Abstract
Diagnostic medical bacteriology is a conservative discipline. When busy house officers scribble 'M, C & S' on a form, they are requesting two techniques - microscopy and culture of microorganisms - that date back to the late 17th and late 19th century, respectively. The third technique - antibiotic susceptibility testing - has changed little in a half a century. Relatively few front-line diagnostic bacteriology laboratories have embraced molecular methods; on my own campus, the hospital's medical microbiology department does not even possess a thermal cycler!
And yet in the research arena, genome sequencing has transformed almost every corner of the biomedical sciences, including the study of bacterial pathogens Furthermore, over the past 5 years, high-throughput (or 'next-generation') sequencing technologies have delivered a step change in our ability to sequence microbial genomes [1]. Since arriving in the market place, these technologies have experienced sustained technical improvement, which, twinned with lively competition between alternative platforms, has placed sequencing in a state of 'permanent revolution'.
At last, it seems that genomics has come up with a game-changer, a killer app, a disruptive technology that even those long wedded to the Gram stain and the agar plate can no longer ignore. Does this mean we are on the brink of a revolution in diagnostic and public health microbiology, in which high-throughput sequencing usurps the traditional 'M, C & S', or will the discipline's innate conservatism stand firm for decades to come?
Similar content being viewed by others
Genomic epidemiology of bacteria: promise and potential pitfalls
Let's start with the positive. The signs are good that rapid, cost-effective whole-genome sequencing will replace most existing molecular typing methods within public health reference laboratories over the next 5 to 10 years [1]. In this setting, genome sequencing represents the ultimate epidemiological typing method - a universally applicable, digital, 'library' typing method, portable internationally and across time. Its key selling point is that it is far more informative than any other approach, capable of distinguishing strains that differ by as little as a millionth of a genome [2]. On top of that, it is clear that microbial genome sequencing can shed light on the evolution of virulence or the molecular basis of antimicrobial resistance [3, 4].
Proof-of-principle studies now exist as to the value and utility of such approaches in real-world, real-time situations, including pandemic influenza and the recent German Escherichia coli O104:H4 outbreak [4–6]. A heady combination of rapid sequencing, prompt data release and a willingness of scientists across the world to collaborate in crowd-sourced analyses has given birth to an exciting new paradigm of 'open-source genomics' or 'Public Health 2.0'.
However, caveats remain. Bacterial genomic epidemiology is currently overly reliant on the identification of single nucleotide polymorphisms (SNPs) in draft genome sequences, largely because of the historical limitations of short-read technologies. It remains an open question whether for every bacterial lineage, SNP calling across the whole genome will always prove more informative than probing variation in the highly dynamic repetitive regions sampled by existing typing methods. The adoption of single-molecule long-read approaches such as that offered by Pacific Biosciences may also help wean us off a dependence on SNPs and reveal more large-scale genomic changes [6].
Although billed as a one-size-fits-all approach, the comparability and reliability of draft genome analyses remain critically dependent on the sequencing technology and analytical pipelines that are used; a draft genome sequenced today in Europe on one platform may not be easily compared with a draft genome sequenced half a world away on a different instrument in a few years time. And how easy will it be to redeploy staff in heavily unionized public health laboratories employed to use traditional approaches and re-equip them for the era of whole-genome sequencing?
Another important lesson comes from the deliberate release of anthrax into the US postal system in 2001 and the investigation that followed. Solving this case relied primarily on detection of rare colonial morphotypes in culture; genome sequencing had only a subsidiary role [7]. Crucially, this incident highlights the potential for apparently clonal bacterial cultures to contain mixtures of closely related but distinct genotypes. Imagine the following scenario. Patients A and B both carry an identical mixed population of genotypes X and Y. From patient A's sample, you pick a single colony representing genotype X, whereas from patient B you propagate a colony from the Y genotype. In such a situation, you might draw erroneous conclusions as to the relationship between the two infections and chains of transmission between these and other patients. This also highlights the problem that up until now genomic epidemiology has relied on isolation of organisms in pure culture.
Towards a culture-independent approach
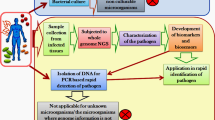
Can we progress to a culture-independent approach? Can we apply high-throughput sequencing not just to epidemiology, but also to the detection, and even discovery, of microbial pathogens? The answer is a qualified yes, with four approaches jostling for our attention.
Firstly, high-throughput sequencing has already breathed new life into well-established community-profiling approaches that exploit amplification of molecular bar codes, such as 16S ribosomal RNA gene sequences [8]. This is delivering ever more detailed surveys of the various human- and animal-associated microbiomes. However, such techniques often fail to distinguish pathogenic species or strains from their closest non-pathogenic relatives (such as Shigella from E. coli; Streptococcus pneumoniae from Streptococcus mitis; enterovirulent E. coli from commensal strains), so, in diagnostic terms, this approach is probably best seen as a way-station to more informative methods that will emerge in the future.
Secondly, we can use metagenomics for diagnostic purposes. This approach involves extracting and sequencing all the DNA from a sample. Clinical specimens will contain variable amounts of human DNA, which may create genetic privacy issues. Human DNA may also swamp the microbial DNA, although with extremely high depth of coverage, sufficient microbial DNA sequences could, at least theoretically, be recovered to reconstruct genomes. However, using current sequencing technologies, metagenomics is still a long way from providing genome-scale information for each member of a microbial community equivalent to that obtained from microorganisms isolated in pure culture. For this approach to come of age, we need a sequencing platform that combines speed and cost-effectiveness with very long read lengths and extremely high throughput - a plausible, but not certain, prospect for the coming decade.
Metatranscriptomics - extracting RNA and then creating and sequencing cDNA - provides a third way forward. This approach has already proven successful in the discovery of new viral pathogens and can also be used to investigate the activities of bacterial communities [9]. Human RNA will also be found in clinical samples sent for microbiological investigation. However, instead of dismissing this as mere contamination, analysis of human transcripts in such samples is likely to provide additional information about a patient's condition. For example, a transcriptional profile associated with inflammation might provide evidence of infection rather than just colonization at a given body site.
A fourth option is to abandon attempts to sequence nucleic acids isolated from whole populations, but instead to use cell sorting or allied techniques to isolate and genome-sequence sub-populations or, in extremis, single cells. Such approaches have proven successful in the research environment, but seem a remote prospect in the clinical laboratory [10]. How soon before we can obtain a chlamydial genome sequence from a urine sample or a mycobacterial genome sequence from a few millilitres of cerebrospinal fluid?
So, will high-throughput sequencing render culture-based approaches redundant? Here, we face another chorus of caveats, centred on the fact that it is not always possible to predict phenotype from genotype. In some cases - for example, sensitivity testing in tuberculosis - we already have a good evidence base on which to judge whether a given mutation is likely to lead to resistance. Similarly, detection of sequences encoding enzymes associated with virulence (such as Shiga toxin) or resistance (such as an extended-spectrum beta-lactamase) will have reasonable predictive value. But one has to remember that many differences in phenotype rely on subtle changes in expression, often of multiple genes, and that a single base-pair change in a promoter or coding sequence can ablate the function of a gene or its associated protein. Therefore, it would be foolish to imagine that sequencing will replace culture for every application in clinical bacteriology. Instead, just as radio survived the advent of TV, culture will remain a part of the discipline, but widespread adoption of sequence-based approaches might mean that it becomes restricted to a limited number of settings.
In conclusion, there are many rivers to cross before medical microbiology becomes simply a branch of genomic medicine, but perhaps the promised land is in sight.
Abbreviations
- MC & S:
-
microscopy, culture and sensitivity
- SNP:
-
single nucleotide polymorphism.
References
Pallen MJ, Loman NJ, Penn CW: High-throughput sequencing and clinical microbiology: progress, opportunities and challenges. Curr Opin Microbiol. 2010, 13: 625-631. 10.1016/j.mib.2010.08.003.
Lewis T, Loman NJ, Bingle L, Jumaa P, Weinstock GM, Mortiboy D, Pallen MJ: High-throughput whole-genome sequencing to dissect the epidemiology of Acinetobacter baumannii isolates from a hospital outbreak. J Hosp Infect. 2010, 75: 37-41. 10.1016/j.jhin.2010.01.012.
Hornsey M, Loman N, Wareham DW, Ellington MJ, Pallen MJ, Turton JF, Underwood A, Gaulton T, Thomas CP, Doumith M, Livermore DM, Woodford N: Whole-genome comparison of two Acinetobacter baumannii isolates from a single patient, where resistance developed during tigecycline therapy. J Antimicrob Chemother. 2011, 66: 1499-1503. 10.1093/jac/dkr168.
Rohde H, Qin J, Cui Y, Li D, Loman NJ, Hentschke M, Chen W, Pu F, Peng Y, Li J, Xi F, Li S, Li Y, Zhang Z, Yang X, Zhao M, Wang P, Guan Y, Cen Z, Zhao X, Christner M, Kobbe R, Loos S, Oh J, Yang L, Danchin A, Gao GF, Song Y, Li Y, Yang H, Wang J, Xu J, Pallen MJ, Wang J, Aepfelbacher M, Yang R: Open-source genomic analysis of shiga-toxin-producing E. coli O104:H4. N Engl J Med. 2011
Trifonov V, Khiabanian H, Greenbaum B, Rabadan R: The origin of the recent swine influenza A(H1N1) virus infecting humans. Euro Surveill. 2009, 14: 19193-
Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, Paxinos EE, Sebra R, Chin CS, Iliopoulos D, Klammer A, Peluso P, Lee L, Kislyuk AO, Bullard J, Kasarskis A, Wang S, Eid J, Rank D, Redman JC, Steyert SR, Frimodt-Moller J, Struve C, Petersen AM, Krogfelt KA, Nataro JP, Schadt EE, Waldor MK: Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med. 2011
Rasko DA, Worsham PL, Abshire TG, Stanley ST, Bannan JD, Wilson MR, Langham RJ, Decker RS, Jiang L, Read TD, Phillippy AM, Salzberg SL, Pop M, Van Ert MN, Kenefic LJ, Keim PS, Fraser-Liggett CM, Ravel J: Bacillus anthracis comparative genome analysis in support of the Amerithrax investigation. Proc Natl Acad Sci USA. 2011, 108: 5027-5032. 10.1073/pnas.1016657108.
Dethlefsen L, Huse S, Sogin ML, Relman DA: The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008, 6: e280-10.1371/journal.pbio.0060280.
Tang P, Chiu C: Metagenomics for the discovery of novel human viruses. Future Microbiol. 2010, 5: 177-189. 10.2217/fmb.09.120.
Ishoey T, Woyke T, Stepanauskas R, Novotny M, Lasken RS: Genomic sequencing of single microbial cells from environmental samples. Curr Opin Microbiol. 2008, 11: 198-204. 10.1016/j.mib.2008.05.006.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
MJP was the recipient of an Ion Torrent Personal Genome Machine and associated hardware and software in a recent Europe-wide competition.
Rights and permissions
About this article
Cite this article
Pallen, M.J., Loman, N.J. Are diagnostic and public health bacteriology ready to become branches of genomic medicine?. Genome Med 3, 53 (2011). https://doi.org/10.1186/gm269
Published:
DOI: https://doi.org/10.1186/gm269