Abstract
Recent proteomic studies in Saccharomyces cerevisiae have identified nearly 200 proteins, other than the structural ribosomal proteins, that participate in the assembly of ribosomal subunits and their transport from the nucleus. In a separate line of research, proteomic studies of mature plant ribosomes have revealed considerable variability in the protein composition of individual ribosomes.
Similar content being viewed by others
The synthesis of ribosomes is a major metabolic task in eukaryotic cells. Transcription of all the genes involved requires the coordinated activities of all three RNA polymerases and consumes more than half of the cellular resources allocated to transcription [1]. Although eukaryotic ribosomes are composed of only four ribosomal RNAs (rRNAs) and around 80 ribosomal proteins, many other proteins are recruited to help deliver ribosomal subunits to the cytoplasm - at the rate of 2,000 or so ribosomes each minute in a growing yeast cell, for example [2, 3]. In the past five years, the extensive use of tandem affinity purification (TAP) of tagged proteins [4] has provided a detailed inventory of nearly 200 auxiliary proteins associated with pre-ribosomal particles [5–7]. These auxiliary proteins include RNases, RNA-modification and - remodeling enzymes, transport factors, and many others whose function is unclear at present. In addition, the protein content of eukaryotic ribosomes has been determined in several proteomics studies, revealing an unexpected variability from ribosome to ribosome that stems from the presence of ribosomal protein isoforms and their post-translational modifications [8, 9].
Regulation of ribosome synthesis at multiple levels
In eukaryotes, a polycistronic 35S pre-rRNA is transcribed in the nucleolus and cleaved into precursors (pre-rRNAs) of mature 18S and 5.8S rRNAs, as well as 25S or 28S rRNAs in yeast and higher eukaryotes, respectively [3]. These pre-rRNAs are subject to covalent nucleotide modifications before they assemble with around 80 ribosomal proteins and the independently transcribed 5S rRNA. Given the demand for equimolar amounts of rRNA and ribosomal proteins during ribosome synthesis, it is essential that the transcription of rRNAs and of the mRNAs for ribosomal proteins is coordinated [1]. Over the past few years, high-throughput experiments have provided evidence that the transcription of the auxiliary proteins involved in ribosome synthesis are also co-regulated. A network of transcription factors has been identified that collectively regulates the expression of rRNA, ribosomal protein genes and trans-acting ribosome biosynthesis factors (so-called ribi factors) [10–13].
Because of the extremely high energy cost of ribosome synthesis for the cell, the various activities are coordinated spatio-temporally for efficiency. A recent further proof of such coordination is the finding that rRNA transcription and rRNA processing are coordinated through a subset of proteins shared by the two processes [14]. In addition, a recent electron microscopy study has shown that 40S-subunit processing proteins associate with and compact the rRNA within seconds of completion of rRNA transcription [15]. These findings confirm the existence of a fine-tuned molecular assembly line where tasks are performed sequentially and without intervening delays.
Surprises in the ribosome maturation pathway
Classic work in the early 1970s identified a large 90S pre-ribosome, which is eventually converted into the precursors of the 40S and 60S subunits. It was also shown that disruptions of either the large or small subunit synthesis pathway do not necessarily impact on the cytoplasmic export of the unaffected subunit [3]. Until 2001, however, most of our knowledge about auxiliary ribosome synthesis factors was based on genetic studies and biochemical experiments, providing what turned out to be a limited picture of the ribosome maturation pathway (for reviews see [2, 3]).
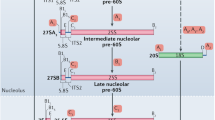
The introduction of TAP techniques and the tagging of proteins known to be involved in ribosome synthesis revolutionized the field by capturing multiple snapshots of this complex process [16–22]. Many additional proteins were placed in newly defined ribosome-assembly maps, including putative enzymes other than the expected nucleases [5, 6]. Although most of the work was done on budding yeast, because of the relative ease of genetic and biochemical studies in this organism, it is safe to assume that most of the observations also apply to higher eukaryotes, as nearly all the proteins involved in ribosome assembly are conserved between yeast and human. The U3 small nucleolar RNA (snoRNA), which is needed for the initial cleavage of nascent rRNA, was found in complexes with the 35S pre-rRNA and more than 30 essential trans-acting proteins [19, 21]. These complexes, probably representing subsets of the early 90S pre-ribosomal particle, included mostly 40S-subunit processing factors and were almost completely devoid of the 60S ribosomal proteins and trans-acting factors [21]. In contrast, separately characterized pre-60S complexes contained precursors of 5.8S and 25S rRNAs but little 35S pre-rRNA, and lacked all known 40S processing factors [17, 20]. Although this striking demarcation between the precursors of the 40S and 60S particles was unexpected, the fact that the large and small ribosomal subunits are synthesized independently fits with early experimental data [3]. According to the current model of ribosome assembly, the processing factors involved in 40S-subunit synthesis assemble co-transcriptionally onto the 35S pre-rRNA as soon as the future 18S rRNA, located towards the 5' end of the 35S rRNA, is transcribed [6, 7, 14]. The 60S-subunit processing machinery is recruited later, after the release of the 40S precursor from the 90S particle and the completion of rRNA transcription (Figure 1). Despite this clear division between the assembly of the 40S and 60S subunits, there are a few processing factors, for example, Rrp5p and the export protein Rrp12p [3, 23], that function in the processing pathways of both subunits.
The ribosome synthesis pathway in eukaryotes. The initial stages of ribosome synthesis take place in the nucleolus. The first step is the association of newly synthesized 35S rRNA with 40S processing proteins and 40S ribosomal proteins which form a complex with the future 18S rRNA sequence even before the transcript is completed (the co-transcriptional assembly stage). After completion of rRNA transcription, the 35S rRNA and its associated proteins form a 90S pre-ribosome particle which contains numerous 40S processing factors and 40S ribosomal proteins but very few 60S processing proteins or 60S ribosomal proteins. RNA cleavage releases U3 snoRNP and separates the 90S particle into 40S and 60S pre-ribosome particles. The latter recruits 60S processing proteins and 60S ribosomal proteins, and the separate pre-ribosome complexes are exported out of the nucleolus into the nucleus and cytoplasm. Most of our knowledge about this pathway has been compiled from studies in budding yeast; see text for further details.
Heterogeneity in mature ribosomes
The experimental task of determining the protein composition of ribosomes is perfectly suited to a proteomics approach. Ribosomes are naturally produced in large amounts and are reasonably stable after maturation; by contrast, pre-ribosome complexes are more dynamic and harder to define precisely. Over the past decade, considerable progress has been made in the characterization of cytosolic ribosomes, and the protein composition of ribosomes has been studied in yeast [24, 25], rats [26] and humans [27]. These studies benefitted from the fact that systematic analyses of eukaryotic gene sequences had identified around 80 conserved ribosomal proteins, making it possible to predict the protein content of ribosomes from various species even before they are experimentally characterized.
As a general rule, ribosomal protein genes are present in eukaryotes as multiple, non-identical copies, which include pseudogenes. In Arabidopsis thaliana, for example, sequence analyses identified 249 genes and 19 pseudogenes for ribosomal proteins, the majority of which appear to be expressed, judging by the analysis of expressed sequence tags (ESTs) [28]. This abundance of ribosomal proteins and their EST variations suggested a heterogeneity in the protein content of the resulting plant ribosomes. Two recent experimental studies have confirmed this prediction and provided further insights into the molecular diversity of eukaryotic ribosomes [8, 9]. Working independently, two research groups purified cytosolic ribosomes from A. thaliana and subjected them to two-dimensional gel electrophoresis and mass spectrometry [8, 9]. A common conclusion from both studies was that roughly half of the ribosomal proteins were found in two or more spots on the gel. This result was expected, given that most ribosomal protein genes in the A. thaliana genome are duplicated and encode three or four variants that would give rise to unique tryptic peptides [28]. A similar trend was also observed in other eukaryotic ribosomes [25–27]. Strikingly, for about 25% of ribosomal protein families, the same gene product was found in multiple spots, indicating post-translational modifications that changed the protein's mass or apparent charge [8, 9]. Although protein degradation could in principle be the reason for this discrepancy, it is unlikely that it would selectively affect only some families of ribosomal proteins. Instead, most of these differences are likely to be the result of specific covalent modifications such as phosphorylation, methylation and amino-terminal acetylation. In one of the studies, Chang et al. [8] confirmed directly that ribosomal protein S6 can be decorated with between one and four phosphates at Ser238 and Ser241. Previous studies have already shown that differential phosphorylation of S6 has a regulatory role [29]. The role of other covalent modifications is less clear, however. There are some indications that ribosome heterogeneity may be linked to specific growth conditions, tissue types or developmental stages [28], and the findings reported in these two papers provide ample material for future studies.
In addition to ribosomal proteins, Chang et al. [8] and Giavalisco et al. [9] identified a small number of non-ribosomal proteins that were relatively stably associated with 80S ribosomes. Although one would expect similar results from both groups when it comes to ribosomal proteins, given the same overall approach, it is comforting that their results overlapped for non-ribosomal proteins as well. In particular, both groups identified a protein with WD-repeat domain, RACK1, homologs of which are found in all eukaryotes but not in bacteria. The association of RACK1 with the 40S subunit has already been observed in yeast and human ribosomes [24], indicating that its function is conserved across the eukaryotic kingdom. A recent cryo-electron microscopy study has placed RACK1 on the head region of the 40S subunit, next to the mRNA exit channel [30]; this location exposes the large surface of the WD-repeats for potential interactions with other proteins that are recruited to the ribosome. Finally, both studies reiterated the known limitations of the approach that combines purification by two-dimensional gel electrophoresis with mass spectrometry. Out of an expected 79 ribosomal proteins, Chang et al. [8] did not detect five and Giavalisco et al. [9] did not detect 19. Most of the missing proteins are of low molecular weight and were predicted to produce a small number of useful peptides after digestion with trypsin. A similar problem has already been observed with the 80S ribosomes of yeast [24]. In addition, the effective separation of ribosomal proteins on two-dimensional gels is hampered by their extreme positive charge or their post-translational modifications. These problems can be surmounted by additional purification steps that include liquid chromatography, as is evident from the larger number of identified proteins that are obtained after using this step [8].
Ribosome synthesis and other regulatory networks
The sheer complexity and dominance of the ribosome synthesis pathway in the cell provides many opportunities for potential intersections with other major cellular processes. Recent studies have shown that ribosome synthesis in budding yeast is intimately linked with transcription, mRNA turnover, proteasome biogenesis, cell growth and the regulation of cell cycle [31–33]. In particular, several trans-acting ribosomal assembly factors have been shown to have essential functions outside ribosome synthesis. For example, the proteins Nob1p, Dim2p/Rrp20p and Cic1p are known components of pre-ribosomes, and are also required for proteasome function (see [34] for a review). Noc3p, Nop7p/Yph1p, Nop15p and Sda1p are required for various aspects of 60S subunit synthesis, yet their individual depletion also inhibits the initiation of DNA replication (Noc3p), progression into S phase of the cell cycle (Nop7p/Yph1p), cytokinesis (Nop15p) and actin cytoskeleton organization (Sda1p; see [31] for a review). In addition, depletion of trans-acting proteins found in early 40S pre-ribosomes [19] causes an arrest in the G1 phase of the cell cycle [35]. Although all details of the mechanism of this link between ribosome synthesis and the cell cycle are not yet understood, it is clear that the quality and quantity of ribosomes directly determine the growth rate of yeast cells and, by extension, the timing of cell division. Because ribosome synthesis is both up- and down-regulated by transcriptional and post-translational signals from several sources [10–12, 14, 33], it is to be expected that the components of this regulatory network will also contribute to cell-cycle regulation. Because most of the results described here were from genetic experiments, it is often difficult to make sense of the dual roles observed for trans-acting proteins. For several trans-acting proteins, however, such as, Nob1p, Dim2p, Nop15p and Cic1p, one can argue for a direct role in ribosome synthesis because they are confidently predicted to contain protein domains that are associated with RNA metabolism [34]. It will be necessary to characterize all the domains found in these proteins in order to understand their individual contributions to various regulatory networks.
In conclusion, we have learned a great deal about ribosomes in this decade, in large part because of high-resolution crystal structures that revealed the molecular details of peptide-bond formation and the RNA-driven nature of the ribosome's catalytic activity [36]. Almost three decades after the first identification of 90S pre-ribosomes, these particles have been purified and characterized [19, 21]. Subsequently, the compilation of around 200 trans-acting proteins involved in ribosome synthesis has prompted numerous genetic and biochemical studies aimed at their characterization (see [6, 7, 31] for reviews). Remarkably, most of these auxiliary proteins are essential in budding yeast, indicating relatively low tolerance of cells for incomplete or defective ribosomes. Furthermore, the majority of trans-acting proteins are conserved from yeast to humans, strongly suggesting that the overall pathway of ribosome synthesis is conserved among eukaryotes. The future challenge will be to decipher the exact cellular functions of all trans-acting proteins. One way to realize this goal is through the combination of computational predictions and experiments [23, 34]. Ultimately, molecular details of their functions will be deduced from structural studies [15, 37].
Although high-resolution crystal structures of ribosomes are available, they represent only static snapshots of this complex molecular machine. The variability of protein content of individual ribosomes and their post-translational modifications are likely to be important for optimal function under continually changing environmental and developmental conditions. Recent studies that combine two-dimensional gel electrophoresis and mass spectrometry have provided the catalog of protein modifications and identified the general degree of ribosome variability [8, 9, 24, 25, 27]. These studies represent only a first step, however, as more sophisticated methods will be necessary if we are to capture the dynamics of ribosome content under different intra- and extra-cellular constraints. The quality and quantity of synthesized ribosomes are important litmus tests of the overall health of the cell, and ribosome synthesis provides important signals for the global regulatory circuitry [11, 33]. It will be exciting to probe further how ribosomes and ribosome-associated factors interact with, and modulate the functions of, other cellular pathways.
References
Warner JR: The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999, 24: 437-440. 10.1016/S0968-0004(99)01460-7.
Kressler D, Linder P, de La Cruz J: Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1999, 19: 7897-7912.
Venema J, Tollervey D: Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet. 1999, 33: 261-311. 10.1146/annurev.genet.33.1.261.
Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B: A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999, 17: 1030-1032. 10.1038/13732.
Fatica A, Tollervey D: Making ribosomes. Curr Opin Cell Biol. 2002, 14: 313-318. 10.1016/S0955-0674(02)00336-8.
Fromont-Racine M, Senger B, Saveanu C, Fasiolo F: Ribosome assembly in eukaryotes. Gene. 2003, 313: 17-42. 10.1016/S0378-1119(03)00629-2.
Granneman S, Baserga SJ: Ribosome biogenesis: of knobs and RNA processing. Exp Cell Res. 2004, 296: 43-50. 10.1016/j.yexcr.2004.03.016.
Chang IF, Szick-Miranda K, Pan S, Bailey-Serres J: Proteomic characterization of evolutionarily conserved and variable proteins of Arabidopsis cytosolic ribosomes. Plant Physiol. 2005, 137: 848-862. 10.1104/pp.104.053637.
Giavalisco P, Wilson D, Kreitler T, Lehrach H, Klose J, Gobom J, Fucini P: High heterogeneity within the ribosomal proteins of the Arabidopsis thaliana 80S ribosome. Plant Mol Biol. 2005, 57: 577-591. 10.1007/s11103-005-0699-3.
Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M: Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002, 297: 395-400. 10.1126/science.1070850.
Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M: A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004, 18: 2491-2505. 10.1101/gad.1228804.
Marion RM, Regev A, Segal E, Barash Y, Koller D, Friedman N, O'Shea EK: Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc Natl Acad Sci USA. 2004, 101: 14315-14322. 10.1073/pnas.0405353101.
Rudra D, Zhao Y, Warner JR: Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J. 2005, 24: 533-542. 10.1038/sj.emboj.7600553.
Gallagher JE, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, Baserga SJ: RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 2004, 18: 2506-2517. 10.1101/gad.1226604.
Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL: Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol Cell. 2004, 16: 943-954. 10.1016/j.molcel.2004.11.031.
Baßler J, Grandi P, Gadal O, Lessmann T, Petfalski E, Tollervey D, Lechner J, Hurt E: Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol Cell. 2001, 8: 517-529. 10.1016/S1097-2765(01)00342-2.
Harnpicharnchai P, Jakovljevic J, Horsey E, Miles T, Roman J, Rout M, Meagher D, Imai B, Guo Y, Brame CJ, et al: Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol Cell. 2001, 8: 505-515. 10.1016/S1097-2765(01)00344-6.
Saveanu C, Bienvenu D, Namane A, Gleizes PE, Gas N, Jacquier A, Fromont-Racine M: Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 2001, 20: 6475-6484. 10.1093/emboj/20.22.6475.
Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, et al: A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002, 417: 967-970. 10.1038/nature00769.
Fatica A, Cronshaw AD, Dlakic M, Tollervey D: Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol Cell. 2002, 9: 341-351. 10.1016/S1097-2765(02)00458-6.
Grandi P, Rybin V, Baßler J, Petfalski E, Strauss D, Marzioch M, Schafer T, Kuster B, Tschochner H, Tollervey D, et al: 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol Cell. 2002, 10: 105-115. 10.1016/S1097-2765(02)00579-8.
Nissan TA, Baßler J, Petfalski E, Tollervey D, Hurt E: 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002, 21: 5539-5547. 10.1093/emboj/cdf547.
Oeffinger M, Dlakic M, Tollervey D: A pre-ribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes Dev. 2004, 18: 196-209. 10.1101/gad.285604.
Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR: Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999, 17: 676-682. 10.1038/10890.
Lee SW, Berger SJ, Martinovic S, Pasa-Tolic L, Anderson GA, Shen Y, Zhao R, Smith RD: Direct mass spectrometric analysis of intact proteins of the yeast large ribosomal subunit using capillary LC/FTICR. Proc Natl Acad Sci USA. 2002, 99: 5942-5947. 10.1073/pnas.082119899.
Louie DF, Resing KA, Lewis TS, Ahn NG: Mass spectrometric analysis of 40 S ribosomal proteins from Rat-1 fibroblasts. J Biol Chem. 1996, 271: 28189-28198. 10.1074/jbc.271.45.28189.
Odintsova TI, Muller EC, Ivanov AV, Egorov TA, Bienert R, Vladimirov SN, Kostka S, Otto A, Wittmann-Liebold B, Karpova GG: Characterization and analysis of posttranslational modifications of the human large cytoplasmic ribosomal subunit proteins by mass spectrometry and Edman sequencing. J Protein Chem. 2003, 22: 249-258. 10.1023/A:1025068419698.
Barakat A, Szick-Miranda K, Chang IF, Guyot R, Blanc G, Cooke R, Delseny M, Bailey-Serres J: The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol. 2001, 127: 398-415. 10.1104/pp.127.2.398.
Williams AJ, Werner-Fraczek J, Chang IF, Bailey-Serres J: Regulated phosphorylation of 40S ribosomal protein S6 in root tips of maize. Plant Physiol. 2003, 132: 2086-2097. 10.1104/pp.103.022749.
Sengupta J, Nilsson J, Gursky R, Spahn CM, Nissen P, Frank J: Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat Struct Mol Biol. 2004, 11: 957-962. 10.1038/nsmb822.
Dez C, Tollervey D: Ribosome synthesis meets the cell cycle. Curr Opin Microbiol. 2004, 7: 631-637. 10.1016/j.mib.2004.10.007.
Granneman S, Baserga SJ: Crosstalk in gene expression: coupling and co-regulation of rDNA transcription, pre-ribosome assembly and pre-rRNA processing. Curr Opin Cell Biol. 2005, 17: 281-286. 10.1016/j.ceb.2005.04.001.
Rudra D, Warner JR: What better measure than ribosome synthesis?. Genes Dev. 2004, 18: 2431-2436. 10.1101/gad.1256704.
Fatica A, Tollervey D, Dlakic M: PIN domain of Nob1p is required for D-site cleavage in 20S pre-rRNA. RNA. 2004, 10: 1698-1701. 10.1261/rna.7123504.
Bernstein KA, Baserga SJ: The small subunit processome is required for cell cycle progression at G1. Mol Biol Cell. 2004, 15: 5038-5046. 10.1091/mbc.E04-06-0515.
Steitz TA: On the structural basis of peptide-bond formation and antibiotic resistance from atomic structures of the large ribosomal subunit. FEBS Lett. 2005, 579: 955-958. 10.1016/j.febslet.2004.11.053.
Nissan TA, Galani K, Maco B, Tollervey D, Aebi U, Hurt E: A pre-ribosome with a tadpole-like structure functions in ATP-dependent maturation of 60S subunits. Mol Cell. 2004, 15: 295-301. 10.1016/j.molcel.2004.06.033.
Acknowledgements
This work was supported in part by NIH Grant P20 RR16455-05 from the INBRE-BRIN Program of the National Center for Research Resources.
Author information
Authors and Affiliations
Corresponding author
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
About this article
Cite this article
Dlakić, M. The ribosomal subunit assembly line. Genome Biol 6, 234 (2005). https://doi.org/10.1186/gb-2005-6-10-234
Published:
DOI: https://doi.org/10.1186/gb-2005-6-10-234