Abstract
Key components of the programmed cell death pathway are conserved between Caenorhabditis elegans, Drosophila melanogaster and humans. The search for additional homologs has been facilitated by the availability of the entire genomic sequence for each of these organisms.
Similar content being viewed by others
Programmed cell death is a conserved, gene-directed mechanism for the elimination of unnecessary or dangerous cells from an organism. The core cell death pathway was first defined through genetic analysis in the nematode Caenorhabditis elegans and has since been found to be conserved in species as diverse as Drosophila melanogaster and Homo sapiens. The rapid progress of the various genome sequencing projects has greatly accelerated the discovery of homologs for key components of the cell death pathway as well as for its regulators. The C. elegans genome sequence was completed two years ago [1], and both the Drosophila [2] and human genomes are essentially completely sequenced at this point. Although many components of the programmed cell death pathway are conserved between species, there are some important differences as well.
The core cell death pathway
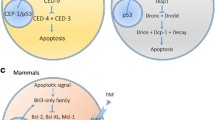
Programmed cell death has long been known to be a part of normal development, but it was not until the discovery of the first genes essential for the phenomenon that our understanding of the events leading up to the deliberate elimination of a cell began to take shape. Before this time, programmed cell death was defined by a set of specific morphological characteristics, including chromatin condensation, nuclear shrinkage and blebbing of the plasma membrane that could be observed in dying cells; the term apoptosis was coined to distinguish this type of cell death from necrotic deaths resulting from injury [3,4]. Four genes make up the core programmed cell death pathway in C. elegans, three of which, egl-1 (egg laying abnormal), ced-3 (cell death abnormal), and ced-4, are pro-apoptotic and one, ced-9, is anti-apoptotic (reviewed in [5]). There was considerable excitement in the field when potential mammalian and Drosophila homologs for ced-3, ced-4, ced-9, and egl-1 were discovered.
The CED-3 protein is one of a continuously growing family of specific cysteine proteases, termed caspases, that are thought to be the executioners of programmed cell death. At least fourteen mammalian caspases have been identified, and they are grouped into two classes on the basis of their proteolytic specificities (reviewed in [6]). Class 1 caspases are mainly involved in cytokine maturation, while Class 2 caspases act mainly in apoptosis. Class 2 has been further subdivided into two groups: upstream or initiator caspases (group 1), and downstream or effector caspases (group 2). Initiator caspases are thought to be at the beginning of a proteolytic cascade that amplifies the cell death signal and results in the activation of the effector caspases. Initiator caspases usually have long pro-domains, while effector caspases have short pro-domains. Drosophila has at least eight caspases, five of which have been at least partially characterized (Dcp-1, Dcp-2/Dredd, drICE, Dronc, and Decay) and three uncharacterized ones found encoded in the genome sequence [7]. C. elegans appears to have only a single caspase - CED-3 - that is essential for all developmental cell death, despite having three other caspases in its genome [8,9]. Three Drosophila caspases - Dcp-2/Dredd, Dronc, and one known only as a sequence in the genome database - have long pro-domains and are thus likely initiator caspases; another four - Dcp-1, drICE, Decay, and another one found in the genome database - have short pro-domains and are probably effector caspases. In addition, Dcp-1 has a substrate specificity that is very similar to that of two effector caspases, mammalian caspase 3 and C. elegans CED-3 [10]. Interestingly, Dronc appears to have a substrate specificity that is so far unique among caspases: while all other known caspases have only been shown to cleave after aspartate residues, Dronc can also cleave after glutamate residues [11]. This unusual substrate specificity may explain why Dronc is resistant to inhibition by the pan-caspase inhibitor p35.
On the basis of current data, it seems that CED-4 functions to help activate the caspase CED-3, and CED-9 blocks this activation through physical interaction with CED-4. Endogenous CED-4 is normally localized to the mitochondria by CED-9, unless EGL-1 is expressed [12,13]. If EGL-1 is expressed, the interaction between CED-4 and CED-9 ceases, and CED-4 translocates to the nuclear membrane where it activates CED-3, resulting in programmed cell death. Only one mammalian CED-4 homolog, Apaf-1, has been extensively characterized to date, but it too aids in caspase activation. Like CED-4, Apaf-1 requires dATP for caspase activation, but Apaf-1 requires cytochrome c in addition [14]. Knock-out studies have shown that mice deficient for Apaf-1 have reduced cell death in certain tissues, further supporting a role for Apaf-1 in programmed cell death. Drosophila has recently been shown also to have a CED-4/Apaf-1 homolog, named Dark/HAC-1/Dapaf-1 (reviewed in [15]). The Drosophila homolog is more similar to Apaf-1 than to CED-4: it has the WD repeats found in Apaf-1, which potentially function in binding regulatory proteins such as cytochrome c. Like Apaf-1 and CED-4, loss of function mutations in dark/hac-1/dapaf-1 result in a reduction in developmental programmed cell death. Intriguingly, the transcription of the Drosophila CED-4/Apaf-1 homolog is upregulated in response to both X-ray and ultraviolet irradiation, suggesting that death-inducing stimuli can feed into the cell death pathway at this step [16].
CED-9 and EGL-1 belong to a large family of proteins related to the mammalian anti-apoptotic protein Bcl-2. This family - with at least 19 members, divided into pro- and anti-apoptotic subgroups - has been largely defined by protein-protein interactions among its members (reviewed in [17]). CED-9 has mammalian counterparts in the anti-apoptotic subgroup of Bcl-2 family members, while EGL-1 has mammalian counterparts in the pro-apoptotic subgroup. Besides binding to Apaf-1, just as CED-9 binds and thereby regulates CED-4 activity, mammalian Bcl-2 family members have been proposed to regulate mitochondrial homeostasis and the release of pro-apoptotic factors such as cytochrome c (reviewed in [17]). The rate of mitochondrial release of cytochrome c in mammalian cells undergoing apoptosis was recently measured and found to be rapid [18]. Indeed, most mitochondria released all their cytochrome c within a one minute period in a temperature-independent manner, suggesting that an enzymatic transport mechanism is probably not involved. There is no clear evidence for cytochrome c release during apoptosis in C. elegans or Drosophila, however. Nevertheless, a specific epitope is uncovered on cytochrome c in Drosophila mitochondria during apoptosis and this may be sufficient to help activate Dark/HAC-1/Dapaf-1; and death-inducing stimuli result in an increase in cytochrome c in Drosophila cultured-cell lysates [19,20]. There is no apparent need for cytochrome c release in C. elegans, since CED-4 does not require it to activate CED-3.
Recently, a pro-apoptotic Drosophila Bcl-2 family member was identified with the help of the database of genomic sequence; this gene is most closely related to mammalian Bok and potentially regulates apoptosis in the fly [21,22,23,24]. The Drosophila Bok homolog interacts with several anti-apoptotic, but not with several pro-apoptotic, Bcl-2 family members and, therefore, possibly functions by antagonizing pro-survival proteins. Ectopic expression of Bok protein promotes apoptosis in transgenic flies as well as in cultured cells. While one group reports that the caspase inhibitor p35 can block apoptosis induced by Drosophila Bok [22], another group did not see p35 inhibition [23]. This difference may be due to the slightly different constructs used in the experiments and the different assay systems. A third group shows that peptide caspase inhibitors can block Drosophila Bok-mediated apoptosis in cell culture, lending support to a model in which expression of Bok protein leads to caspase activation [24]. Interestingly, ectopic expression of Drosophila Bok sensitizes the developing eye to cell death induced by ultraviolet irradiation [21]. Genetically, this Drosophila Bcl-2 family member appears to function upstream of, or in parallel to, the Drosophila ced-4/apaf-1 homolog and downstream of, or in parallel to, the Drosophila regulators of apoptosis, reaper, hid, and grim [22]. The core components of the cell death pathway are illustrated in Figure 1.
Regulation of programmed cell death
Studies of Drosophila have yielded great insights into how programmed cell death is regulated (reviewed in [25]). In addition to the core pathway, several essential regulators have been characterized in Drosophila. A region in the genome which contains the genes reaper, head involution defective (hid), and grim has been shown to be essential for virtually all developmental cell death in the Drosophila embryo. The three proteins encoded by these genes have no significant homology to any other known proteins except for a small stretch of similarity at the amino terminus that is shared by all three but no other known proteins. Each of these three genes can induce programmed cell death when overexpressed in the developing Drosophila eye or in various tissue-culture assays. In addition, Reaper, Hid and Grim are able to induce apoptosis in heterologous systems, suggesting that vertebrate homologs of these proteins exist (reviewed in [26]).
Further evidence for the existence of vertebrate homologs of Reaper, Hid, and Grim come from observations that these three proteins are able to bind a class of inhibitors of apoptosis, or IAPs. IAPs were first discovered in baculovirus, but have since been shown to play a vital role in blocking apoptosis in Drosophila as well as in mammals (reviewed in [27]). Also, Hid has been shown to be negatively regulated by survival signals mediated by receptor tyrosine kinases through the Ras and mitogen-activated protein (MAP) kinase pathway, which is also well conserved in mammals [28,29]. More recently, reports of a potential functional mammalian analog of Reaper, Hid, and Grim have been published [30,31]. Although Diablo/Smac shares no sequence homology with Reaper, Hid, or Grim, it too can bind IAPs, preventing them from inhibiting caspase activation, and thereby inducing apoptosis. Furthermore, Diablo/Smac interacts through its amino terminus with the BIR (baculovirus IAP repeat) domains of IAPs, similar to Reaper, Hid, and Grim [32,33]. An important difference between the function of Diablo/Smac and Reaper, Hid, and Grim is that Diablo/Smac is not pro-apoptotic by itself when overexpressed, but instead requires a death-inducing stimulus to promote apoptosis, unlike the three Drosophila proteins. Table 1 provides a summary of the conserved regulators of cell death across species.
In contrast to cell death effectors such as caspases, the cell death regulators Reaper and Grim are specifically expressed several hours before apoptosis in cells destined to die (reviewed in [26]). The expression of reaper has been shown to be regulated by distinct stimuli, including X-irradiation, steroid hormone signaling and a block in cell differentiation ([34,35,36] and A.-F. Lambin and H.S., unpublished observations). Recently, a Drosophila p53 ortholog was identified by searching the genome database (reviewed in [37]), and it was shown to bind a specific region of the reaper promoter, thereby potentially regulating reaper transcription in response to ionizing radiation. Furthermore, the reaper promoter was also shown to contain a response element for the complex made up of the steroid hormone ecdysone and its receptor [36]. The observation that transcription of both reaper and dark/hac-1/dapaf-1 can be induced by ionizing radiation suggests that these death-inducing signals can feed into the cell death pathway at multiple points. Unlike reaper and grim, hid appears to be regulated not only transcriptionally but also post-translationally by the Ras-MAP kinase pathway [28,29]. This additional level of control for hid could explain the observation that hid is expressed in cells that are doomed to die as well as in cells that continue to survive [38].
A second class of key apoptotic regulators in Drosophila is the IAP family. IAPs contain at least one, and up to three, BIR domains, which are important for protein-protein interactions, and often a carboxy-terminal RING domain (reviewed in [27]). While several IAPs have been implicated in inhibiting apoptosis, proteins with single BIR domains can also have functions in cell cycle regulation and cytokinesis [39,40]. In fact, Saccharomyces cerevisiae contains neither caspases nor an apoptotic program, but it does have an IAP-like protein with a single BIR domain that functions in regulating cell division [41], suggesting that the IAPs are diverse family with only some members being anti-apoptotic: hence this protein family is more accurately referred to as BIR-containing proteins (BIRPs). Three Drosophila BIRPs have been shown to be inhibitors of apoptosis, Diap1, Diap2, and Deterin [42,43,44,45]. Of these, Diap1 has been most extensively characterized; it can block cell death caused by the ectopic expression of reaper, hid, and grim (reviewed in [26]). In addition, Diap1 is able to block the cell death induced by the Drosophila caspases Dcp-1 or drICE in Saccharomyces, as well as apoptosis induced by the expression of drICE in insect cell culture [46,47,48]. Loss-of-function mutants in diap1 have a very striking early embryonic phenotype: virtually all cells display an apoptotic morphology, as well as displaying DNA fragmentation seen by TUNEL labeling, four to five hours into development [47,49,50]. This phenotype is especially noteworthy since it manifests itself several hours before the first developmental cell deaths are normally observed in the Drosophila embryo, and it clearly shows that diap1 is vital for normal development.
In general, IAPs are thought to function at the caspase activation step in the cell death pathway, binding to the inactive, pro-domain-containing caspase zymogen and preventing it from being processed into the active enzyme (reviewed in [27]). Reaper, Hid, Grim, and Diablo/Smac can physically interact with IAPs, thereby inhibiting the anti-apoptotic activity of IAPs and allowing caspase activation. Further insight into a possible mechanism for IAP function was recently gained when IAPs were observed to have ubiquitin ligase activity [51,52]. It was shown that certain cellular IAPs can be degraded in a proteasome-dependent manner prior to thymocyte apoptosis. These IAPs are also capable of auto-ubiquitination, an activity shown to require the RING domain [52]. In addition, IAPs can mono-ubiquitinate several caspases in vitro, possibly marking them for degradation, and again, this activity requires the RING domain [51]. The exact role of the ubiquitination pathway in regulating apoptosis is still unclear. Even the role of the RING domain in IAPs is open to debate: mutations in the RING domain of endogenous diap1 can be either pro- or anti-apoptotic, depending on the death-inducing stimulus ([49] and J. Agapite, L. Goyal, K. McCall and H.S., unpublished observations).
Strengths and limitations of genomic analysis
The completed genomes of C. elegans and Drosophila have already yielded a wealth of information. New cell death gene homologs are being described at a very rapid pace, often by several laboratories at once. Using information from the genome project, C. elegans was shown to contain several potential caspases in addition to CED-3, but none of these has been shown to function in programmed cell death [9]. In addition, while C. elegans has two BIR-domain-containing proteins, neither appears to regulate cell death: BIR-1 has been demonstrated to be essential for cytokinesis instead [39]. Hence, C. elegans seems to have a mechanism for regulating caspase activation that differs from that of mammals and Drosophila, in that it relies solely on the two Bcl-2 family members [53]. For Drosophila, the genome project played a key role in the identification of the Bcl-2 family member(s) and the CED-4/Apaf-1 homolog, showing that the core cell death pathway is conserved in Drosophila as well. Various other homologs for cell death genes, including three additional caspase-like sequences and a second Bcl-2 family member have been identified in the Drosophila genome sequence [7] and will undoubtedly be published in the near future. And analysis of the human genomic sequence is sure to lead to a better understanding of the number of mammalian homologs for the various components of the cell death pathway.
While it is clear that the search for homologs of known cell death genes has been greatly facilitated by the availability of the entire genome sequence for C. elegans, Drosophila, and humans, there are limitations to this strategy. For instance, the small size of the Reaper protein - only of 65 amino acids - makes it extremely difficult to identify homologs on the basis of genomic sequence information in other species: current gene-finding programs typically require an open reading frame of at least 70 amino acids. Furthermore, there is considerable divergence even among cell death proteins with clearly homologous functions, such as CED-4/Apaf-1 and even CED-9/Bcl-2, and sequence homology between these genes was recognized only after the genes and their products had been functionally characterized. Nevertheless, in combination with traditional approaches including genetics and biochemistry, knowledge of the genome sequence for model organisms will undoubtedly continue to drive our rapidly increasing insights into the molecular basis of programmed cell death.
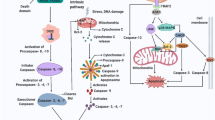
The core pathway of programmed cell death. Multiple pathways lead to the activation of the executioners of death, the caspases (reviewed in [53]). IAPs (inhibitors of apoptosis) have been shown to block the conversion of pro-caspases into active enzymes, and Reaper, Hid, Grim, and Diablo/Smac prevent IAPs from carrying out this protective function. Caspases can also be activated with the aid of Apaf-1, which in turn appears to be regulated by cytochrome c and dATP. The Bcl-2 family appears to function in regulating the release of pro-apoptotic components from mitochondria as well as by possibly inhibiting Apaf-1 directly. This pathway integrates knowledge gained in multiple species, showing that apoptosis appears to be regulated in a similar manner regardless of the organism. One notable exception is that the C. elegans homologs of IAPs do not appear to function in programmed cell death.
References
The C. elegans sequencing consortium: Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998, 282: 2012-2018. 10.1126/science.282.5396.2012.
Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al: The genome sequence of Drosophila melanogaster. Science. 2000, 287: 2185-2195. 10.1126/science.287.5461.2185.
Wyllie AH: Cell death: the significance of apoptosis. Int Rev Cytol. 1980, 68: 251-306.
Kerr JFR, Wyllie AH, Currie AR: Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972, 26: 239-257.
Metzstein MM, Stanfield GM, Horvitz HR: Genetics of programmed cell death in C. elegans : past, present and future. Trends Genet. 1998, 14: 410-416. 10.1016/S0168-9525(98)01573-X.
Nicholson DW: Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ. 1999, 6: 1028-1042. 10.1038/sj/cdd/4400598.
Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, et al: Comparative genomics of the eukaryotes. Science. 2000, 287: 2204-2215. 10.1126/science.287.5461.2204.
Ellis HM, Horvitz HR: Genetic control of programmed cell death in the nematode C. elegans. Cell. 1986, 44: 817-829. 10.1016/0092-8674(86)90004-8.
Shaham S: Identification of multiple Caenorhabditis elegans caspases and their potential roles in proteolytic cascades. J Biol Chem. 1998, 273: 35109-35117. 10.1074/jbc.273.52.35109.
Song Z, Guan B, Bergmann A, Nicholson DW, Thornberry NA, Peterson EP, Steller H: Biochemical and genetic interactions between Drosophila caspases and the proapoptotic genes rpr, hid, and grim. Mol Cell Biol. 2000, 20: 2907-2914. 10.1128/MCB.20.8.2907-2914.2000.
Hawkins CJ, Yoo SJ, Peterson EP, Wang SL, Vernooy SY, Hay BA: The Drosophila caspase DRONC is a glutamate/aspartate protease whose activity is regulated by DIAP1, HID, and GRIM. J Biol Chem. 2000, 275: 27084-27093.
Chen F, Hersh BM, Conradt B, Zhou Z, Riemer D, Gruenbaum Y, Horvitz HR: Translocation of C. elegans CED-4 to nuclear membranes during programmed cell death. Science. 2000, 287: 1485-1489. 10.1126/science.287.5457.1485.
del Peso L, González VM, Inohara N, Ellis RE, Núñez G: Disruption of the CED-9/CED-4 complex by EGL-1 is a critical step for programmed cell death in C. elegans. J Biol Chem. 2000, 275: 27205-27211.
Cecconi F: Apaf1 and the apoptotic machinery. Cell Death Differ. 1999, 6: 1087-1098. 10.1038/sj/cdd/4400602.
White K: Cell death: Drosophila Apaf-1- no longer in the (d)Ark. Curr Biol. 2000, 10: R167-R169. 10.1016/S0960-9822(00)00333-X.
Zhou L, Song Z, Tittel J, Steller H: HAC-1, a Drosophila homolog of APAF-1 and CED-4, functions in developmental and radiation-induced apoptosis. Mol Cell. 1999, 4: 745-755. 10.1016/S1097-2765(00)80385-8.
Gross A, McDonnell JM, Korsmeyer SJ: BCL-2 family members and the mitochondria in apoptosis. Genes Dev. 1999, 13: 1899-1911.
Goldstein JC, Waterhouse NJ, Juin P, Evan GI, Green DR: The coordinate release of cytochrome c during apoptosis is rapid, complete and kinetically invariant. Nat Cell Biol. 2000, 2: 156-162. 10.1038/35004029.
Kanuka H, Sawamoto K, Inohara N, Matsuno K, Okano H, Miura M: Control of the cell death pathway by Dapaf-1, a Drosophila Apaf-1/CED-4-related caspase activator. Mol Cell. 1999, 4: 757-769. 10.1016/S1097-2765(00)80386-X.
Varkey J, Chen P, Jemmerson R, Abrams JM: Altered cytochrome c display precedes apoptotic cell death in Drosophila. J Cell Biol. 1999, 144: 701-710. 10.1083/jcb.144.4.701.
Brachmann CB, Jassim OW, Wachsmuth BD, Cagan RL: The Drosophila Bcl-2 family member dBorg-1 functions in the apoptotic response to UV-irradiation. Curr Biol. 2000, 10: 547-550. 10.1016/S0960-9822(00)00474-7.
Colussi PA, Quinn LM, Huang DCS, Coombe M, Read SH, Richardson H, Kumar S: Debcl, a proapoptotic Bcl-2 homologue, is a component of the Drosophila melanogaster cell death machinery. J Cell Biol. 2000, 148: 703-714. 10.1083/jcb.148.4.703.
Igaki T, Kanuka H, Inohara N, Sawamoto K, Núñez G, Okano H, Miura M: Drob-1, a Drosophila member of the Bcl-2/CED-9 family that promotes cell death. Proc Natl Acad Sci USA. 2000, 97: 662-667. 10.1073/pnas.97.2.662.
Zhang H, Huang Q, Ke N, Matsuyama S, Hammock B, Godzik A, Reed JC: Drosophila pro-apoptotic Bcl-2/Bax homologue reveals evolutionary conservation of cell death mechanisms. J Biol Chem. 2000, 275: 27303-27306.
Bangs P, White K: Regulation and execution of apoptosis during Drosophila development. Dev Dyn. 2000, 218: 68-79. 10.1002/(SICI)1097-0177(200005)218:1<68::AID-DVDY6>3.3.CO;2-0.
Bergmann A, Agapite J, Steller H: Mechanisms and control of programmed cell death in invertebrates. Oncogene. 1998, 17: 3215-3223. 10.1038/sj/onc/1202586.
Deveraux QL, Reed JC: IAP family proteins - suppressors of apoptosis. Genes Dev. 1999, 13: 239-252.
Bergmann A, Agapite J, McCall K, Steller H: The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell . 1998, 95: 331-341. 10.1016/S0092-8674(00)81765-1.
Kurada P, White K: Ras promotes cell survival in Drosophila by downregulating hid expression. Cell. 1998, 95: 319-329. 10.1016/S0092-8674(00)81764-X.
Du C, Fang M, Li Y, Li L, Wang X: Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000, 102: 33-42. 10.1016/S0092-8674(00)00008-8.
Verhagen AM, Ekert PG, Pakusch M, Silke J, Connolly LM, Reid GE, Moritz RL, Simpson RJ, Vaux DL: Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell. 2000, 102: 43-53. 10.1016/S0092-8674(00)00009-X.
Srinivasula SM, Datta P, Fan XJ, Fernandes-Alnemri T, Huang Z, Alnemri ES: Molecular determinants of the caspase-promoting activity of Smac/DIABLO and its role in the death receptor pathway. J Biol Chem . 2000,
Chai J, Du C, Wu J-W, Wang X, Shi Y: Structural and biochemical basis of apoptotic activation by Smac/DIABLO. Nature. 2000, 406: 855-862. 10.1038/35022514.
Nordstrom W, Chen P, Steller H, Abrams JM: Activation of the reaper gene during ectopic cell killing in Drosophila. Dev Biol. 1996, 180: 213-226. 10.1006/dbio.1996.0296.
Robinow S, Draizen TA, Truman JW: Genes that induce apoptosis: transcriptional regulation in identified, doomed neurons of the Drosophila CNS. Dev Biol. 1997, 190: 206-213. 10.1006/dbio.1997.8696.
Jiang C, Lamblin A-F, Steller H, Thummel CS: A steroid-triggered transcriptional hierarchy controls salivary gland cell death during Drosophila metamorphosis. Mol Cell . 2000, 5: 445-455. 10.1016/S1097-2765(00)80439-6.
Steller H: Drosophila p53: meeting the Grim Reaper. Nat Cell Biol. 2000, 2: E100-E102. 10.1038/35014093.
Grether ME, Abrams JM, Agapite J, White K, Steller H: The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995, 9: 1694-1708.
Fraser AG, James C, Evan GI, Hengartner MO: Caenorhabditis elegans inhibitor of apoptosis protein (IAP) homologue BIR-1 plays a conserved role in cytokinesis. Curr Biol. 1999, 9: 292-301. 10.1016/S0960-9822(99)80137-7.
Li F, Ambrosini G, Chu EY, Plescia J, Tognin S, Marchisio PC, Altieri DC: Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998, 396: 580-584. 10.1038/25141.
Li F, Flanary PL, Altieri DC, Dohlman HG: Cell division regulation by BIR1, a member of the inhibitor of apoptosis family in yeast. J Biol Chem. 2000, 275: 6707-6711. 10.1074/jbc.275.10.6707.
Duckett CS, Nava VE, Gedrich RW, Clem RJ, Van Dongen JL, Gilfillan MC, Shiels H, Hardwick JM, Thompson CB: A conserved family of cellular genes related to the baculovirus iap gene and encoding apoptosis inhibitors. EMBO J. 1996, 15: 2685-2694.
Hay BA, Wassarman DA, Rubin GM: Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995, 83: 1253-1262. 10.1016/0092-8674(95)90150-7.
Uren AG, Pakusch M, Hawkins CJ, Puls KL, Vaux DL: Cloning and expression of apoptosis inhibitory protein homologs that function to inhibit apoptosis and/or bind tumor necrosis factor receptor-associated factors. Proc Natl Acad Sci USA. 1996, 93: 4974-4978. 10.1073/pnas.93.10.4974.
Jones G, Jones D, Zhou L, Steller H, Chu Y: Deterin, a new inhibitor of apoptosis from Drosophila melanogaster. J Biol Chem. 2000, 275: 22157-22165. 10.1074/jbc.M000369200.
Kaiser WJ, Vucic D, Miller LK: The Drosophila inhibitor of apoptosis D-IAP1 suppresses cell death induced by the caspase drICE. FEBS Lett. 1998, 440: 243-248. 10.1016/S0014-5793(98)01465-3.
Wang SL, Hawkins CJ, Yoo SJ, Müller H-AJ, Hay BA: The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 1999, 98: 453-463. 10.1016/S0092-8674(00)81974-1.
Hawkins CJ, Wang SL, Hay BA: A cloning method to identify caspases and their regulators in yeast: identification of Drosophila IAP1 as an inhibitor of the Drosophila caspase DCP-1. Proc Natl Acad Sci USA. 1999, 96: 2885-2890. 10.1073/pnas.96.6.2885.
Lisi S, Mazzon I, White K: Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics. 2000, 154: 669-678.
Goyal L, McCall K, Agapite J, Hartwieg E, Steller H: Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 2000, 19: 589-597. 10.1093/emboj/19.4.589.
Huang H-K, Joazeiro CAP, Bonfoco E, Kamada S, Leverson JD, Hunter T: The inhibitor of apoptosis, cIAP1, functions as a ubiquitin-protein ligase and promotes in vitro mono-ubiquitination of caspases-3 and -7. J Biol Chem. 2000, 275: 26661-26664.
Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD: Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000, 288: 874-877. 10.1126/science.288.5467.874.
Song Z, Steller H: Death by design: mechanism and control of apoptosis. Trends Cell Biol. 1999, 9: M49-M52. 10.1016/S0962-8924(99)01670-0.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tittel, J.N., Steller, H. A comparison of programmed cell death between species. Genome Biol 1, reviews0003.1 (2000). https://doi.org/10.1186/gb-2000-1-3-reviews0003
Published:
DOI: https://doi.org/10.1186/gb-2000-1-3-reviews0003