Abstract
The telomeres that cap the ends of eukaryotic chromosomes serve a dual role in protecting the chromosome ends and in intracellular signaling for regulating cell proliferation. A complex of six telomere-associated proteins has been identified - the telosome or shelterin complex - that is crucial for both the maintenance of telomere structure and its signaling functions.
Similar content being viewed by others
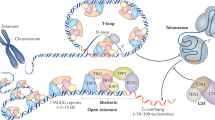
Telomeres are specialized structures at the ends of eukaryotic chromosomes that help to maintain genome integrity in eukaryotes by preventing chromosomal rearrangements or chromosomes fusing to each other, and by enabling complete replication of the ends of the linear DNA molecules. Telomeric DNA is composed of a series of sequence repeats and terminates in a 3' single-stranded (ss) DNA overhang. At each round of DNA replication the telomeric DNA becomes shorter, but it can be regenerated by the enzyme telomerase, an RNA-containing DNA polymerase. Both the double and single-stranded telomeric DNA is bound and protected by DNA-binding proteins that in turn associate with other signaling proteins/complexes to achieve telomere-end protection and length control. The length of telomeric DNA is maintained by the enzyme telomerase, but in addition, six telomere-associated proteins - TRF1, TRF2, POT1, RAP1, TIN2 and TPP1 in mammalian cells - have been shown to form a complex known as the telosome, or shelterin complex, that is essential for telomere function [1–10]. Here we will briefly review the composition of the telosome, its role in telomere maintenance, and its connections with intracellular signaling pathways.
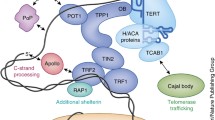
Telomere repeat factor-1 (TRF1) and -2 (TRF2) are related proteins that share a number of sequence and organizational similarities, and along with protection of telomeres-1 (POT1), they interact directly with telomeric DNA. RAP1 (the human homolog of the yeast telomeric protein Rap1), TRF1-interacting protein 2 (TIN2), and TPP1 (also known as TINT1/PTOP/PIP1) associate with these DNA-binding proteins to form the core telosome (Figure 1). Various signaling pathways originate from these core telomeric proteins and their subcomplexes, and from this it has been possible to deduce a telomere 'interactome' [11]. In this interactome, the telosome serves as the core building block, coordinating protein-protein interactions and protein complex cross-talk on the telomeres.
TRF1 and TRF2 and their interaction networks
TRF1 and TRF2 each bind telomeric double stranded (ds) DNA as homodimers, with dimerization mediated by the TRF-homology (TRFH) domain [3, 4, 12]. TRF1 homodimers are postulated to monitor telomere length, whereas TRF2 homodimers serve to stabilize telomeric loop (t-loop) formation and protect the telomere end (t-loops are structures that appear to form as a result of the 3' overhang invading the duplex telomeric repeats). TRF1 and TRF2 interactions with a number of proteins within the interactome have also been mapped to their respective TRFH domains [13]. TRF1 has a propensity for binding long tracts of dsDNA whereas TRF2 binds the ds/ssDNA junction [14]. Both TRF1 and TRF2 have carboxy-terminal Myb domains, which are essential for binding directly to telomere duplex DNA [3, 4].
Human TRF1 and TRF2 differ from each other at their amino terminus, which comprises an acidic region in TRF1 and a basic region in TRF2. The function of these regions is poorly understood, although recent studies suggest that the basic amino-terminal domain of TRF2 is important for binding of the ds/ssDNA junction and for the supercoiling of telomeric DNA, and may regulate the formation and stabilization of the t-loop structure [15–17]. Deletion of the basic region of TRF2 does not affect its targeting or binding to telomeres in vivo; the overexpression of this truncated protein does, however, lead to disruption of telomere end protection and the induction of cellular senescence and apop-tosis [18, 19]. Overexpression of a TRF2 construct lacking both the basic and the Myb domains leads to an increased occurrence of chromosomal fusions and interchromosomal bridging [20].
As illustrated in Figure 1, TRF1 and TRF2 also function as protein-interaction hubs within the telomere signaling network, interacting directly with the other members of the telosome and with a diverse array of proteins and protein complexes that are involved in the cell cycle and in DNA repair and recombination, to maintain telomere structure and length [2, 21–28]. TRF1 has been postulated to modulate the length of telomere repeats primarily via its interaction with the telosome proteins TIN2, TPP1 and POT1, and with PINX1, an inhibitor of telomerase [7, 9, 10, 29–37]. For example, the direct interaction of TRF1 with PINX1 provides a possible mechanism for how TRF1 could regulate telomere length [34]. PINX1 may be recruited to the telomeres through its interaction with TRF1 and negatively regulate telomere length by directly inhibiting telomerase.
TIN2 was identified on the basis of its ability to interact with TRF1 in yeast two-hybrid assays [30]. TIN2 is a key component of the telosome, and associates with both TRF1 and TRF2 [1, 38, 39]. TRF1-TIN2 interaction occurs through the TRFH domain and the TIN2 carboxy-terminal domain [13, 30]. TIN2 is a negative regulator of telomere length and is essential for bringing together the DNA-binding proteins within the telosome complex.
TPP1 interacts with both TIN2 and POT1, and is the link that connects the activities of the dsDNA-binding TRF1 to those of the ssDNA-binding POT1 [8–10]. POT1 binds ssDNA, regulates telomere length, and helps to stabilize the T-loop and protect the telomere end. TIN2 is the major TRF1-interacting protein, and TPP1 is the major POT1-interacting protein, so TPP1 links these two DNA-binding activities assembled on the telomeres. The TRF1-TIN2-TPP1-POT1 association illustrates an important path through which signals are communicated along a telomere. The functions of POT1 and TPP1 are discussed in more detail later.
In addition to the interactions described above, TRF1 can associate with tankyrase, a protein with poly(ADP-ribose) polymerase activity [33], end-binding protein 1 (EB1) [40], the nucleolar protein nucleostemin [41], and the F-box protein FBX4, which participates in protein ubiquitination [42] (Figure 1). Human EB1 is able to interact with and target the tumor suppressor protein adenomatous polyposis coli (APC) to microtubules in a cell-cycle-dependent manner. Tankyrase has been implicated in the control of spindle structure [43] and sister-chromatid cohesion [44], and thus through interactions with tankyrase and EB1, TRF1 could be involved in cell-cycle dependent regulation of telomere function. Levels of TRF1 protein can be controlled by tankyrase, FBX4 and nucleostemin [41, 42, 45]. TRF1 can be poly-ADP ribosylated by tankyrases [33], which may lead to its ubiquitination and subsequent degradation [45], while FBX4 is an E3 ligase specific for TRF1 ubiquitination via the Cul1-containing SCF complex [42], which leads to proteasomal TRF1 degradation. Nucleostemin enhances TRF1 degradation by a ubiquitination-independent pathway [41].
Both TRF1 and TRF2 can be sumoylated by the SUMO ligase MMS21, a component of the SMC5/6 complex, which is involved in DNA repair and recombination [46]. A number of human tumors and tumor cell lines have a telomerase-independent mechanism for telomere elongation that involves homologous recombination, and which is referred to as 'alternative lengthening of telomeres' (ALT) [47]. As demonstrated in cells that display ALT, sumoylation of TRF1 and TRF2 helps to promote the recruitment of telomeres to intranuclear macromolecular complexes called APBs (the equivalent of PML bodies in other cells) and promote telomere lengthening through homologous recombination. However, it remains to be determined whether TRF1 and TRF2 are similarly modified in other cell types.
TRF1 and TRF2 and DNA damage response pathways
Both TRF1 and TRF2 are intimately linked with DNA damage response pathways. The ss/dsDNA structure at the telomere could be perceived by the cell as DNA damage, and TRF1 and TRF2 appear to be part of the mechanism that prevents a damage response being generated. TRF1 co-immunoprecipitates with the protein kinase ATM (ataxia telangiectasia mutated), a sensor of DNA damage, and can be phosphorylated by ATM both in vivo and in vitro [48, 49]. Phosphorylation of TRF1 by ATM leads to impairment of TRF1's capacity to interact with DNA [49], and the expression of phosphorylationsite mutant TRF1 induces mitotic entry and apoptosis [40]. The MRN complex, functioning together with ATM, is also important for regulating TRF1 activity [49]. The MRN complex appears to be required for ATM-mediated phosphorylation of TRF1.
Numerous studies have demonstrated the essential role of TRF2 in telomere end protection. In addition to ATM, TRF2 also recruits a variety of other DNA damage-sensing and DNA repair proteins to the telomere, such as nucleases ERCC1/XPF [50] and Apollo [51, 52]; the DNA repair MRN complex [53, 54]; the helicases BLM [55] and WRN [55]; Ku70/Ku86 [54, 56], and poly-ADP ribose polymerases PARP1/2 [54, 57, 58] (Figure 1). The recruitment of these proteins presumably functions to prevent telomere ends being recognized as DNA breaks or to sensitize the cell to damage to the telomeres. It is equally possible that TRF2-associated complexes of 'damage proteins' are different in composition or modification state from the canonical complexes involved in repairing radiation-induced double-strand DNA breaks, given that the TRF2-based complexes normally do not evoke a cell-cycle checkpoint response [59, 60]. It should be noted that TRF2 has been shown to localize to sites of high-energy radiation-induced DNA damage outside the telomeres [61, 62]. Therefore, the association of TRF2 with DNA damage response proteins may have a role beyond telomere protection.
TRF2 mediates its protective function partly through heterodimerization with the telosome component RAP1, which contains a Myb domain [6]. In human cells, inhibition of RAP1 or dominant-negative expression of RAP1 truncation mutants led to elongated telomeres and loss of telomere heterogeneity [54, 63]. TRF2 has also recently been shown to associate with the origin replication protein ORC1 [64], which implicates the origin recognition complex (ORC) in facilitating telomere replication. Despite the critical role of the ORC complex in eukaryotic DNA replication, how it is recruited to origins of replication is poorly understood. Sequence-specific DNA-binding proteins or epigenetic factors may play a role. In this case, the specific interaction between TRF2 and subunits of the ORC complex point to a possible mechanism for targeting the ORC complex to the telomeres. However, whether TRF2 does have a role in telomere replication remains to be determined.
Recent studies suggest that the TRFH domains are the first modular domains identified in telomere proteins that can recognize linear peptide sequences [13]. And those findings have further solidified TRF1 and TRF2 as the major hubs within the telomere interactome. The TRFH domains of TRF1 and TRF2 display distinct specificities and affinities for their targets, suggesting a new avenue of research for probing the function of TRF1 and TRF2, and deciphering how players from diverse pathways are recruited to the telomeres.
POT1 and TPP1 and their functions
While the telosome forms a platform to which additional players can be recruited (Figure 1), complicated interactions are also at play within the protein complex itself [11]. TIN2 and TPP1 are critical to its assembly [65], and the ssDNA-binding protein POT1 serves as the effector of the complex in its role of maintaining telomere integrity. Both POT1 and TPP1 contain one or more oligonucleotide/oligosaccharide-binding folds (OB folds) [66–70]. Recent work has highlighted the evolutionary conservation in both structure and function among OB-fold-containing proteins participating in telomere maintenance and integrity, such as POT1 and TPP1, compared with those involved in DNA protection, such as the heterotrimeric replication protein A (RPA) complex [66, 67, 71–77].
Genetic studies in yeast, Tetrahymena, plants, humans and mice support an essential role for POT1 in maintaining telomere integrity [7, 78–82]. Unlike humans, mice contain two isoforms of POT1 - POT1a and POT1b [79, 80]. Recent work has shown the functional dichotomy of these two isoforms, and provided much-needed insight into the evolutionary divergence and conservation of POT1 homologs in different species [78–80, 83]. Conditional knockout studies of POT1a and POT1b suggest that both are needed for complete protection and maintenance of the telomeres [79, 80, 83]. While the two proteins have overlapping functions and each may compensate to some extent for the loss of the other, they are not interchangeable. In particular, POT1a is essential for suppressing DNA damage responses at telomere termini, whereas POT1b regulates the 3' ssDNA overhangs [79, 80, 83]. When the ends of chromosomes are not coated and protected by proteins, the telomeres (with or without the overhang) may be recognized as DNA damage, eliciting DNA damage response pathways. POT1b appears important for protecting the 3' overhangs from degrading nucleases. This functional difference may be achieved, in part, through the interaction of POT1a and POT1b with different sets of proteins in the telomere interactome. For example, one potential target of POT1b could be nuclease(s) that are involved in processing the 3' overhang [79].
Although both TRF2 and POT1 bind telomere DNA and are required for telomere capping, recent studies indicate that they regulate distinct signaling pathways [84, 85]. Loss of function of TRF2 in a number of mammalian cell types (tumor and primary cell lines), and in cells from conditional TRF2-knockout mice, elicits DNA damage responses mediated mainly through the ATM pathway, whereas POT1 knockout triggers the DNA damage response pathway initiated by the protein kinase ATR (ataxia telangiectasia related) [84]. These results are consistent with the telomere interactome map (Figure 1), where TRF2 interacts with the MRN complex and DNA-PK, proteins that mediate repair of double-strand breaks, with which ATM is preferentially associated [53, 54, 56, 86]. In addition, the repression of ATR activity by POT1 is probably a result of POT1 binding telomere ssDNA and inhibiting ATR activation by blocking access of the single-strand binding protein RPA, by which ATR is recruited, to the telomere [84, 87]. As shown in the interactome map, few proteins are known to bind directly to POT1. How POT1 signals through pathways other than the ATR pathway merits further investigation.
In the cilate Oxytricha nova, heterodimers of the OB-fold telomere end-binding proteins TEBP-α and TEBP-β are bound to the TTTTGGGG repeats of telomeric DNA. TEBP-α contains three OBfolds, two of which are involved in ssDNA recognition while the third interacts with TEBP-β [68]. Human POT1 is a homolog of ciliate TEBP-α. Although TPP1 lacks an obvious OB fold, careful biochemical, structural and molecular studies have revealed that it does indeed contain an OB-fold structure, and that it is a functional homolog of the ciliate TEBP-β[66, 67]. Whereas TPP1 exhibits little or no telomere ssDNA-binding activities in gel-shift experiments, a POT1-TPP1-DNA ternary complex can form in these assays. TPP1 has also been shown to enhance POT1 DNA-binding activity [66, 67], supporting the model that POT1 may interact with DNA in the form of a heterodimer with TPP1. The TPP1-POT1 heterodimer has been postulated to modulate telomerase access to the telomeres.
Inciliates, TEBP-β canalso promote G-quadruplex formation [88]. G-quadruplexes are tetrads of hydrogen-bonded guanine bases that can form in G-rich DNA and RNA sequences, and upon which higher-order structures can be built. Folding of telomere DNA into G-quadruplexes appears to inhibit telomerase access. This activity is unlikely to be conserved in TPP1, as TPP1 lacks the basic domain of TEBP-βthat is responsible for G-quadruplex-stimulatingactivity. In contrast, POT1 has been shown to inhibit G-quadruplex structure [89], suggesting evolutionary divergence in G-quadruplex control mechanisms. The core telomere proteins TIN2, TRF1 and TRF2 are not found in ciliates. These proteins seem to have evolved for telomere homeostasis in vertebrates, and may provide additional mechanisms for regulating telomere G-quadruplex formation.
Both TPP1 and POT1 are critical for regulating telomere length, and POT1 is the only telomere protein identified so far that binds to telomere ssDNA. TPP1 has been shown to be able to interact with telomerase both in vitro and in cells [66, 67] and its putative OB fold is required for telomerase recruitment. In addition to direct binding, the POT1-TPP1 complex appears to enhance the processivity of the telomerase component TERT in vitro [67]. Consistent with this finding, expressing a TPP1 mutant lacking the OB fold resulted in modest telomere shortening in human cells compared to parental cells or cells expressing full-length TPP1 [66]. Because TPP1 on its own does not bind ssDNA, this probably means that POT1 and TPP1 function together to recruit telomerase to telomeric ssDNA through the TPP1 OB fold, in addition to protecting telomere ends and negatively regulating telomerase access. Generally, telomeres only become accessible to the telomerase during the S phase of the cell cycle. This is achieved through multiple mechanisms, including regulation of telomerase expression and activity, sequestering of telomeres, and coating of telomeres with telomere-binding proteins such as POT1, which presumably serves to block telomerase access.
The realization that there are two classes of OB-fold-containing proteins with distinct functions has in turn helped to establish a unified model regarding the function of OB-fold-containing proteins in telomere overhang binding (Table 1) [66–70]. While much conservation exists between the various OB-fold-containing complexes, differences such as DNA-binding specificities, domain structures, and interaction partners help to set these proteins apart. From yeast to human, RPA-like or TEBP heteromultimeric complexes may have evolved for the more specialized function of ssDNA protection at the telomeres [66, 67, 71–77].
Compartmentalization of telomeric protein complexes
Structural, temporal and developmental variation greatly impact on the assembly and disassembly of the various sub-complexes that make up the dynamic telomere interactome. While numerous studies have been carried out to elucidate protein-protein interactions and telomere localizations of multiple factors within the interactome (for example, TRF1, TIN2 and TRF2), surprisingly little is known regarding the subcellular localization and regulated targeting of core telomere proteins.
Proteins of the telosome have been found in cellular locations other than the telomeres. For example, TRF2 and RAP1 have been shown to associate with the Epstein-Barr virus origin of replication [90], and TRF2 can be recruited to intra-satellite double-strand breaks when the damage level is high [91]. The growth status of human cells may influence the localization of TIN2 [92]. In growth-arrested epithelial cells, TIN2 was found to migrate into non-telomeric domains that contained the protein HP1, a marker of heterochromatin. It is possible that different complexes may form under these different conditions.
Recent studies have indicated for the first time the importance of nuclear export and spatial control of telomeric proteins in telomere maintenance in mammalian cells, as endogenous TIN2, TPP1 and POT1 have been found to localize in both the cytoplasm and the nucleus [93]. In addition, as determined by bimolecular fluorescence complementation assays [93, 94], different pairs of telomeric proteins appear to interact with each other in different cellular compartments. Whereas TIN2-TRF2 interaction takes place exclusively in the nucleus (including at telomeres), TIN2-TPP1 and TPP1-POT1 interactions occur in both the cytoplasm and nucleus. These results suggested telomere protein subcomplex formation in the cytoplasm. Interestingly, a nuclear export signal (NES) has been identified on TPP1 that directly controls the amount of TPP1 and POT1 in the nucleus. This NES resides next to the POT1-recruitment domain on TPP1, raising the possibility that interaction and nuclear localization of the TPP1-POT1 complex may be linked.
Binding of TIN2 to TPP1 promotes nuclear localization of TPP1 and POT1, by a mechanism yet to be determined [93]. The finding that TIN2 promotes nuclear retention of TPP1 and POT1 suggests that TIN2 plays a dual role in telosome assembly. While acting as a molecular tether for telosome subunits, TIN2 also ensures nuclear targeting and assembly of the entire complex. It would be of great interest to determine whether there exist other signaling pathways that control the nuclear import and export of telomeric complexes. Unexpectedly, disrupting TPP1 nuclear export can result in telomeric DNA damage response and telomere length disregulation [93]. This underlines the importance of spatial control of telomeric complexes, such that too much TPP1 in the nucleus may be detrimental to cells, and TPP1 nuclear export may regulate the concentration of TPP1-POT1 in the nucleus. These findings suggest that coordinated interactions among TIN2, TPP1 and POT1 in the cytoplasm could regulate the assembly and function of the telosome in the nucleus.
References
Liu D, O'Connor MS, Qin J, Songyang Z: Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem. 2004, 279: 51338-51342. 10.1074/jbc.M409293200.
de Lange T: Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005, 19: 2100-2110. 10.1101/gad.1346005.
Broccoli D, Smogorzewska A, Chong L, de Lange T: Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet. 1997, 17: 231-235. 10.1038/ng1097-231.
Bilaud T, Brun C, Ancelin K, Koering CE, Laroche T, Gilson E: Telomeric localization of TRF2, a novel human telobox protein. Nat Genet. 1997, 17: 236-239. 10.1038/ng1097-236.
Shen M, Haggblom C, Vogt M, Hunter T, Lu KP: Characterization and cell cycle regulation of the related human telomeric proteins Pin2 and TRF1 suggest a role in mitosis. Proc Natl Acad Sci USA. 1997, 94: 13618-13623. 10.1073/pnas.94.25.13618.
Li B, Oestreich S, de Lange T: Identification of human Rap1: implications for telomere evolution. Cell. 2000, 101: 471-483. 10.1016/S0092-8674(00)80858-2.
Baumann P, Cech TR: Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001, 292: 1171-1175. 10.1126/science.1060036.
Houghtaling BR, Cuttonaro L, Chang W, Smith S: A Dynamic Molecular Link between the Telomere Length Regulator TRF1 and the Chromosome End Protector TRF2. Curr Biol. 2004, 14: 1621-1631. 10.1016/j.cub.2004.08.052.
Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T: POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004, 18: 1649-1654. 10.1101/gad.1215404.
Liu D, Safari A, O'Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z: PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol. 2004, 6: 673-680. 10.1038/ncb1142.
Songyang Z, Liu D: Inside the mammalian telomere interactome: regulation and regulatory activities of telomeres. Crit Rev Eukaryot Gene Expr. 2006, 16: 103-118.
Fairall L, Chapman L, Moss H, de Lange T, Rhodes D: Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol Cell. 2001, 8: 351-361. 10.1016/S1097-2765(01)00321-5.
Chen Y, Yang Y, van Overbeek M, Donigian JR, Baciu P, de Lange T, Lei M: A Shared Docking Motif in TRF1 and TRF2 Used for Differential Recruitment of Telomeric Proteins. Science. 2008, 319: 1092-1096. 10.1126/science.1151804.
Stansel RM, de Lange T, Griffith JD: T-loop assembly in vitro involves binding of TRF2 near the 3' telomeric overhang. EMBO J. 2001, 20: 5532-5540. 10.1093/emboj/20.19.5532.
Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T: Mammalian telomeres end in a large duplex loop. Cell. 1999, 97: 503-514. 10.1016/S0092-8674(00)80760-6.
Fouche N, Cesare AJ, Willcox S, Ozgur S, Compton SA, Griffith JD: The basic domain of TRF2 directs binding to DNA junctions irrespective of the presence of TTAGGG repeats. J Biol Chem. 2006, 281: 37486-37495. 10.1074/jbc.M608778200.
Amiard S, Doudeau M, Pinte S, Poulet A, Lenain C, Faivre-Moskalenko C, Angelov D, Hug N, Vindigni A, Bouvet P, Paoletti J, Gilson E, Giraud-Panis MJ: A topological mechanism for TRF2-enhanced strand invasion. Nat Struct Mol Biol. 2007, 14: 147-154. 10.1038/nsmb1192.
Wang RC, Smogorzewska A, de Lange T: Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004, 119: 355-368. 10.1016/j.cell.2004.10.011.
Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T: p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999, 283: 1321-1325. 10.1126/science.283.5406.1321.
van Steensel B, Smogorzewska A, de Lange T: TRF2 protects human telomeres from end-to-end fusions. Cell. 1998, 92: 401-413. 10.1016/S0092-8674(00)80932-0.
Blackburn EH: Switching and signaling at the telomere. Cell. 2001, 106: 661-673. 10.1016/S0092-8674(01)00492-5.
Wright WE, Shay JW: Cellular senescence as a tumor-protection mechanism: the essential role of counting. Curr Opin Genet Dev. 2001, 11: 98-103. 10.1016/S0959-437X(00)00163-5.
Maser RS, DePinho RA: Connecting chromosomes, crisis, and cancer. Science. 2002, 297: 565-569. 10.1126/science.297.5581.565.
Kim SH, Kaminker P, Campisi J: Telomeres, aging and cancer: in search of a happy ending. Oncogene. 2002, 21: 503-511. 10.1038/sj.onc.1205077.
Baumann P: Are mouse telomeres going to pot?. Cell. 2006, 126: 33-36. 10.1016/j.cell.2006.06.027.
Blasco MA: The epigenetic regulation of mammalian telomeres. Nat Rev Genet. 2007, 8: 299-309. 10.1038/nrg2047.
Verdun RE, Karlseder J: Replication and protection of telomeres. Nature. 2007, 447: 924-931. 10.1038/nature05976.
Longhese MP: DNA damage response at functional and dysfunctional telomeres. Genes Dev. 2008, 22: 125-140. 10.1101/gad.1626908.
van Steensel B, de Lange T: Control of telomere length by the human telomeric protein TRF1. Nature. 1997, 385: 740-743. 10.1038/385740a0.
Kim SH, Kaminker P, Campisi J: TIN2, a new regulator of telomere length in human cells. Nat Genet. 1999, 23: 405-412. 10.1038/70508.
Smith S, de Lange T: Tankyrase promotes telomere elongation in human cells. Curr Biol. 2000, 10: 1299-1302. 10.1016/S0960-9822(00)00752-1.
Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T: Control of human telomere length by TRF1 and TRF2. Mol Cell Biol. 2000, 20: 1659-1668. 10.1128/MCB.20.5.1659-1668.2000.
Smith S, Giriat I, Schmitt A, de Lange T: Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998, 282: 1484-1487. 10.1126/science.282.5393.1484.
Zhou XZ, Lu KP: The Pin2/TRF1-interacting protein PinX1 is a potent telomerase inhibitor. Cell. 2001, 107: 347-359. 10.1016/S0092-8674(01)00538-4.
Cong YS, Wright WE, Shay JW: Human telomerase and its regulation. Microbiol Mol Biol Rev. 2002, 66: 407-425. 10.1128/MMBR.66.3.407-425.2002.
Loayza D, De Lange T: POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003, 424: 1013-1018. 10.1038/nature01688.
Lillard-Wetherell K, Machwe A, Langland GT, Combs KA, Behbehani GK, Schonberg SA, German J, Turchi JJ, Orren DK, Groden J: Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum Mol Genet. 2004, 13: 1919-1932. 10.1093/hmg/ddh193.
Kim SH, Beausejour C, Davalos AR, Kaminker P, Heo SJ, Campisi J: TIN2 mediates functions of TRF2 at human telomeres. J Biol Chem. 2004, 279: 43799-43804. 10.1074/jbc.M408650200.
Ye JZ, Donigian JR, Van Overbeek M, Loayza D, Luo Y, Krutchinsky AN, Chait BT, De Lange T: TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J Biol Chem. 2004, 279: 47264-47271. 10.1074/jbc.M409047200.
Nakamura M, Zhou XZ, Kishi S, Lu KP: Involvement of the telomeric protein Pin2/TRF1 in the regulation of the mitotic spindle. FEBS Lett. 2002, 514: 193-198. 10.1016/S0014-5793(02)02363-3.
Zhu Q, Yasumoto H, Tsai RY: Nucleostemin Delays Cellular Senescence and Negatively Regulates TRF1 Protein Stability. Mol Cell Biol. 2006, 26: 9279-9290. 10.1128/MCB.00724-06.
Lee TH, Perrem K, Harper JW, Lu KP, Zhou XZ: The F-box protein FBX4 targets PIN2/TRF1 for ubiquitin-mediated degradation and regulates telomere maintenance. J Biol Chem. 2006, 281: 759-768. 10.1074/jbc.M509855200.
Chang P, Coughlin M, Mitchison TJ: Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat Cell Biol. 2005, 7: 1133-1139. 10.1038/ncb1322.
Dynek JN, Smith S: Resolution of sister telomere association is required for progression through mitosis. Science. 2004, 304: 97-100. 10.1126/science.1094754.
Chang W, Dynek JN, Smith S: TRF1 is degraded by ubiquitin-mediated proteolysis after release from telomeres. Genes Dev. 2003, 17: 1328-1333. 10.1101/gad.1077103.
Potts PR, Yu H: The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat Struct Mol Biol. 2007, 14: 581-590. 10.1038/nsmb1259.
Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR: Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997, 3: 1271-1274. 10.1038/nm1197-1271.
Kishi S, Zhou XZ, Ziv Y, Khoo C, Hill DE, Shiloh Y, Lu KP: Telomeric protein Pin2/TRF1 as an important ATM target in response to double strand DNA breaks. J Biol Chem. 2001, 276: 29282-29291. 10.1074/jbc.M011534200.
Wu Y, Xiao S, Zhu XD: MRE11-RAD50-NBS1 and ATM function as co-mediators of TRF1 in telomere length control. Nat Struct Mol Biol. 2007, 14: 832-840. 10.1038/nsmb1286.
Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T: ERCC1/XPF removes the 3' overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol Cell. 2003, 12: 1489-1498. 10.1016/S1097-2765(03)00478-7.
van Overbeek M, de Lange T: Apollo, an Artemis-related nuclease, interacts with TRF2 and protects human telomeres in S phase. Curr Biol. 2006, 16: 1295-1302. 10.1016/j.cub.2006.05.022.
Lenain C, Bauwens S, Amiard S, Brunori M, Giraud-Panis MJ, Gilson E: The Apollo 5' exonuclease functions together with TRF2 to protect telomeres from DNA repair. Curr Biol. 2006, 16: 1303-1310. 10.1016/j.cub.2006.05.021.
Zhu XD, Kuster B, Mann M, Petrini JH, de Lange T: Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat Genet. 2000, 25: 347-352. 10.1038/77139.
O'Connor MS, Safari A, Liu D, Qin J, Songyang Z: The human Rap1 protein complex and modulation of telomere length. J Biol Chem. 2004, 279: 28585-28591. 10.1074/jbc.M312913200.
Opresko PL, von Kobbe C, Laine JP, Harrigan J, Hickson ID, Bohr VA: Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J Biol Chem. 2002, 277: 41110-41119. 10.1074/jbc.M205396200.
Song K, Jung D, Jung Y, Lee SG, Lee I: Interaction of human Ku70 with TRF2. FEBS Lett. 2000, 481: 81-85. 10.1016/S0014-5793(00)01958-X.
Dantzer F, Giraud-Panis MJ, Jaco I, Amé JC, Schultz I, Blasco M, Koering CE, Gilson E, Ménissier-de Murcia J, de Murcia G, Schreiber V: Functional interaction between poly(ADP-Ribose) polymerase 2 (PARP-2) and TRF2: PARP activity negatively regulates TRF2. Mol Cell Biol. 2004, 24: 1595-1607. 10.1128/MCB.24.4.1595-1607.2004.
Gomez M, Wu J, Schreiber V, Dunlap J, Dantzer F, Wang Y, Liu Y: PARP1 Is a TRF2-associated Poly(ADP-Ribose)Polymerase and Protects Eroded Telomeres. Mol Biol Cell. 2006, 17: 1686-1696. 10.1091/mbc.E05-07-0672.
Verdun RE, Crabbe L, Haggblom C, Karlseder J: Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol Cell. 2005, 20: 551-561. 10.1016/j.molcel.2005.09.024.
Verdun RE, Karlseder J: The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006, 127: 709-720. 10.1016/j.cell.2006.09.034.
Bradshaw PS, Stavropoulos DJ, Meyn MS: Human telomeric protein TRF2 associates with genomic double-strand breaks as an early response to DNA damage. Nat Genet. 2005, 37: 193-197. 10.1038/ng1506.
Williams ES, Stap J, Essers J, Ponnaiya B, Luijsterburg MS, Krawczyk PM, Ullrich RL, Aten JA, Bailey SM: DNA double-strand breaks are not sufficient to initiate recruitment of TRF2. Nat Genet. 2007, 39: 696-698. 10.1038/ng0607-696. author reply 698-699.
Li B, de Lange T: Rap1 affects the length and heterogeneity of human telomeres. Mol Biol Cell. 2003, 14: 5060-5068. 10.1091/mbc.E03-06-0403.
Atanasiu C, Deng Z, Wiedmer A, Norseen J, Lieberman PM: ORC binding to TRF2 stimulates OriP replication. EMBO Rep. 2006, 7: 716-721. 10.1038/sj.embor.7400730.
O'Connor MS, Safari A, Xin H, Liu D, Songyang Z: A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc Natl Acad Sci USA. 2006, 103: 11874-11879. 10.1073/pnas.0605303103.
Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z: TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007, 445: 559-562. 10.1038/nature05469.
Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M: The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007, 445: 506-510. 10.1038/nature05454.
Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC: Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998, 95: 963-974. 10.1016/S0092-8674(00)81720-1.
Lei M, Podell ER, Baumann P, Cech TR: DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature. 2003, 426: 198-203. 10.1038/nature02092.
Lei M, Podell ER, Cech TR: Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004, 11: 1223-1229. 10.1038/nsmb867.
Martin V, Du LL, Rozenzhak S, Russell P: Protection of telomeres by a conserved Stn1-Ten1 complex. Proc Natl Acad Sci USA. 2007, 104: 14038-14043. 10.1073/pnas.0705497104.
Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V: RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol. 2007, 14: 208-214. 10.1038/nsmb1205.
Bochkarev A, Bochkareva E: From RPA to BRCA2: lessons from single-stranded DNA binding by the OB-fold. Curr Opin Struct Biol. 2004, 14: 36-42. 10.1016/j.sbi.2004.01.001.
Iftode C, Daniely Y, Borowiec JA: Replication protein A (RPA): the eukaryotic SSB. Crit Rev Biochem Mol Biol. 1999, 34: 141-180. 10.1080/10409239991209255.
Miyoshi T, Kanoh J, Saito M, Ishikawa F: Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science. 2008, 320: 1341-1344. 10.1126/science.1154819.
Lee J, Mandell EK, Tucey TM, Morris DK, Lundblad V: The Est3 protein associates with yeast telomerase through an OB-fold domain. Nat Struct Mol Biol. 2008, 15: 990-997. 10.1038/nsmb.1472.
Young Yu E, Wang F, Lei M, Lue NF: A proposed OB-fold with a protein-interaction surface in Candida albicans telomerase protein Est3. Nat Struct Mol Biol. 2008, 15: 985-989. 10.1038/nsmb.1471.
Shakirov EV, Surovtseva YV, Osbun N, Shippen DE: The Arabidopsis Pot1 and Pot2 proteins function in telomere length homeostasis and chromosome end protection. Mol Cell Biol. 2005, 25: 7725-7733. 10.1128/MCB.25.17.7725-7733.2005.
Hockemeyer D, Daniels JP, Takai H, de Lange T: Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell. 2006, 126: 63-77. 10.1016/j.cell.2006.04.044.
Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, Deng Y, Behringer RR, Chang S: Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006, 126: 49-62. 10.1016/j.cell.2006.05.037.
Churikov D, Wei C, Price CM: Vertebrate POT1 restricts G-overhang length and prevents activation of a telomeric DNA damage checkpoint but is dispensable for overhang protection. Mol Cell Biol. 2006, 26: 6971-6982. 10.1128/MCB.01011-06.
Jacob NK, Lescasse R, Linger BR, Price CM: Tetrahymena POT1a regulates telomere length and prevents activation of a cell cycle checkpoint. Mol Cell Biol. 2007, 27: 1592-1601. 10.1128/MCB.01975-06.
He H, Multani AS, Cosme-Blanco W, Tahara H, Ma J, Pathak S, Deng Y, Chang S: POT1b protects telomeres from end-to-end chromosomal fusions and aberrant homologous recombination. EMBO J. 2006, 25: 5180-5190. 10.1038/sj.emboj.7601294.
Denchi EL, de Lange T: Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007, 448: 1068-1071. 10.1038/nature06065.
Guo X, Deng Y, Lin Y, Cosme-Blanco W, Chan S, He H, Yuan G, Brown EJ, Chang S: Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. EMBO J. 2007, 26: 4709-4719. 10.1038/sj.emboj.7601893.
Zhou BB, Elledge SJ: The DNA damage response: putting checkpoints in perspective. Nature. 2000, 408: 433-439. 10.1038/35044005.
Zou L, Elledge SJ: Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003, 300: 1542-1548. 10.1126/science.1083430.
Fang G, Cech TR: The beta subunit of Oxytricha telomere-binding protein promotes G-quartet formation by telomeric DNA. Cell. 1993, 74: 875-885. 10.1016/0092-8674(93)90467-5.
Zaug AJ, Podell ER, Cech TR: Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci USA. 2005, 102: 10864-10869. 10.1073/pnas.0504744102.
Deng Z, Atanasiu C, Burg JS, Broccoli D, Lieberman PM: Telomere repeat binding factors TRF1, TRF2, and hRAP1 modulate replication of Epstein-Barr virus OriP. J Virol. 2003, 77: 11992-12001. 10.1128/JVI.77.22.11992-12001.2003.
Bradshaw PS, Stavropoulos DJ, Meyn MS: Human telomeric protein TRF2 associates with genomic double-strand breaks as an early response to DNA damage. Nat Genet. 2005, 37: 193-197. 10.1038/ng1506.
Kaminker P, Plachot C, Kim SH, Chung P, Crippen D, Petersen OW, Bissell MJ, Campisi J, Lelievre SA: Higher-order nuclear organization in growth arrest of human mammary epithelial cells: a novel role for telomere-associated protein TIN2. J Cell Sci. 2005, 118: 1321-1330. 10.1242/jcs.01709.
Chen LY, Liu D, Songyang Z: Telomere maintenance through spatial control of telomeric proteins. Mol Cell Biol. 2007, 27: 5898-5909. 10.1128/MCB.00603-07.
Hu CD, Kerppola TK: Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat Biotechnol. 2003, 21: 539-545. 10.1038/nbt816.
Acknowledgements
This work is supported by NIH grants CA84208 and GM69572 and the Welch Foundation. DL is supported in part by the American Heart Association. ZS is a Leukemia and Lymphoma Society Scholar.
Author information
Authors and Affiliations
Corresponding author
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
About this article
Cite this article
Xin, H., Liu, D. & Songyang, Z. The telosome/shelterin complex and its functions. Genome Biol 9, 232 (2008). https://doi.org/10.1186/gb-2008-9-9-232
Published:
DOI: https://doi.org/10.1186/gb-2008-9-9-232