Abstract
Background
Organic anion transporting polypeptide (OATP) 1B1, OATP1B3, and OATP2B1 (encoded by SLCO1B1, SLCO1B3, SLCO2B1) mediate the hepatic uptake of endogenous compounds like bile acids and of drugs, for example, the lipid-lowering atorvastatin, thereby influencing hepatobiliary elimination. Here we systematically elucidated the contribution of SLCO variants on expression of the three hepatic OATPs under consideration of additional important covariates.
Methods
Expression was quantified by RT-PCR and immunoblotting in 143 Caucasian liver samples. A total of 109 rare and common variants in the SLCO1B3-SLCO1B1 genomic region and the SLCO2B1 gene were genotyped by MALDI-TOF mass spectrometry and genome-wide SNP microarray technology. SLCO1B1 haplotypes affecting hepatic OATP1B1 expression were associated with pharmacokinetic data of the OATP1B1 substrate atorvastatin (n = 82).
Results
Expression of OATP1B1, OATP1B3, and OATP2B1 at the mRNA and protein levels showed marked interindividual variability. All three OATPs were expressed in a coordinated fashion. By a multivariate regression analysis adjusted for non-genetic and transcription covariates, increased OATP1B1 expression was associated with the coding SLCO1B1 variant c.388A > G (rs2306283) even after correction for multiple testing (P = 0.00034). This held true for haplotypes harboring c.388A > G but not the functional variant c.521T > C (rs4149056) associated with statin-related myopathy. c.388A > G also significantly affected atorvastatin pharmacokinetics. SLCO variants and non-genetic and regulatory covariates together accounted for 59% of variability of OATP1B1 expression.
Conclusions
Our results show that expression of OATP1B1, but not of OATP1B3 and OATP2B1, is significantly affected by genetic variants. The SLCO1B1 variant c.388A > G is the major determinant with additional consequences on atorvastatin plasma levels.
Similar content being viewed by others
Background
The organic anion transporting polypeptides 1B1 (OATP1B1, encoded by the SLCO1B1 gene), OATP1B3 (SLCO1B3), and OATP2B1 (SLCO2B1) are major uptake transporters on the sinusoidal membrane of human hepatocytes. They mediate the influx of endogenous compounds such as bile salts, bilirubin glucuronides, thyroid hormones and steroid hormone metabolites, and clinically frequently used drugs like statins, HIV protease inhibitors, and the anti-cancer agents irinotecan or methotrexate [1–5]. The importance of OATP transporters for hepatobiliary uptake is emphasized by the Rotor syndrome, which is a two-gene disorder caused by the complete combined deficiency of OATP1B1 and OATP1B3 [6].
Numerous clinical studies support the relevance of common but also rare SLCO1B1 missense variants altering either the pharmacokinetics or drug response of OATP1B1 substrates [4, 7, 8]. The common variant c.521T > C (rs4149056; Val174Ala) is highlighted by a genome-wide association study (GWAS) suggesting an increased risk for simvastatin-induced myopathy in variant carriers [9]. Reduced hepatic uptake of the OATP1B1 substrates atorvastatin and rosuvastatin is supported by in vitro experiments using cell lines stably expressing the c.521T > C variant [2, 10]. Moreover, the in vivo disposition of endogenous and/or xenobiotic substances, including drugs, is also affected by SLCO1B3 and SLCO2B1 variants [11].
Irrespective of well-established data on functional consequences of SLCO variants, the contribution of SLCO variants to the interindividual variability of hepatic expression of OATP transporters is still unknown. Although cholestasis has been recognized as an additional determinant of OATP1B1 and OATP1B3 expression [12], a comprehensive analysis including SLCO variants as well as non-genetic and regulatory covariates is presently lacking. Similar to recent work related to the hepatic uptake transporters OCT1 and OCT3 [13], we therefore investigated the impact of > 100 SLCO variants as well as non-genetic and regulatory covariates on the interindividual variability of hepatic OATP1B1, OATP1B3, and OATP2B1 expression. The transcription factors hepatocyte nuclear factor (HNF)1α, farnesoid X receptor (FXR), liver X receptor (LXR)α, specificity protein 1 (Sp1), aryl hydrocarbon receptor (AhR), constitutive androstane receptor (CAR), and HNF3β were selected because literature data indicate they are involved in regulation of the three hepatic SLCO genes with potential consequences on variability of expression [14–21]. The novel finding that the SLCO1B1 missense variant Asn130Asp (rs2306283) alters hepatic protein expression most effectively is supported by pharmacokinetic data of atorvastatin in a study of healthy volunteers. SLCO variants and non-genetic and regulatory covariates accounted for 59% of variability of hepatic OATP1B1 expression.
Materials and methods
Human liver samples
Liver tissue and corresponding blood samples were collected from patients undergoing liver surgery at the Department of General, Visceral, and Transplantation Surgery (University Medical Center Charité, Berlin, Germany) as described previously [13, 22, 23]. Tissue samples had been examined by a pathologist; only histologically normal liver tissue was used for further studies. For each patient, detailed information was available regarding age, sex, smoking status, alcohol consumption, presurgery medication, indication for liver resection, and presurgery liver serum parameters. Samples from patients with hepatitis, cirrhosis, or chronic alcohol use were excluded. A total of 143 liver samples from which high quality RNA and complete documentation could be obtained were finally included (Table S1 in Additional file 1). The study was approved by the ethics committees of the Charité, Humboldt University (Berlin, Germany) and the University of Tübingen (Tübingen, Germany) in accordance with the principles of the Declaration of Helsinki. Written informed consent was obtained from each patient.
Atorvastatin pharmacokinetics in vivo
Atorvastatin pharmacokinetic variables were obtained from 82 healthy volunteers (Table S2 in Additional file 1) as previously described [24–26]. Briefly, volunteers ingested a single 20 mg dose of atorvastatin (Lipitor; Pfizer/Gödecke, Karlsruhe, Germany). No other drugs or grapefruit products were consumed prior to atorvastatin administration [25]. The study was approved by the Ethics Committee of the Hospital District of Helsinki and Uusimaa, Finland. Written informed consent was obtained from the participants.
Selection of genetic variants and genotyping strategies
Genomic DNA was purified from EDTA blood samples with the QIAmp DNA Blood MiniKit (Qiagen, Hilden, Germany). All samples were genotyped for 109 genetic variants (Figure S1 and Table S3 in Additional file 1). We selected 58 genetic variants of the SLCO1B3-SLCO1B1 genomic region (chromosome 12) and the SLCO2B1 gene (chromosome 11) from the National Center for Biotechnology Information database (dbSNP build 129) based on functional criteria and/or frequency distribution. Genotyping was performed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) using the MassARRAY Compact system (Sequenom, San Diego, CA, USA) or by 5'-nuclease assays (ABI Prism 7900 Sequence Detection System, Applied Biosystems/Life Technologies, Carlsbad, CA, USA). Moreover, 51 additional variants of the SLCO1B3-SLCO1B1 genomic region and the SLCO2B1 gene were genotyped using the HumanHap300v1.1 chip data set (Illumina, San Diego, CA, USA) as described previously [27] (NCBI Gene Expression Omnibus series GSE39036, GSE32504). For MALDI-TOF MS and TaqMan analysis, about 10% of samples were re-genotyped as quality control resulting in 100% concordance. Missing calls in MALDI-TOF MS and TaqMan genotyping were re-typed, resulting in final call rates of 99%. Laboratory staff were blinded to the case status of liver samples. Details of primers and genotyping assays are available upon request.
RNA isolation and quantification
High-quality total RNA was extracted from liver samples and reverse-transcribed as described [13]. mRNA was quantified by TaqMan technology (Additional file 1).
Quantification of OATPs in human liver samples
OATP proteins were quantified in membrane fractions from the liver samples by immunoblot analyses using previously characterized antibodies [28, 29] (Additional file 1).
Generation of HEK cells stably expressing OATP2B1 and missense variants
Human embryonic kidney (HEK) cells were transfected with constructs encoding human SLCO2B1 or the missense variants c.601G > A (rs35199625; GenBank: NM_007256.4 as reference), c.935G > A (rs12422149), and c.1457C > T (rs2306168) (Additional file 1).
Immunofluorescence microscopy
Cryosections of liver samples were immunostained for OATP1B1 and OATP2B1 using previously characterized antibodies [29, 30]. Images were taken with a confocal laser scanning microscope (Additional file 1).
Transport studies and prediction of functional effects
Transport of atorvastatin, rosuvastatin, and estrone sulfate by OATP2B1 and variants was measured using stably-transfected HEK cells (Additional file 1). Functional effects of OATP2B1 missense variants were calculated using four different algorithms (Additional file 1).
Statistics
Hardy-Weinberg equilibrium calculations [31, 32] were used to compare observed and expected allele and genotype frequencies. Linkage disequilibrium analysis of the SLCO1B3-SLCO1B1 genomic region and the SLCO2B1 gene was performed with Haploview [31] using results from our study population of 143 Caucasians. Haplotype analysis was performed with R-package haplo.stats-1.4.4 [33] (R-2.13.0).
R-Package SNPassoc-1.6-0 was applied to study associations between each variant and OATP expression, with correction for non-genetic (Table 1) and transcription factors (HNF1α, Sp1, AhR, LXRα, FXR, CAR, HNF3β). Multivariate linear models and step-wise model selection were used to determine the fraction of variance in OATP expression explained by non-genetic, genetic, and transcription covariates. All statistical tests were two-tailed and statistical significance was defined as P < 0.05. Where indicated, P-values were adjusted for multiple testing according to Holm [34] (see Additional file 1 for detailed information).
Results
Hepatic OATP expression
OATP1B1, OATP1B3, and OATP2B1 mRNA and protein expression varied considerably within the 143 liver samples and were not normally distributed (Figure 1; Figure S2 in Additional file 1). mRNA and protein expression weakly correlated for SLCO1B1/OATP1B1 and SLCO1B3/OATP1B3, but not for SLCO2B1/OATP2B1 (Figure 1b-d). Marked interindividual variability was also obtained when only non-cholestatic liver samples (n = 117) were analyzed (Table S4A in Additional file 1). Moreover, of interest, SLCO1B1, SLCO1B3, and SLCO2B1 mRNA levels were significantly correlated with each other, which held true to a lesser extent for protein levels (Figure 1e, f).
Hepatic expression of OATP1B1, OATP1B3 and OATP2B1 varies between individuals, but is coordinately regulated within one individual. (a) Representative immunoblot of liver membrane fractions (20 μg) quantified relative to a standard liver sample. (b-d) Correlation analyses of OATP protein and SLCO mRNA levels in human liver samples. (e, f) Correlation analyses of all three OATPs showed a coordinated expression of SLCO1B1, SLCO1B3, and SLCO2B1 mRNA and OATP1B1, OATP1B3, and OATP2B1 protein levels in the non-cholestatic liver sample set. rS, Spearman rank-order correlation coefficient.
Impact of non-genetic factors on OATP expression
Multivariate linear regression analyses showed no association between OATP expression and sex, age, smoking habit, presurgery medication, bilirubin levels, or γ-glutamyl transferase levels (Table 1). Alcohol consumption was significantly associated with reduced SLCO1B1 and SLCO2B1 mRNA levels. Pathophysiologically increased C-reactive protein levels were associated with reduced SLCO1B1 mRNA levels. As expected from previous studies [12], OATP1B1 and OATP1B3 protein levels were significantly lower in cholestatic liver samples. When analyzing only the non-cholestatic samples, alcohol consumption was again significantly associated with reduced SLCO2B1 mRNA levels (Table S5 in Additional file 1).
SLCO genetic variants and frequencies
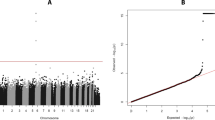
Table S3 in Additional file 1 specifies all 109 common and rare variants genotyped, including additional information on location and allele frequencies. We detected 83 variants in the 143 liver samples, 66 variants in the SLCO1B3-SLCO1B1 genomic region and 17 variants in the SLCO2B1 gene (Figure S1 in Additional file 1). No deviations from Hardy-Weinberg equilibrium were observed. Notably, nearly complete linkage disequilibrium was observed for several variants in SLCO1B1 (Figure 2a).
SLCO genetic variants affect expression of hepatic OATPs. (a, b) Pairwise linkage disequilibrium map of the SLCO1B3-SLCO1B1 genomic region (a) and the SLCO2B1 gene (b) including all variants detected in the 143 livers. Coloring corresponds to the standard Haploview (D'/LOD) [31]. (c,d) SLCO1B1 haplotypes, calculated based on the four missense variants present in the liver cohort (c.388A > G, c.463C > A, c.521T > C, c.1929A > C) and also previously described as key variants [8, 80], affect OATP1B1 protein expression in the non-cholestatic liver samples (n = 117). Only haplotypes with frequencies ≥ 2% are given. The effect sizes indicate differences of OATP1B1 expression compared to reference haplotype SLCO1B1*1a. Boldface: significant P-values. (d) SLCO1B1 genotypes (percentage genotype frequencies in brackets) are sorted by median OATP1B1 protein expression. SLCO1B1 alleles harboring only variant c.388A > G and not c.521T > C (*1b, *14, *35, boldface) confer considerably higher OATP1B1 expression in a gene-dose dependent manner. Horizontal line indicates median; boxes indicate the 25th to 75th percentiles; whiskers indicate non-outlier range. (e) Percentage of interindividual variability of expression of OATPs in the non-cholestatic liver samples apportioned to non-genetic factors (gray), regulatory factors (blue), and SLCO variants (orange), and to the combination of all three categories (red) was calculated using multivariate linear regression analyses and stepwise model selection.
SLCO/OATP genotype-phenotype correlation analyses
In order to exclude confounding by cholestasis, all subsequent analyses were performed with the non-cholestatic liver samples (n = 117). Multivariate linear regression models were used to analyze associations between variants with frequencies ≥ 1% in the SLCO1B3-SLCO1B1 genomic region and the SLCO2B1 gene (Figure S1 in Additional file 1) and the expression of OATP1B1, OATP1B3, and OATP2B1. Models were corrected for eight non-genetic covariates (Table S6 in Additional file 1) and the regulatory factors HNF1, LXR, FXR, HNF3, Sp1, CAR, and AhR previously suggested by literature data as transcriptional regulators of at least one SLCO gene [14–21]. Interindividual variability of the expression of these transcription factors is given in Table S4B in Additional file 1.
Considering the additive genetic model, 38 variants in the SLCO1B3-SLCO1B1 genomic region correlated with OATP1B1 or OATP1B3 expression, and 3 variants in the SLCO2B1 gene with OATP2B1 expression (Table S6 in Additional file 1). After multiple testing correction, 16 variants in the SLCO1B3-SLCO1B1 genomic region were still significantly associated with OATP1B1 expression. Notably, for the OATP1B1 protein, variant c.388A > G (Asn130Asp, rs2306283) showed the lowest P-value (P = 0.00034) whereas the variant c.521T > C (Val174Ala, rs4149056), associated with statin-induced myopathy [9, 35, 36], did not show any association. Stratification of baseline characteristics by carriers of c.388A > G (dominant model) did not show any significance.
Additionally, we calculated haplotypes for the non-cholestatic liver samples using different strategies. Firstly, haplotypes were calculated for genetic variants of SLCO1B1, SLCO1B3, and SLCO2B1 detected in the liver samples (Table S3 in Additional file 1). Only variants with a frequency ≥ 1% were included. Significant associations with substantial effect sizes between haplotypes and expression were only found for OATP1B1 protein and SLCO2B1 mRNA expression, even after correction for multiple testing (Tables S7 to S9 in Additional file 1). Secondly, previously reported SLCO1B1 haplotypes containing only the variants c.388A > G, c.463C > A, c.521T > C and c.1929A > C were calculated (Figure 2c). The haplotypes *1b (P = 0.0037), *14 (P = 1.8 × 10-5), and *35 (P = 0.0004), including the c.388A > G but not the c.521T > C variant, were significantly associated with increased OATP1B1 protein expression, even after multiple testing correction. These haplotypes exhibited substantial effect sizes (*1b, 0.43; *14, 0.53; *35, 0.56) and clustered with samples of highest median OATP1B1 expression (Figure 2d). OATP1B1 protein levels also increased with the number of *1b, *14, and *35 alleles (Figure S3A in Additional file 1).
Thirdly, we calculated haplotypes covering all variants of the SLCO1B3-SLCO1B1 genomic region (Table S10 in Additional file 1). Two haplotypes, which included the SLCO1B1 variant c.388A > G, were significantly associated with increased OATP1B1 protein levels and showed substantial effect sizes for the correlation (H-cluster04, 0.62; H-cluster12, 0.59).
Contribution of genetic, non-genetic, and transcription factors to variable OATP expression
Interindividual variability of hepatic OATP1B1 protein expression was substantially explained by SLCO1B1 variants (50%) while non-genetic (8%) and transcription factors (7%) contributed only to a minor extent (Figure 2e). Notably, 59% of variability could be apportioned to the combination of all three categories (that is, genetic, non-genetic, transcription factors). Total variance of OATP1B3 protein expression was explained to 55% by these categories but only 24% of variance of OATP2B1 protein expression could be apportioned to the three categories.
Atorvastatin pharmacokinetics
SLCO1B1 haplotypes for all 82 volunteers were calculated considering the SLCO1B1 variants c.388A > G, c.463C > A, c.521T > C and c.1929A > C. Seven different haplotypes with a frequency distribution ≥ 2% were detected (Figure 3a; Table S11 in Additional file 1). A linear regression analysis including body weight as covariate indicated that the haplotypes *1b and *14, both harboring c.388A > G but not c.521T > C, were associated with decreased atorvastatin area under the plasma concentration-time curve (AUC) compared to the reference haplotype (*1a), reaching statistical significance for *14 (P = 0.0126). Notably, homozygous carriers of *1b and *14 showed the lowest atorvastatin AUC (Figure 3b). In contrast, haplotype *15, harboring c.388A > G and c.521T > C, was associated with an increased atorvastatin AUC (P = 2.7 × 10-6; Figure 3a) and an increased atorvastatin Cmax (P = 0.031; Table S11 in Additional file 1). Here, homozygous carriers for *15 showed highest AUC for the total study cohort (Figure 3b). Atorvastatin AUC also decreased with the number of *1b and *14 alleles (Figure S3B in Additional file 1).
SLCO1B1 haplotypes affect atorvastatin pharmacokinetics in 82 healthy volunteers. (a) SLCO1B1 haplotypes calculated on the presence of the four previously described key variants and change in atorvastatin area under the plasma concentration-time curve (AUC), corrected for body weight, in comparison with the reference haplotype SLCO1B1*1a. Haplotypes are given with a frequency > 2%. (b) SLCO1B1 genotypes (percentage genotype frequencies in brackets) are sorted by median atorvastatin AUC. SLCO1B1 alleles harboring only variant c.388A > G and not c.521T > C (boldface) confer considerably lower AUCs with lowest levels in homozygous variant or compound heterozygous carriers for the SLCO1B1*14 or *1b alleles. Horizontal line indicates the median; boxes indicate 25th to 75th percentiles; whiskers indicate non-outlier range.
In order to identify whether the effects of variant c.388A > G may be confounded by variant c.521T > C, we conducted multivariate models including both variants simultaneously. The variant c.388A > G was significantly associated with decreased AUC for atorvastatin (P = 0.00066, recessive model) whereas the variant c.521T > C resulted in an increased AUC (P = 2.6 × 10-10, additive model). Of note, these data held true when the interaction effect of c.388A > G and c.521T > C was included into the model (c.388A > G, P = 0.00535; c.521T > C, P = 3.0 × 10-7; interaction effect, P = 0.67).
Statin transport by OATP2B1 and its missense variants
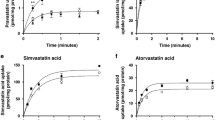
The SLCO2B1 missense variants c.601G > A, c.935G > A and c.1457C > T were the only ones present in our liver cohort (Table S3 in Additional file 1). Although they had no effect on expression (Figure 2e), functional consequences of these variants could not be excluded with certainty. We therefore stably expressed the three variants separately in HEK cells. The variant transfectants showed correct immunolocalization of OATP2B1 in the plasma membrane (Figure S4A in Additional file 1). Only c.1457C > T but not c.601G > A or c.935G > A showed reduced uptake of atorvastatin, rosuvastatin and the prototypical substrate estrone sulfate (Figure S5 in Additional file 1). The calculated maximal uptake rate of atorvastatin was significantly lower for c.1457C > T and tended to be lower for rosuvastatin (Table S12 in Additional file 1). In contrast, the Km value of c.935G > A was significantly higher for atorvastatin (Table S12 in Additional file 1). Sinusoidal hepatocyte membrane immunostaining of OATP2B1 in liver cryosections from homozygous variant carriers for c.935AA was similar to OATP2B1-reference (Figure S4B in Additional file 1). Moreover, four independent computational tools did not correctly predict experimental uptake data (Table S13 in Additional file 1). Altogether, the uptake studies indicate some functional consequences, which needs validation by in vivo studies.
Discussion
This first systematic analysis of the human membrane transporters OATP1B1, OATP1B3, and OATP2B1 on the interindividual variability of hepatic expression revealed a major contribution of SLCO1B1 variants on OATP1B1 protein expression. Expression of OATP1B3 or OATP2B1 was apparently not influenced by genetic variants. Most strikingly, the SLCO1B1 missense variant c.388A > G resulted in the strongest association with increased OATP1B1 protein expression whereas the c.521T > C variant, linked to statin-related myopathy [9, 35, 36], did not alter protein levels. This was observed for both the non-cholestatic livers (n = 117; Figure 2c, d) as well as the total liver set of 143 samples (Table S14 in Additional file 1), indicating that c.388A > G alters OATP1B1 expression independent of cholestasis. The observation that variant c.388A > G is associated with highest OATP1B1 protein expression is corroborated by our haplotype analysis since homozygous or compound heterozygous carriers of haplotypes *1b, *14, or *35, harboring c.388A > G but not c.521T > C, showed also the highest OATP1B1 protein levels. A mislocalization of OATP1B1 protein in genotyped livers for variant c.388A > G could be excluded by immunolocalization analyses (Figure S6 in Additional file 1).
The results of the liver study are confirmed by functional data of the pharmacokinetic healthy volunteer study since the *14 haplotype was associated with a significantly decreased atorvastatin plasma AUC in contrast to the *15 haplotype, harboring both c.388A > G and c.521T > C, and resulting in an increased AUC. Thus, the SLCO1B1 variant c.388A > G is an important determinant of OATP1B1 expression and function, independent of the c.521T > C variant. In line with in vitro reports [37, 38], one may suggest that the increased clearance by OATP1B1 c.388A > G is mainly caused by a higher protein expression. The increased hepatic atorvastatin uptake in c.388A > G carriers is also supported by haplotype data from previous studies indicating that only c.388A > G was associated with significant lower AUCs for the OATP1B1 substrates pravastatin and repaglinide [39–41]. Moreover, variant c.388A > G carriers showed a significantly improved response to statins measured by low-density lipoprotein cholesterol levels [42, 43] and were even protected from statin-induced side effects [42]. Of note, however, the effect of the c.388A > G variant seems to be substrate-specific since the AUC was not altered, for example, for rosuvastatin [44], underscoring the requirement to perform pharmacokinetic studies for each OATP1B1 drug separately. Mechanistically, we suggest that variant c.388A > G modifies mRNA secondary structure, resulting in an alteration of translation efficiency as previously shown for the CFTR membrane protein [45]. Indeed, computational theoretical modeling provided evidence that the SLCO1B1 mRNA secondary structures are altered only by haplotypes *1b, *14, and *35 harboring the c.388A > G variant (Figure S7 in Additional file 1).
In line with the localization of the OATP1B1*15 protein in the cell membrane using stably expressing HEK cells [38], a mislocalization of the OATP1B1 protein in livers genotyped for *15 haplotype was not found. The increased atorvastatin AUC in carriers of the *15 haplotype in our pharmacokinetic study revealed impaired atorvastatin uptake, indicating that the variant OATP1B1*15 protein is associated with decreased intrinsic transport activity. This observation is supported by previous studies using oocytes that revealed significantly reduced atorvastatin uptake by OATP1B1*15 in comparison with the reference sequence [46].
Another novel observation of our study is the coordinated expression of OATP1B1, OATP1B3, and OATP2B1 in human liver. The coordinate expression may be explained by regulatory networks considering previously identified transcription factors like HNF1α, Sp1, AhR, LXRα, FXR, CAR, and HNF3β [14–21]. Since HNF1α confers basal promoter activity of SLCO1B1 and SLCO1B3 and functional binding sites have been detected in the promoter regions of both genes [14, 17], we analyzed the impact of HNF1α on SLCO mRNA expression. Because exon_1e transcripts of SLCO2B1 are highly abundant in human liver [47] and the exon_1e promoter region has so far not been analyzed, we identified three high-scoring putative HNF1α binding sites in the exon_1e promoter (Methods and Figure S8A in Additional file 1) by in silico analysis. Electromobility shift assays confirmed in vitro binding of HNF1α to each of the three identified motifs, thereby indicating that the exon_1e transcript of SLCO2B1 is regulated by HNF1α in liver. Binding of HNF1α to any of the three sites in the exon_1e promoter of SLCO2B1 appeared to be weaker than binding to the respective motifs in the SLCO1B1 and SLCO1B3 gene promoters (Figure S8B in Additional file 1).
Among several clinical and demographic factors, including age, sex, alcohol consumption, smoking habit, and presurgery medication, only cholestasis resulted in significantly reduced OATP1B1 and OATP1B3 protein levels considering the total set of 143 liver samples (Table 1). This is in line with previous data since cholestasis in cases of primary biliary cirrhosis or progressive familial intrahepatic cholestasis is associated with reduced OATP1B1 and OATP1B3 expression [12] and the concerted down-regulation of both bile salt uptake transporters is considered as a protective mechanism against hepatocellular injury caused by cytotoxic bile acids. Cholestasis-related down-regulation of OATP1B1 and OATP1B3 might be of clinical relevance regarding drug response to several well-established OATP substrates, for example, lopinavir [48] and irinotecan [49].
Moreover, one aim of our study was to elucidate systematically the contribution of genetic, non-genetic and regulatory factors for the prediction of the interindividual variability of the expression of hepatic OATP transporters. Generally, the interindividual variability of expression of hepatic OATPs is considerably higher than variation of the hepatic intrinsic clearance of drugs that are substrates for OATP transport proteins as reported by clinical studies [50]. This discrepancy is, however, not surprising since the hepatic clearance is the sum of a number of processes and factors - for example, involvement of different drug transporters for one substrate drug, short-term regulation of transporters (for example, by protein kinases), differences in retrieval of transporters from the plasma membrane, and multiple binding sites on transporters. Our multivariate analyses only using non-cholestatic liver samples revealed that the combination of genetic, non-genetic, and transcription factors explained 59% and 55% of variability of OATP1B1 and OATP1B3 protein expression, respectively, while OATP2B1 protein variance remained largely unaffected (24%). Several reasons might explain the poor prediction of interindividual variability of hepatic OATP2B1 expression, although systematic ascertainment of covariates in our liver bank was guaranteed [51] (Table S1 in Additional file 1). Other non-genetic factors like cytokines [52] may affect OATP2B1 expression. Moreover, unidentified transcription factors, genetic variation in those genes as well as epigenetics such as DNA methylation [53] or regulation by microRNA [54] may contribute to the interindividual variability of OATP2B1.
Conclusions
In our comprehensive work on the expression of hepatic OATPs we show for the first time a major contribution of SLCO1B1 genetics to the interindividual variability of OATP1B1 protein expression. The SLCO1B1 variant c.388A > G strongly affects OATP1B1 expression with additional functional consequences on atorvastatin plasma levels. Further work is warranted to identify underlying mechanisms for the so far unexplained interindividual variability of hepatic OATP1B3 and OATP2B1 expression.
Abbreviations
- AhR:
-
aryl hydrocarbon receptor
- AUC:
-
area under the plasma concentration-time curve
- CAR:
-
constitutive androstane receptor
- FXR:
-
farnesoid X receptor
- HEK:
-
human embryonic kidney cells
- HNF:
-
hepatocyte nuclear factor
- LXR:
-
liver X receptor
- MALDI-TOF MS:
-
matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
- OATP:
-
organic anion transporting polypeptide
- SLC:
-
solute carrier
- Sp1:
-
specificity protein 1.
References
Hagenbuch B, Meier PJ: Organic anion transporting polypeptides of the OATP/SLC21 family: phylogenetic classification as OATP/SLCO superfamily, new nomenclature and molecular/functional properties. Pflugers Arch. 2004, 447: 653-665. 10.1007/s00424-003-1168-y.
Ho RH, Kim RB: Transporters and drug therapy: implications for drug disposition and disease. Clin Pharmacol Ther. 2005, 78: 260-277. 10.1016/j.clpt.2005.05.011.
Giacomini KM, Huang SM, Tweedie DJ, Benet LZ, Brouwer KL, Chu X, Dahlin A, Evers R, Fischer V, Hillgren KM, Hoffmaster KA, Ishikawa T, Keppler D, Kim RB, Lee CA, Niemi M, Polli JW, Sugiyama Y, Swaan PW, Ware JA, Wright SH, Yee SW, Zamek-Gliszczynski MJ, Zhang L: Membrane transporters in drug development. Nat Rev Drug Discov. 2010, 9: 215-236. 10.1038/nrd3028.
Niemi M, Pasanen MK, Neuvonen PJ: Organic anion transporting polypeptide 1B1: a genetically polymorphic transporter of major importance for hepatic drug uptake. Pharmacol Rev. 2011, 63: 157-181. 10.1124/pr.110.002857.
Nies AT, Schwab M, Keppler D: Interplay of conjugating enzymes with OATP uptake transporters and ABCC/MRP efflux pumps in the elimination of drugs. Expert Opin Drug Metab Toxicol. 2008, 4: 545-568. 10.1517/17425255.4.5.545.
van de Steeg E, Stranecky V, Hartmannova H, Noskova L, Hrebicek M, Wagenaar E, van Esch A, de Waart DR, Oude Elferink RP, Kenworthy KE, Sticová E, al-Edreesi M, Knisely AS, Kmoch S, Jirsa M, Schinkel AH: Complete OATP1B1 and OATP1B3 deficiency causes human Rotor syndrome by interrupting conjugated bilirubin reuptake into the liver. J Clin Invest. 2012, 122: 519-528. 10.1172/JCI59526.
Pasanen MK, Neuvonen M, Neuvonen PJ, Niemi M: SLCO1B1 polymorphism markedly affects the pharmacokinetics of simvastatin acid. Pharmacogenet Genomics. 2006, 16: 873-879. 10.1097/01.fpc.0000230416.82349.90.
Ramsey LB, Bruun GH, Yang W, Trevino LR, Vattathil S, Scheet P, Cheng C, Rosner GL, Giacomini KM, Fan Y, Sparreboom A, Mikkelsen TS, Corydon TJ, Pui CH, Evans WE, Relling MV: Rare versus common variants in pharmacogenetics: SLCO1B1 variation and methotrexate disposition. Genome Res. 2012, 22: 1-8. 10.1101/gr.129668.111.
SEARCH Collaborative Group: SLCO1B1 variants and statin-induced myopathy -- a genomewide study. N Engl J Med. 2008, 359: 789-799.
Kameyama Y, Yamashita K, Kobayashi K, Hosokawa M, Chiba K: Functional characterization of SLCO1B1 (OATP-C) variants, SLCO1B1*5, SLCO1B1*15 and SLCO1B1*15+C1007G, by using transient expression systems of HeLa and HEK293 cells. Pharmacogenet Genomics. 2005, 15: 513-522. 10.1097/01.fpc.0000170913.73780.5f.
Clarke JD, Cherrington NJ: Genetics or environment in drug transport: the case of organic anion transporting polypeptides and adverse drug reactions. Expert Opin Drug Metab Toxicol. 2012, 8: 349-360. 10.1517/17425255.2012.656087.
Wagner M, Zollner G, Trauner M: New molecular insights into the mechanisms of cholestasis. J Hepatol. 2009, 51: 565-580. 10.1016/j.jhep.2009.05.012.
Nies AT, Koepsell H, Winter S, Burk O, Klein K, Kerb R, Zanger UM, Keppler D, Schwab M, Schaeffeler E: Expression of organic cation transporters OCT1 (SLC22A1) and OCT3 (SLC22A3) is affected by genetic factors and cholestasis in human liver. Hepatology. 2009, 50: 1227-1240. 10.1002/hep.23103.
Jung D, Hagenbuch B, Gresh L, Pontoglio M, Meier PJ, Kullak-Ublick GA: Characterization of the human OATP-C (SLC21A6) gene promoter and regulation of liver-specific OATP genes by hepatocyte nuclear factor 1 alpha. J Biol Chem. 2001, 276: 37206-37214. 10.1074/jbc.M103988200.
Jung D, Podvinec M, Meyer UA, Mangelsdorf DJ, Fried M, Meier PJ, Kullak-Ublick GA: Human organic anion transporting polypeptide 8 promoter is transactivated by the farnesoid X receptor/bile acid receptor. Gastroenterology. 2002, 122: 1954-1966. 10.1053/gast.2002.33583.
Vavricka SR, Jung D, Fried M, Grutzner U, Meier PJ, Kullak-Ublick GA: The human organic anion transporting polypeptide 8 (SLCO1B3) gene is transcriptionally repressed by hepatocyte nuclear factor 3beta in hepatocellular carcinoma. J Hepatol. 2004, 40: 212-218. 10.1016/j.jhep.2003.10.008.
Ohtsuka H, Abe T, Onogawa T, Kondo N, Sato T, Oshio H, Mizutamari H, Mikkaichi T, Oikawa M, Rikiyama T, Katayose Y, Unno M: Farnesoid X receptor, hepatocyte nuclear factors 1alpha and 3beta are essential for transcriptional activation of the liver-specific organic anion transporter-2 gene. J Gastroenterol. 2006, 41: 369-377. 10.1007/s00535-006-1784-3.
Maeda T, Hirayama M, Higashi R, Sato M, Tamai I: Characterization of human OATP2B1 (SLCO2B1) gene promoter regulation. Pharm Res. 2006, 23: 513-520. 10.1007/s11095-006-9572-6.
Jigorel E, Le VM, Boursier-Neyret C, Parmentier Y, Fardel O: Differential regulation of sinusoidal and canalicular hepatic drug transporter expression by xenobiotics activating drug-sensing receptors in primary human hepatocytes. Drug Metab Dispos. 2006, 34: 1756-1763. 10.1124/dmd.106.010033.
Furihata T, Satoh T, Yamamoto N, Kobayashi K, Chiba K: Hepatocyte nuclear factor 1 alpha is a factor responsible for the interindividual variation of OATP1B1 mRNA levels in adult Japanese livers. Pharm Res. 2007, 24: 2327-2332. 10.1007/s11095-007-9458-2.
Meyer zu Schwabedissen HE, Bottcher K, Chaudhry A, Kroemer HK, Schuetz EG, Kim RB: Liver X receptor alpha and farnesoid X receptor are major transcriptional regulators of OATP1B1. Hepatology. 2010, 52: 1797-1807. 10.1002/hep.23876.
Meier Y, Pauli-Magnus C, Zanger UM, Klein K, Schaeffeler E, Nussler AK, Nussler N, Eichelbaum M, Meier PJ, Stieger B: Interindividual variability of canalicular ATP-binding-cassette (ABC)-transporter expression in human liver. Hepatology. 2006, 44: 62-74.
Klein K, Winter S, Turpeinen M, Schwab M, Zanger UM: Pathway-targeted pharmacogenomics of CYP1A2 in human liver. Front Pharmacol. 2010, 1: 129-
Pasanen MK, Fredrikson H, Neuvonen PJ, Niemi M: Different effects of SLCO1B1 polymorphism on the pharmacokinetics of atorvastatin and rosuvastatin. Clin Pharmacol Ther. 2007, 82: 726-733. 10.1038/sj.clpt.6100220.
Keskitalo JE, Kurkinen KJ, Neuvoneni PJ, Niemi M: ABCB1 haplotypes differentially affect the pharmacokinetics of the acid and lactone forms of simvastatin and atorvastatin. Clin Pharmacol Ther. 2008, 84: 457-461. 10.1038/clpt.2008.25.
Keskitalo JE, Zolk O, Fromm MF, Kurkinen KJ, Neuvonen PJ, Niemi M: ABCG2 polymorphism markedly affects the pharmacokinetics of atorvastatin and rosuvastatin. Clin Pharmacol Ther. 2009, 86: 197-203. 10.1038/clpt.2009.79.
Schröder A, Klein K, Winter S, Schwab M, Bonin M, Zell A, Zanger UM: Genomics of ADME gene expression: mapping expression quantitative trait loci relevant for absorption, distribution, metabolism and excretion of drugs in human liver. Pharmacogenomics J. 2011, 13: 12-20.
Cui Y, König J, Nies AT, Pfannschmidt M, Hergt M, Franke WW, Alt W, Moll R, Keppler D: Detection of the human organic anion transporters SLC21A6 (OATP2) and SLC21A8 (OATP8) in liver and hepatocellular carcinoma. Lab Invest. 2003, 83: 527-538.
Kullak-Ublick GA, Ismair MG, Stieger B, Landmann L, Huber R, Pizzagalli F, Fattinger K, Meier PJ, Hagenbuch B: Organic anion-transporting polypeptide B (OATP-B) and its functional comparison with three other OATPs of human liver. Gastroenterology. 2001, 120: 525-533. 10.1053/gast.2001.21176.
Gui C, Miao Y, Thompson L, Wahlgren B, Mock M, Stieger B, Hagenbuch B: Effect of pregnane X receptor ligands on transport mediated by human OATP1B1 and OATP1B3. Eur J Pharmacol. 2008, 584: 57-65. 10.1016/j.ejphar.2008.01.042.
Barrett JC, Fry B, Maller J, Daly MJ: Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005, 21: 263-265. 10.1093/bioinformatics/bth457.
Wigginton JE, Cutler DJ, Abecasis GR: A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005, 76: 887-893. 10.1086/429864.
The R project for statistical computing. [http://www.r-project.org]
Holm S: A simple sequentially rejective multiple test procedure. Scand J Statist. 1979, 6: 65-70.
Voora D, Shah SH, Spasojevic I, Ali S, Reed CR, Salisbury BA, Ginsburg GS: The SLCO1B1*5 genetic variant is associated with statin-induced side effects. J Am Coll Cardiol. 2009, 54: 1609-1616. 10.1016/j.jacc.2009.04.053.
Brunham LR, Lansberg PJ, Zhang L, Miao F, Carter C, Hovingh GK, Visscher H, Jukema JW, Stalenhoef AF, Ross CJ, Carleton BC, Kastelein JJ, Hayden MR: Differential effect of the rs4149056 variant in SLCO1B1 on myopathy associated with simvastatin and atorvastatin. Pharmacogenomics J. 2011, 12: 237-
Nozawa T, Nakajima M, Tamai I, Noda K, Nezu J, Sai Y, Tsuji A, Yokoi T: Genetic polymorphisms of human organic anion transporters OATP-C (SLC21A6) and OATP-B (SLC21A9): allele frequencies in the Japanese population and functional analysis. J Pharmacol Exp Ther. 2002, 302: 804-813. 10.1124/jpet.302.2.804.
Iwai M, Suzuki H, Ieiri I, Otsubo K, Sugiyama Y: Functional analysis of single nucleotide polymorphisms of hepatic organic anion transporter OATP1B1 (OATP-C). Pharmacogenetics. 2004, 14: 749-757. 10.1097/00008571-200411000-00006.
Mwinyi J, Johne A, Bauer S, Roots I, Gerloff T: Evidence for inverse effects of OATP-C (SLC21A6) 5 and 1b haplotypes on pravastatin kinetics. Clin Pharmacol Ther. 2004, 75: 415-421. 10.1016/j.clpt.2003.12.016.
Maeda K, Ieiri I, Yasuda K, Fujino A, Fujiwara H, Otsubo K, Hirano M, Watanabe T, Kitamura Y, Kusuhara H, Sugiyama Y: Effects of organic anion transporting polypeptide 1B1 haplotype on pharmacokinetics of pravastatin, valsartan, and temocapril. Clin Pharmacol Ther. 2006, 79: 427-439. 10.1016/j.clpt.2006.01.011.
Kalliokoski A, Backman JT, Neuvonen PJ, Niemi M: Effects of the SLCO1B1*1B haplotype on the pharmacokinetics and pharmacodynamics of repaglinide and nateglinide. Pharmacogenet Genomics. 2008, 18: 937-942. 10.1097/FPC.0b013e32830d733e.
Donnelly LA, Doney AS, Tavendale R, Lang CC, Pearson ER, Colhoun HM, McCarthy MI, Hattersley AT, Morris AD, Palmer CN: Common nonsynonymous substitutions in SLCO1B1 predispose to statin intolerance in routinely treated individuals with type 2 diabetes: a go-DARTS study. Clin Pharmacol Ther. 2011, 89: 210-216. 10.1038/clpt.2010.255.
Rodrigues AC, Perin PM, Purim SG, Silbiger VN, Genvigir FD, Willrich MA, Arazi SS, Luchessi AD, Hirata MH, Bernik MM, Dorea EL, Santos C, Faludi AA, Bertolami MC, Salas A, Freire A, Lareu MV, Phillips C, Porras-Hurtado L, Fondevila M, Carracedo A, Hirata RD: Pharmacogenetics of OATP transporters reveals that SLCO1B1 c.388A > G variant is determinant of increased atorvastatin response. Int J Mol Sci. 2011, 12: 5815-5827. 10.3390/ijms12095815.
Lee E, Ryan S, Birmingham B, Zalikowski J, March R, Ambrose H, Moore R, Lee C, Chen Y, Schneck D: Rosuvastatin pharmacokinetics and pharmacogenetics in white and Asian subjects residing in the same environment. Clin Pharmacol Ther. 2005, 78: 330-341. 10.1016/j.clpt.2005.06.013.
Bartoszewski RA, Jablonsky M, Bartoszewska S, Stevenson L, Dai Q, Kappes J, Collawn JF, Bebok Z: A synonymous single nucleotide polymorphism in DeltaF508 CFTR alters the secondary structure of the mRNA and the expression of the mutant protein. J Biol Chem. 2010, 285: 28741-28748. 10.1074/jbc.M110.154575.
Choi MK, Shin HJ, Choi YL, Deng JW, Shin JG, Song IS: Differential effect of genetic variants of Na(+)-taurocholate co-transporting polypeptide (NTCP) and organic anion-transporting polypeptide 1B1 (OATP1B1) on the uptake of HMG-CoA reductase inhibitors. Xenobiotica. 2011, 41: 24-34. 10.3109/00498254.2010.523736.
Pomari E, Nardi A, Fiore C, Celeghin A, Colombo L, Valle LD: Transcriptional control of human organic anion transporting polypeptide 2B1 gene. J Steroid Biochem Mol Biol. 2009, 115: 146-152. 10.1016/j.jsbmb.2009.04.003.
Seminari E, Gentilini G, Galli L, Hasson H, Danise A, Carini E, Dorigatti F, Soldarini A, Lazzarin A, Castagna A: Higher plasma lopinavir concentrations are associated with a moderate rise in cholestasis markers in HIV-infected patients. J Antimicrob Chemother. 2005, 56: 790-792. 10.1093/jac/dki314.
Raymond E, Boige V, Faivre S, Sanderink GJ, Rixe O, Vernillet L, Jacques C, Gatineau M, Ducreux M, Armand JP: Dosage adjustment and pharmacokinetic profile of irinotecan in cancer patients with hepatic dysfunction. J Clin Oncol. 2002, 20: 4303-4312. 10.1200/JCO.2002.03.123.
Kusuhara H, Sugiyama Y: Pharmacokinetic modeling of the hepatobiliary transport mediated by cooperation of uptake and efflux transporters. Drug Metab Rev. 2010, 42: 539-550. 10.3109/03602530903491824.
Wolbold R, Klein K, Burk O, Nussler AK, Neuhaus P, Eichelbaum M, Schwab M, Zanger UM: Sex is a major determinant of CYP3A4 expression in human liver. Hepatology. 2003, 38: 978-988.
Le Vee M, Jouan E, Stieger B, Lecureur V, Fardel O: Regulation of drug transporter expression by oncostatin M in human hepatocytes. Biochem Pharmacol. 2011, 82: 304-311. 10.1016/j.bcp.2011.04.017.
Schaeffeler E, Hellerbrand C, Nies AT, Winter S, Kruck S, Hofmann U, van der Kuip H, Zanger UM, Koepsell H, Schwab M: DNA methylation is associated with downregulation of the organic cation transporter OCT1 (SLC22A1) in human hepatocellular carcinoma. Genome Med. 2011, 3: 82-10.1186/gm298.
Borel F, Han R, Visser A, Petry H, van Deventer SJ, Jansen PL, Konstantinova P: Adenosine triphosphate-binding cassette transporter genes up-regulation in untreated hepatocellular carcinoma is mediated by cellular microRNAs. Hepatology. 2012, 55: 821-832. 10.1002/hep.24682.
Lang T, Klein K, Fischer J, Nussler AK, Neuhaus P, Hofmann U, Eichelbaum M, Schwab M, Zanger UM: Extensive genetic polymorphism in the human CYP2B6 gene with impact on expression and function in human liver. Pharmacogenetics. 2001, 11: 399-415. 10.1097/00008571-200107000-00004.
König J, Cui Y, Nies AT, Keppler D: A novel human organic anion transporting polypeptide localized to the basolateral hepatocyte membrane. Am J Physiol Gastrointest Liver Physiol. 2000, 278: G156-G164.
Ho RH, Tirona RG, Leake BF, Glaeser H, Lee W, Lemke CJ, Wang Y, Kim RB: Drug and bile acid transporters in rosuvastatin hepatic uptake: function, expression, and pharmacogenetics. Gastroenterology. 2006, 130: 1793-1806. 10.1053/j.gastro.2006.02.034.
Nies AT, Herrmann E, Brom M, Keppler D: Vectorial transport of the plant alkaloid berberine by double-transfected cells expressing the human organic cation transporter 1 (OCT1, SLC22A1) and the efflux pump MDR1 P-glycoprotein (ABCB1). Naunyn Schmiedebergs Arch Pharmacol. 2008, 376: 449-461. 10.1007/s00210-007-0219-x.
Kopplow K, Letschert K, König J, Walter B, Keppler D: Human hepatobiliary transport of organic anions analyzed by quadruple-transfected cells. Mol Pharmacol. 2005, 68: 1031-1038. 10.1124/mol.105.014605.
Nies AT, Hofmann U, Resch C, Schaeffeler E, Rius M, Schwab M: Proton pump inhibitors inhibit metformin uptake by organic cation uptake transporters (OCTs). PLoS ONE. 2011, 6: e22163-10.1371/journal.pone.0022163.
Locker J, Ghosh D, Luc PV, Zheng J: Definition and prediction of the full range of transcription factor binding sites--the hepatocyte nuclear factor 1 dimeric site. Nucleic Acids Res. 2002, 30: 3809-3817. 10.1093/nar/gkf484.
Tegude H, Schnabel A, Zanger UM, Klein K, Eichelbaum M, Burk O: Molecular mechanism of basal CYP3A4 regulation by hepatocyte nuclear factor 4alpha: evidence for direct regulation in the intestine. Drug Metab Dispos. 2007, 35: 946-954. 10.1124/dmd.106.013565.
Zukunft J, Lang T, Richter T, Hirsch-Ernst KI, Nussler AK, Klein K, Schwab M, Eichelbaum M, Zanger UM: A natural CYP2B6 TATA box polymorphism (-82T-- > C) leading to enhanced transcription and relocation of the transcriptional start site. Mol Pharmacol. 2005, 67: 1772-1782. 10.1124/mol.104.008086.
Geick A, Eichelbaum M, Burk O: Nuclear receptor response elements mediate induction of intestinal MDR1 by rifampin. J Biol Chem. 2001, 276: 14581-14587. 10.1074/jbc.M010173200.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR: A method and server for predicting damaging missense mutations. Nat Methods. 2010, 7: 248-249. 10.1038/nmeth0410-248.
PolyPhen-2. [http://genetics.bwh.harvard.edu/pph2]
Kumar P, Henikoff S, Ng PC: Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009, 4: 1073-1081. 10.1038/nprot.2009.86.
SIFT. [http://sift.jcvi.org]
Ferrer-Costa C, Orozco M, de lC X: Sequence-based prediction of pathological mutations. Proteins. 2004, 57: 811-819. 10.1002/prot.20252.
PMut. [http://mmb.pcb.ub.es/PMut]
Yue P, Melamud E, Moult J: SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006, 7: 166-10.1186/1471-2105-7-166.
SNPs3D. [http://www.snps3d.org]
Mathews DH, Sabina J, Zuker M, Turner DH: Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999, 288: 911-940. 10.1006/jmbi.1999.2700.
Zuker M: Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31: 3406-3415. 10.1093/nar/gkg595.
MFold. [http://mfold.rit.albany.edu/?q=mfold]
Tietz NW: Clinical Guide to Laboratory Tests. 1995, Philadelphia: WB Saunders
Benichou C: Criteria of drug-induced liver disorders. Report of an international consensus meeting. J Hepatol. 1990, 11: 272-276.
Ho WF, Koo SH, Yee JY, Lee EJ: Genetic variations of the SLCO1B1 gene in the Chinese, Malay and Indian populations of Singapore. Drug Metab Pharmacokinet. 2008, 23: 476-482. 10.2133/dmpk.23.476.
Schaeffeler E, Zanger UM, Eichelbaum M, Asante-Poku S, Shin JG, Schwab M: Highly multiplexed genotyping of thiopurine S-methyltransferase variants using MALDI-TOF mass spectrometry: reliable genotyping in different ethnic groups. Clin Chem. 2008, 54: 1637-1647. 10.1373/clinchem.2008.103457.
Tirona RG, Leake BF, Merino G, Kim RB: Polymorphisms in OATP-C: identification of multiple allelic variants associated with altered transport activity among European- and African-Americans. J Biol Chem. 2001, 276: 35669-35675. 10.1074/jbc.M103792200.
Acknowledgements
We gratefully acknowledge Dr Peter Fritz, chief pathologist at the Robert-Bosch-Hospital, for expert histological examination of all liver samples, Dr Kathrin Klein for providing selected SLCO microarray data and Rabea Riedel for help in generating OATP2B1 transfectants. We greatly appreciate the expert technical help of Silvia Hübner, Ursula Waldherr, Monika Elbl, Heidi Köhler, and Igor Liebermann. This work was supported by the Robert Bosch Foundation (Stuttgart, Germany), the Federal Ministry for Education and Research (BMBF, Berlin, Germany) grant 03 IS 2061C and 0315755, the FP7-grant PITN-GA-2009-238132, and the Sigrid Jusélius Foundation (Helsinki, Finland).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MS was responsible for study design and drafting the manuscript. ATN and ES were responsible for study design and drafting the manuscript, and collected, analyzed and interpreted data. MN collected, analyzed and interpreted data. OB analyzed and interpreted data. Statistical analysis was performed by SW. UMZ and BS provided samples and reagents and critically revised the manuscript for important intellectual content. All authors have read and approved the manuscript for publication.
Matthias Schwab and Elke Schaeffeler contributed equally to this work.
Electronic supplementary material
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Nies, A.T., Niemi, M., Burk, O. et al. Genetics is a major determinant of expression of the human hepatic uptake transporter OATP1B1, but not of OATP1B3 and OATP2B1. Genome Med 5, 1 (2013). https://doi.org/10.1186/gm405
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1186/gm405