in a mummy. New Engl J Med. 2013, 369: 289-290. 10.1056/NEJMc1302295.
Shakya M, Quince C, Campbell JH, Yang ZK, Schadt CW, Podar M: Comparative metagenomic and rRNA microbial diversity characterization using archaeal and bacterial synthetic communities. Environ Microbiol. 2013, 15: 1882-1899. 10.1111/1462-2920.12086.
Bos KI, Schuenemann VJ, Golding GB, Burbano HA, Waglechner N, Coombes BK, McPhee JB, DeWitte SN, Meyer M, Schmedes S, Wood J, Earn DJ, Herring DA, Bauer P, Poinar HN, Krause J: A draft genome of Yersinia pestis from victims of the Black Death. Nature. 2011, 478: 506-510. 10.1038/nature10549.
Links MG, Dumonceaux TJ, Hemmingsen SM, Hill JE: The chaperonin-60 universal target is a barcode for bacteria that enables de novo assembly of metagenomic sequence data. PLoS One. 2012, 7: e49755-10.1371/journal.pone.0049755.
Wu D, Wu M, Halpern A, Rusch DB, Yooseph S, Frazier M, Venter JC, Eisen JA: Stalking the fourth domain in metagenomic data: searching for, discovering, and interpreting novel, deep branches in marker gene phylogenetic trees. PLoS One. 2011, 6: e18011-10.1371/journal.pone.0018011.
Leng J, Zhong X, Zhu RJ, Yang SL, Gou X, Mao HM: Assessment of protozoa in Yunnan Yellow cattle rumen based on the 18S rRNA sequences. Mol Biol Rep. 2011, 38: 577-585. 10.1007/s11033-010-0143-x.
Sirohi SK, Singh N, Dagar SS, Puniya AK: Molecular tools for deciphering the microbial community structure and diversity in rumen ecosystem. Appl Microbiol Biotechnol. 2012, 95: 1135-1154. 10.1007/s00253-012-4262-2.
Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, Rotter JI, Wang HL, McGovern DP, Brown GD, Underhill DM: Interactions between commensal fungi and the C-Type lectin receptor Dectin-1 influence colitis. Science. 2012, 336: 1314-1317. 10.1126/science.1221789.
Gonzalez JM, Portillo MC, Belda-Ferre P, Mira A: Amplification by PCR artificially reduces the proportion of the rare biosphere in microbial communities. PLoS One. 2012, 7: e29973-10.1371/journal.pone.0029973.
Schloss PD, Gevers D, Westcott SL: Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One. 2011, 6: e27310-10.1371/journal.pone.0027310.
Forney LJ, Zhou X, Brown CJ: Molecular microbial ecology: land of the one-eyed king. Curr Opin Microbiol. 2004, 7: 210-220. 10.1016/j.mib.2004.04.015.
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO: Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004, 304: 66-74. 10.1126/science.1093857.
Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K, Eisen JA, Heidelberg KB, Manning G, Li W, Jaroszewski L, Cieplak P, Miller CS, Li H, Mashiyama ST, Joachimiak MP, van Belle C, Chandonia JM, Soergel DA, Zhai Y, Natarajan K, Lee S, Raphael BJ, Bafna V, Friedman R, Brenner SE, Godzik A, Eisenberg D, Dixon JE, Taylor SS, et al: The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol. 2007, 5: e16-10.1371/journal.pbio.0050016.
Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, Carlson C, Chan AM, Haynes M, Kelley S, Liu H, Mahaffy JM, Mueller JE, Nulton J, Olson R, Parsons R, Rayhawk S, Suttle CA, Rohwer F: The marine viromes of four oceanic regions. PLoS Biol. 2006, 4: e368-10.1371/journal.pbio.0040368.
Ng TF, Manire C, Borrowman K, Langer T, Ehrhart L, Breitbart M: Discovery of a novel single-stranded DNA virus from a sea turtle fibropapilloma by using viral metagenomics. J Virol. 2009, 83: 2500-2509. 10.1128/JVI.01946-08.
Svraka S, Rosario K, Duizer E, van der Avoort H, Breitbart M, Koopmans M: Metagenomic sequencing for virus identification in a public-health setting. J Gen Virol. 2010, 91: 2846-2856. 10.1099/vir.0.024612-0.
Phan TG, Kapusinszky B, Wang C, Rose RK, Lipton HL, Delwart EL: The fecal viral flora of wild rodents. PLoS Pathog. 2011, 7: e1002218-10.1371/journal.ppat.1002218.
Gaynor AM, Nissen MD, Whiley DM, Mackay IM, Lambert SB, Wu G, Brennan DC, Storch GA, Sloots TP, Wang D: Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 2007, 3: e64-10.1371/journal.ppat.0030064.
Smits S, Zijlstra E, Hellemond J, Schapendonk C, Bodewes R, Schürch A, Haagmans B, Osterhaus A: Novel Cyclovirus in human cerebrospinal fluid, Malawi, 2010-2011. Emerging Infect Dis. 2013, doi: 10.3201/eid1909.130404
Xu B, Zhi N, Hu G, Wan Z, Zheng X, Liu X, Wong S, Kajigaya S, Zhao K, Mao Q, Young NS: Hybrid DNA virus in Chinese patients with seronegative hepatitis discovered by deep sequencing. Proc Natl Acad Sci USA. 2013, 110: 10264-10269. 10.1073/pnas.1303744110.
Delwart EL: Viral metagenomics. Rev Med Virol. 2007, 17: 115-131. 10.1002/rmv.532.
Thurber RV, Haynes M, Breitbart M, Wegley L, Rohwer F: Laboratory procedures to generate viral metagenomes. Nat Protoc. 2009, 4: 470-483. 10.1038/nprot.2009.10.
Allander T, Emerson SU, Engle RE, Purcell RH, Bukh J: A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc Natl Acad Sci USA. 2001, 98: 11609-11614. 10.1073/pnas.211424698.
Hamady M, Knight R: Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res. 2009, 19: 1141-1152. 10.1101/gr.085464.108.
Karch H, Meyer T, Russmann H, Heesemann J: Frequent loss of Shiga-like toxin genes in clinical isolates of Escherichia coli upon subcultivation. Infect Immun. 1992, 60: 3464-3467.
Nair S, Alokam S, Kothapalli S, Porwollik S, Proctor E, Choy C, McClelland M, Liu SL, Sanderson KE: Salmonella enterica serovar Typhi strains from which SPI7, a 134-kilobase island with genes for Vi exopolysaccharide and other functions, has been deleted. J Bacteriol. 2004, 186: 3214-3223. 10.1128/JB.186.10.3214-3223.2004.
Dubourg G, Lagier JC, Armougom F, Robert C, Hamad I, Brouqui P, Raoult D: The proof of concept that culturomics can be superior to metagenomics to study atypical stool samples. Eur J Clin Microbiol Infect Dis. 2013, 32: 1099-10.1007/s10096-013-1843-7.
Nakamura K, Oshima T, Morimoto T, Ikeda S, Yoshikawa H, Shiwa Y, Ishikawa S, Linak MC, Hirai A, Takahashi H, Altaf-Ul-Amin M, Ogasawara N, Kanaya S: Sequence-specific error profile of Illumina sequencers. Nucleic Acids Res. 2011, 39: e90-10.1093/nar/gkr344.
Schroder J, Bailey J, Conway T, Zobel J: Reference-free validation of short read data. PLoS One. 2010, 5: e12681-10.1371/journal.pone.0012681.
Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y: A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012, 13: 341-10.1186/1471-2164-13-341.
Li H, Homer N: A survey of sequence alignment algorithms for next-generation sequencing. Brief Bioinform. 2010, 11: 473-483. 10.1093/bib/bbq015.
Moore RA, Warren RL, Freeman JD, Gustavsen JA, Chenard C, Friedman JM, Suttle CA, Zhao Y, Holt RA: The sensitivity of massively parallel sequencing for detecting candidate infectious agents associated with human tissue. PLoS One. 2011, 6: e19838-10.1371/journal.pone.0019838.
Namiki T, Hachiya T, Tanaka H, Sakakibara Y: MetaVelvet: an extension of Velvet assembler to de novo metagenome assembly from short sequence reads. Nucleic Acids Res. 2012, 40: e155-10.1093/nar/gks678.
Soding J: Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005, 21: 951-960. 10.1093/bioinformatics/bti125.
Stenglein MD, Sanders C, Kistler AL, Ruby JG, Franco JY, Reavill DR, Dunker F, Derisi JL: Identification, characterization, and in vitro culture of highly divergent arenaviruses from boa constrictors and annulated tree boas: candidate etiological agents for snake inclusion body disease. mBio. 2012, 3: e00180-00112-
Johansson H, Bzhalava D, Ekstrom J, Hultin E, Dillner J, Forslund O: Metagenomic sequencing of “HPV-negative”condylomas detects novel putative HPV types. Virology. 2013, 440: 1-7. 10.1016/j.virol.2013.01.023.
Bertelli C, Greub G: Rapid bacterial genome sequencing: methods and applications in clinical microbiology. Clin Microbiol Infect. 2013, doi: 10.1111/1469-0691.12217
Lipkin WI: The changing face of pathogen discovery and surveillance. Nat Rev Microbiol. 2013, 11: 133-141. 10.1038/nrmicro2949.
US Food and Drug Administration: Ultra high throughput sequencing for clinical diagnostic applications - approaches to assess analytical validity: report from the public meeting (June 23, 2011). [http://www.fda.gov/downloads/MedicalDevices/NewsEvents/WorkshopsConferences/UCM266607.pdf]
Köser CU, Ellington MJ, Cartwright EJP, Gillespie SH, Brown NM, Farrington M, Holden MTG, Dougan G, Bentley SD, Parkhill J, Peacock SJ: Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog. 2012, 8: e1002824-10.1371/journal.ppat.1002824.
Relman DA: Metagenomics, infectious disease diagnostics, and outbreak investigations: sequence first, ask questions later?. JAMA. 2013, 309: 1531-1532. 10.1001/jama.2013.3678.
Andersson P, Klein M, Lilliebridge RA, Giffard PM: Sequences of multiple bacterial genomes and a Chlamydia trachomatis genotype from direct sequencing of DNA derived from a vaginal swab diagnostic specimen. Clin Microbiol Infect. 2013, doi: 10.1111/1469-0691
Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, Huttenhower C: Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol. 2012, 8: e1002606-10.1371/journal.pcbi.1002606.
The Human Microbiome Consortium: A framework for human microbiome research. Nature. 2012, 486: 215-221. 10.1038/nature11209.
Lemon KP, Armitage GC, Relman DA, Fischbach MA: Microbiota-targeted therapies: an ecological perspective. Sci Transl Med. 2012, 4: 137rv5-10.1126/scitranslmed.3004183.
Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, Gordon JI, Onderdonk AB, Glimcher LH: Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010, 8: 292-300. 10.1016/j.chom.2010.08.004.
Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B, Nielsen J, Backhed F: Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013, 498: 99-103. 10.1038/nature12198.
de Vos WM, Nieuwdorp M: Genomics: a gut prediction. Nature. 2013, 498: 48-49. 10.1038/nature12251.
Gough E, Shaikh H, Manges AR: Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis. 2011, 53: 994-1002. 10.1093/cid/cir632.
Huffnagle GB, Noverr MC: The emerging world of the fungal microbiome. Trends Microbiol. 2013, 21: 334-341. 10.1016/j.tim.2013.04.002.
Faires MC, Pearl DL, Ciccotelli WA, Straus K, Zinken G, Berke O, Reid-Smith RJ, Weese JS: A prospective study to examine the epidemiology of methicillin-resistant Staphylococcus aureus and Clostridium difficile contamination in the general environment of three community hospitals in southern Ontario. Canada. BMC Infect Dis. 2012, 12: 290-10.1186/1471-2334-12-290.
Weber DJ, Rutala WA, Miller MB, Huslage K, Sickbert-Bennett E: Role of hospital surfaces in the transmission of emerging health care-associated pathogens: norovirus, Clostridium difficile, and Acinetobacter species. Am J Infect Control. 2010, 38: S25-S33. 10.1016/j.ajic.2010.04.196.
McLean JS, Lombardo MJ, Badger JH, Edlund A, Novotny M, Yee-Greenbaum J, Vyahhi N, Hall AP, Yang Y, Dupont CL, Ziegler MG, Chitsaz H, Allen AE, Yooseph S, Tesler G, Pevzner PA, Friedman RM, Nealson KH, Venter JC, Lasken RS: Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum. Proc Natl Acad Sci USA. 2013, 110: E2390-E2399. 10.1073/pnas.1219809110.
Hospital Microbiome. [http://hospitalmicrobiome.com/]
Pallen MJ, Loman NJ: Are diagnostic and public health bacteriology ready to become branches of genomic medicine?. Genome Med. 2011, 3: 53-10.1186/gm269.
Gevers D, Pop M, Schloss PD, Huttenhower C: Bioinformatics for the human microbiome project. PLoS Comput Biol. 2012, 8: e1002779-10.1371/journal.pcbi.1002779.
Human Microbiome Project Catalog and Statistics. [http://www.hmpdacc.org/catalog/]
Loman NJ, Constantinidou C, Chan JZ, Halachev M, Sergeant M, Penn CW, Robinson ER, Pallen MJ: High-throughput bacterial genome sequencing: an embarrassment of choice, a world of opportunity. Nat Rev Microbiol. 2012, 10: 599-606. 10.1038/nrmicro2850.
Mokili JL, Rohwer F, Dutilh BE: Metagenomics and future perspectives in virus discovery. Curr Opin Virol. 2012, 2: 63-77. 10.1016/j.coviro.2011.12.004.
Swei A, Russell BJ, Naccache SN, Kabre B, Veeraraghavan N, Pilgard MA, Johnson BJ, Chiu CY: The genome sequence of Lone Star Virus, a highly divergent bunyavirus found in the Amblyomma americanum tick. PLoS One. 2013, 8: e62083-10.1371/journal.pone.0062083.
Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, Baker CC, Di Francesco V, Howcroft TK, Karp RW, Lunsford RD, Wellington CR, Belachew T, Wright M, Giblin C, David H, Mills M, Salomon R, Mullins C, Akolkar B, Begg L, Davis C, Grandison L, Humble M, Khalsa J, NIH HMP Working Group, et al: The NIH Human Microbiome Project. Genome Res. 2009, 19: 2317-2323.
Fredericks DN, Relman DA: Sequence-based identification of microbial pathogens: a reconsideration of Koch’s postulates. Clin Microbiol Rev. 1996, 9: 18-33.
Chiu CY, Yagi S, Lu X, Yu G, Chen EC, Liu M, Dick EJ, Carey KD, Erdman DD, Leland MM, Patterson JL: A novel adenovirus species associated with an acute respiratory outbreak in a baboon colony and evidence of coincident human infection. mBio. 2013, 4: e00084-
Hajishengallis G, Darveau RP, Curtis MA: The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012, 10: 717-725. 10.1038/nrmicro2873.
Ioannidis JP: Why most published research findings are false. PLoS Med. 2005, 2: e124-10.1371/journal.pmed.0020124.
Erlwein O, Robinson MJ, Dustan S, Weber J, Kaye S, McClure MO: DNA extraction columns contaminated with murine sequences. PLoS One. 2011, 6: e23484-10.1371/journal.pone.0023484.
Evans GE, Murdoch DR, Anderson TP, Potter HC, George PM, Chambers ST: Contamination of Qiagen DNA extraction kits with Legionella DNA. J Clin Microbiol. 2003, 41: 3452-3453. 10.1128/JCM.41.7.3452-3453.2003.
Foxman B, Rosenthal M: Implications of the Human Microbiome Project for epidemiology. Am J Epidemiol. 2013, 177: 197-201. 10.1093/aje/kws449.
Schmieder R, Edwards R: Insights into antibiotic resistance through metagenomic approaches. Future Microbiol. 2012, 7: 73-89. 10.2217/fmb.11.135.
Acknowledgments
We thank Mark Pallen for his helpful comments on an earlier version of this manuscript.
This work is supported financially by the British Columbia Centre for Disease Control Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
About this article
Cite this article
Miller, R.R., Montoya, V., Gardy, J.L. et al. Metagenomics for pathogen detection in public health. Genome Med 5, 81 (2013). https://doi.org/10.1186/gm485
Published:
DOI: https://doi.org/10.1186/gm485