Abstract
Introduction
Tacrolimus is an important immunosuppressive drug for organ transplantation patients. It has a narrow therapeutic range, toxic side effects, and a blood concentration with wide intra- and interindividual variability. Hence, it is of the utmost importance to monitor tacrolimus blood concentration, thereby ensuring clinical effect and avoiding toxic side effects. Prediction models for tacrolimus blood concentration can improve clinical care by optimizing monitoring of these concentrations, especially in the initial phase after transplantation during intensive care unit (ICU) stay. This is the first study in the ICU in which support vector machines, as a new data modeling technique, are investigated and tested in their prediction capabilities of tacrolimus blood concentration. Linear support vector regression (SVR) and nonlinear radial basis function (RBF) SVR are compared with multiple linear regression (MLR).
Methods
Tacrolimus blood concentrations, together with 35 other relevant variables from 50 liver transplantation patients, were extracted from our ICU database. This resulted in a dataset of 457 blood samples, on average between 9 and 10 samples per patient, finally resulting in a database of more than 16,000 data values. Nonlinear RBF SVR, linear SVR, and MLR were performed after selection of clinically relevant input variables and model parameters. Differences between observed and predicted tacrolimus blood concentrations were calculated. Prediction accuracy of the three methods was compared after fivefold cross-validation (Friedman test and Wilcoxon signed rank analysis).
Results
Linear SVR and nonlinear RBF SVR had mean absolute differences between observed and predicted tacrolimus blood concentrations of 2.31 ng/ml (standard deviation [SD] 2.47) and 2.38 ng/ml (SD 2.49), respectively. MLR had a mean absolute difference of 2.73 ng/ml (SD 3.79). The difference between linear SVR and MLR was statistically significant (p < 0.001). RBF SVR had the advantage of requiring only 2 input variables to perform this prediction in comparison to 15 and 16 variables needed by linear SVR and MLR, respectively. This is an indication of the superior prediction capability of nonlinear SVR.
Conclusion
Prediction of tacrolimus blood concentration with linear and nonlinear SVR was excellent, and accuracy was superior in comparison with an MLR model.
Similar content being viewed by others
Introduction
Purpose
Tacrolimus blood concentrations demonstrate a wide intra- and interindividual variability. Therefore, monitoring of these concentrations remains an issue of pivotal importance to safeguard therapeutic efficacy and to manage the risk for nephrotoxicity, other toxicities, and rejection in liver transplantation patients [1]. This study examines the feasibility and clinical benefits of using a support vector regression (SVR) algorithm in comparison with a multiple linear regression (MLR) algorithm in predicting tacrolimus blood concentration. Tacrolimus blood concentration is predicted starting from a selected number of clinically relevant input variables.
Background
Hospital information systems in intensive care medicine generate large datasets on a daily basis. These rapidly increasing amounts of data make the task of extracting correct and relevant clinical information from intensive care unit (ICU) patients difficult [2, 3]. Data modeling techniques based on machine learning such as support vector machines (SVMs) can partially reduce workload, aid clinical decision-making, and lower the frequency of human error [4]. Fundamental research in clinical data modeling forms the basis on which later validation can be performed in multicentered clinical trials. This is the first study to use SVM for data modeling in the ICU domain. SVMs have been applied, however, in molecular biology [5–7], bioinformatics [8], as well as in genetics [9] and proteomics [10, 11]. In cancer research, kernel methods (or SVM) have been used to predict malignancy in brain tumors [12, 13] and also in staging certain forms of breast and prostate cancer [14, 15]. In cardiology, heart valve disease has been predicted with SVMs, and in fundamental cardiology research, nucleotide polymorphisms of candidate genes for ischemic heart disease have been modeled by kernel methods [16, 17]. Clinical decision-making has been compared for prospective performance with logistic regression and SVM [18]. In contrast with the absence of data concerning SVM applications in the ICU, artificial neural networks (ANNs) – as a less recent statistical learning technique – have been studied thoroughly in the ICU environment: they have been used for prediction of ICU mortality and prognosis in septic shock [19, 20], clinical decision-making [21], and prediction of plasma drug concentrations [22]. Also, the management of infectious diseases [23], real-time analysis of hemodynamics [24], and research in cardiology [25, 26] and oncology [27, 28] have benefited from recent evolutions in artificial intelligence (AI) and ANN.
Underlying theory
The roots of SVM lie in the statistical learning theory [29], which describes properties of learning machines which enable them to generalize well to unseen data. During the 1990s, SVM was developed by Vapnik and coworkers [30–32] at Bell Labs (formerly AT&T Bell Laboratories, Murray Hill, NJ, USA). A profound overview of the underlying theory and the SVM algorithm itself is given by Guyon and Elisseeff [33]. In the case of SVR [29], the goal is to find a function that predicts the target values of the training data with a deviation of at most ε, while requiring this function to be as flat as possible. The core of the support vector algorithm does this for linear functions f(x) = <w,x> + b, where (w,x) denotes the dot product of vectors w and x, thereby enforcing flatness by minimizing |w| (|w| denotes the Euclidian norm of vector w). By using a dual representation of the minimization problem, the algorithm requires only dot products of the input patterns. This allows the application of nonlinear regression by using a kernel function [34] that represents the dot product of the two transformed vectors. The MLR and the linear support vector algorithm are both linear approaches, but they differ in their underlying theoretical heuristics: the MLR method fits a model using the least-mean-squares heuristic (that is, the sum of the squared distances to the regression line is minimized). The support vector algorithm fits a flat-as-possible function by searching a separating hyperplane (Figure 1). The radial basis function (RBF) SVR method fits a nonlinear function onto the data, again aiming for maximum flatness. The RBF kernel is also often named a Gaussian kernel since the kernel function is the same as the Gaussian distribution function. Smola and Schölkopf [35] give an excellent overview of many details of the SVR procedure.
The support vector algorithm heuristic. In support vector machines, classification of datapoints or prediction of an outcome parameter is done by finding the 'hyperplane' that separates the datapoints by transforming the input variable dataset by a mathematical function into a 'higher dimension' in which separation is much easier (feature map = input variables dataset). The basis of this new heuristic is that classification of a seemingly chaotic input space is possible when one increases dimensionality and thereby finds a separating plane. Copyright permission from V.P. Bioinformatics (Improved Outcomes Software, Kingston, ON, Canada).
Materials and methods
Data
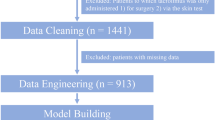
This study received approval from the Ethics Committee of Ghent University Hospital. Fifty patients who had recently undergone liver transplantation in Ghent University Hospital were included, and their medical records were reviewed. Tacrolimus blood concentrations, together with 35 other clinically relevant variables, were extracted from the ICU database. The following input variables were considered to influence tacrolimus blood concentration and were included: gender, age, weight, number of transplantations, number of days after surgery, existence of renal dysfunction (serum creatinine [CR] and urea [UR]) or liver dysfunction (alanine aminotransferase [ALT], aspartate aminotransferase [AST], gamma-glutamyl transpeptidase [GGT], total and conjugated bilirubin, alkaline phosphatise [ALKPHOS], and lactate dehydrogenase [LDH] levels), hematocrit (Hct), albumin, glucose, cholesterol, and six doses of tacrolimus, namely the dose at 8 a.m. and 8 p.m. from the three days (day 1, day 2, and day 3) before the day of the measured tacrolimus blood concentration (day 0). Coadministered medications were not included. Variables cholesterol and albumin were omitted due to too great a percentage of missing data (greater than 99%) (caused by not measuring these variables on a daily basis). The data were reorganized in patient days in which each record contained the following variables: gender, weight, age, days since transplantation, the 12 previously mentioned biochemical variables measured on day 0, the same parameters on day 1, tacrolimus blood concentration on day 1, the last six tacrolimus doses given, and the tacrolimus blood concentration on day 0 as a prediction target. This resulted in a total amount of 35 input variables and 1 output variable. Records in which the output parameter was missing were removed from the data. Patient days in which less than four of the six previous doses were available were also left out. This resulted in 457 records originating from 50 patients and a total of more than 16,000 data values. In these 457 records, 77% were complete, 15% contained a single missing value, and the remaining 7% had a maximum of 3 (of 35) values missing. This resulted in a total of 147/15,995 (0.92%) missing values. This extremely low number of missing values was filled in by means of an expectation maximization method [36].
Data analysis
Data analysis for the linear SVR and the RBF SVR model was performed using software implemented by the authors based on the libSVM 2.82 [37] software package. Analysis for the MLR model was performed in SPSS 12.0 (SPSS Inc., Chicago, IL, USA). A mean absolute difference with the measured tacrolimus blood concentration of maximum 3 ng/ml and a standard deviation (SD) of maximum 5 ng/ml was agreed upon to be acceptable by expert opinion.
Variable selection for the linear SVR and the RBF SVR model
This phase in the SVR model building is analogous with the variable selection phase for the MLR model. Using all 35 variables to construct a data model would result in suboptimal accuracy because different variables may contain overlapping information that disturbs the model-constructing process. Therefore, for each method (linear SVR, RBF SVR, and MLR), variable selection out of this total of 35 variables was done using recursive addition, recursive removal, stepwise addition, and stepwise removal of the input variables. These selection procedures are inspired by the commonly used stepwise regression technique in MLR, first presented by Effroymson [38]. The four selection procedures often result in different variable subsets. The best-performing subset was selected. In linear SVR, 15 features were selected: weight, age, days since transplantation, Hct, UR, ALKPHOS, ALT, total bilirubin, GGT (all on day 0), LDH on day 1, UR on day 1, morning doses of tacrolimus on day 2 and day 3, evening dose of tacrolimus on day 1, and the tacrolimus concentration on day 1. For RBF SVR, only two features sufficed: tacrolimus blood concentration on day 1 and the evening dose of tacrolimus on day 1. To validate a specific variable selection in linear SVR and RBF SVR, fivefold cross-validation was used. In this process, the available data are split into five equally sized parts. The remainder of the procedure is repeated five times. In each iteration, a different one of the five parts is kept apart, while the remaining four parts are used to construct the data model. The part that was kept separate is then used to verify the data model. The reported accuracy is the total of those measured in each of the five iterations, thus covering the total amount of available data.
Variable selection for the MLR model
In the MLR model also, the variable selection was performed with a forward, a backward, and a stepwise algorithm for simple linear regression in SPSS 12.0 and regression coefficients were checked for significance. The significance level was set at α = 0.05. Adjusted R2 values and goodness of fit were compared for the different MLR variable selections in SPSS. After selection of the final variable set for MLR, these variables were tested for correlation and multicollinearity. Variance inflation factor and eigenvalues were determined. For MLR, 16 variables were retained: gender, weight, age, Hct, LDH, UR, ALKPHOS, GGT, CR (all on day 0), AST on day 1, ALT on day 1, morning and evening doses of tacrolimus on day 1, evening dose of tacrolimus on day 2, and the morning dose of tacrolimus on day 3. Gender, weight, and age were included because of their clinical relevancy. After linear regression of this final variable set, normality testing of the residues as well as heteroscedasticity testing were performed. After searching the lambda value for the maximum likelihood with the Box-Cox algorithm, a transformation of the dependent variable (tacrolimus blood concentration) in the MLR model was performed because of heteroscedasticity of the residuals. To validate the final regression model, fivefold cross-validation was used, as in the SVR model.
Parameter selection for the linear SVR and the RBF SVR model
Parameter selection denotes the process of setting data model parameters. These are the parameters that tune a data modeling technique. The MLR method has no such parameters. The linear SVR method has two such parameters: ε and C. Epsilon controls the flatness of the resulting data model, whereas C controls the cost of a prediction error: setting C to high values will result in fewer prediction errors in the training data. The RBF SVR method has three model parameters: the already discussed ε and C and the extra kernel function parameter γ, which determines the degree of nonlinearity: setting γ to high values results in a highly nonlinear data model [39]. The model parameters can be set using theoretical considerations that may assume certain properties of the data. The data, however, are not always perfect: it may contain noise and nonremoved trends. Parameter values obtained in this way are thus suboptimal. Therefore, in this study, the parameters are set using theoretical heuristics after which this initial setting is fine-tuned using pattern search [40]. To validate a specific parameter selection in linear SVR and RBF SVR, again fivefold cross-validation was used.
Statistical analysis
Statistical analysis was carried out with SPSS 12.0. Results are reported as percentages, means, minimums and maximums, ranges, and SDs (as appropriate). A fivefold cross-validation algorithm was applied for validation of the prediction results. The correlation between measured and predicted tacrolimus blood concentrations was analyzed with a Spearman rank correlation coefficient. Differences between linear SVR, RBF SVR, and MLR were analyzed with the Friedman test and Wilcoxon signed rank test. A Bonferroni adjustment was performed for multiple testing. A Bland-Altman plot was used to illustrate significant differences between the three compared methods. Absolute difference as well as signed difference were studied. Mean absolute difference is the absolute difference between predicted and measured values, without its sign, and is an indication of the magnitude of the error, whereas mean signed difference indicates whether a model tends to predict higher or lower values than the measured value. The significance level was set at α = 0.05.
Results
Of the total study population, 58% (29/50) were male, mean age was 54 years (range 22 to 70), and mean weight was 79 kg. Table 1 gives a summary of the mean absolute differences between measured and predicted tacrolimus concentrations for the three models. In the distribution of the prediction errors made by the three methods, it has to be noted that the MLR model has the largest number of outliers (Figure 2). Figures 3 to 5 demonstrate the correlation between the observed tacrolimus blood concentration and the predicted blood concentration for linear SVR (Figure 3), RBF SVR (Figure 4), and MLR (Figure 5). These findings were corroborated by the Spearman rank correlation coefficients, which indicated good correlations for the three methods between the measured and the predicted blood concentrations: 0.762, 0.753, and 0.742 for linear SVR, RBF SVR, and MLR, respectively. Mean absolute difference between measured and predicted blood concentrations was smallest when using linear SVR: this difference between linear SVR and MLR was statistically significant (p < 0.001). Also, when mean signed differences were analyzed, the same significantly better results were observed in linear SVR in comparison with MLR. Even after post hoc analyses (α/3 for multiple testing, thus significance when p < 0.017), the significant difference between linear SVR and MLR remained valid. A Bland-Altman plot (Figure 6) outlines the difference between linear SVR and MLR.
Outliers for the prediction of the tacrolimus blood concentration for the three models. MLR, multiple linear regression; RBF SVR, radial basis function support vector regression (nonlinear support vector regression); SVR, support vector regression. *'s represent extreme values (values more extreme than 3*IQR).
Bland-Altman plot for comparison of linear support vector regression (SVR) (a) and multiple linear regression (MLR) (b): x-axis = difference (linear SVR and MLR) = a - b; y-axis = mean of linear SVR and MLR = (a + b)/2. If two models are the same, all datapoints are represented around one point. However, in this figure, there is a considerable amount of spread between the datapoints of the linear SVR method and the MLR method which is a graphical illustration of the statistical difference found between these two methods.
Discussion
Linear SVR for prediction of tacrolimus blood concentration resulted in a lower mean absolute error in comparison with the MLR model (Table 1). Incorporating nonlinearity in the predictor, however, by using a nonlinear kernel function, resulted in a prediction accuracy that was slightly less than in linear SVR, but this prediction still outweighed the accuracy of the MLR model. It is remarkable that this result was obtained using much fewer variables: only 2 input variables were used instead of 15 and 16 variables by the linear methods. Apparently, these 2 input variables contained more information in a nonlinear way than the other 15 or 16 contained in a linear way. When a linear method (linear SVR or MLR) was examined with only the 2 input variables used by the nonlinear RBF SVR model, the obtained prediction accuracy was lower than when using the nonlinear RBF SVR method. However, the added prediction strength of the extra 13 or 14 input variables in the linear SVR and MLR methods, respectively, is rather small. The nonlinear RBF SVR method is able to extract this extra information from only 2 input variables.
It is worth noting that in each of the three prediction models, the tacrolimus blood concentration on day 1 is incorporated, along with other variables. Obviously, the tacrolimus concentration on day 1 on its own already contains a lot of information about the level to be predicted. To verify the added value of incorporating this extra variable (tacrolimus dose on day 1), an RBF SVR model using only the previous tacrolimus blood concentration was constructed and evaluated using fivefold cross-validation. This model yielded a mean absolute error of 3.23 ng/ml (SD 3.12) and a maximum error of 26.33 ng/ml, indicating that adding the last evening dose of tacrolimus improves performance drastically. Moreover, it should be mentioned that in the linear kernel model a moderate amount of collinearity between the input variables was present (UR on 2 consecutive days, tacrolimus dose on 2 consecutive days) and that collinearity was not present between the two input variables of the RBF SVR model. In the MLR model, there was no problem of multicollinearity after the variable selection phase. It will be very interesting to see whether the results of this SVR model will be corroborated by similar results after testing this new technology on large multicentered ICU databases in future research.
This is the first report in which tacrolimus concentration is modeled by SVR, but a few other studies have already performed prediction of tacrolimus concentration using other AI techniques. Chen and colleagues [22] reported the use of a neural network and a genetic algorithm to predict the tacrolimus blood concentration. This neural network algorithm resulted in an average difference of the observed and predicted tacrolimus concentrations of 1.74 ng/ml with a range from 0.08 to 5.26 ng/ml. Bayesian forecasting as well has been applied in modeling tacrolimus concentrations. Fukudo and colleagues [41] demonstrated that Bayesian prediction of tacrolimus concentrations on the basis of previously acquired population-based pharmacokinetic data in adult patients receiving living-donor liver transplantation was possible within a certain timeframe after liver transplantation. However, a study by Willis and colleagues [42], using a population pharmacokinetic model based on Bayesian forecasting and adapted for individual pharmacokinetic, demographic, and covariate data, resulted in predictions that were too imprecise. In future research, the SVR-based model will be adapted to predict the tacrolimus dose to be given to ICU patients to obtain a predefined window of tacrolimus concentrations. Afterward, a randomized controlled trial will compare the accuracy of intensivists versus this SVR model in daily clinical practice.
Conclusion
Results demonstrate a statistically significant superiority of linear SVR in comparison with MLR as well as a trend toward superiority of nonlinear SVR in comparison with MLR for the prediction of tacrolimus blood concentration in post-liver transplantation patients during ICU stay. The accuracies were all within clinically acceptable ranges. Moreover, nonlinear SVR required only two variables to make the tacrolimus blood concentration predictions. SVM technology has promising possibilities as a clinical decision agent in the ICU environment.
Abbreviations
- AI:
-
artificial intelligence
- ALKPHOS:
-
alkaline phosphatise
- ALT:
-
alanine aminotransferase
- ANN:
-
artificial neural network
- AST:
-
aspartate aminotransferase
- CR:
-
serum creatinine
- GGT:
-
gamma-glutamyl transpeptidase
- Hct:
-
hematocrit
- ICU:
-
intensive care unit
- LDH:
-
lactate dehydrogenase
- MLR:
-
multiple linear regression
- RBF:
-
radial basis function
- RBF SVR:
-
radial basis function support vector regression (nonlinear support vector regression)
- SD:
-
standard deviation
- SVM:
-
support vector machine
- SVR:
-
support vector regression
- UR:
-
urea.
References
Kershner RP, Fitzsimmons WE: Relationship of FK506 whole blood concentrations and efficacy and toxicity after liver and kidney transplantation. Transplantation 1996, 62: 920-926.
Decruyenaere J, De Turck F, Vanhastel S, Vandermeulen F, Demeester P, de Moor G: On the design of a generic and scalable multilayer software architecture for data flow management in the intensive care unit. Methods Inf Med 2003, 42: 79-88.
De Turck F, Decruyenaere J, Thysebaert P, Van Hoecke S, Volckaert B, Danneels C, Colpaert K, De Moor G: Design of a flexible platform for execution of medical decision support agents in the intensive care unit. Comput Biol Med 2007, 37: 97-112.
Colpaert K, Claus B, Somers A, Vandewoude K, Robays H, Decruyenaere J: Impact of computerized physician order entry on medication prescription errors in the intensive care unit: a controlled cross-sectional trial. Crit Care 2006, 10: R21.
Guan CP, Jiang ZR, Zhou YH: Predicting the coupling specificity of GPCRs to G-proteins by support vector machines. Genomics Proteomics Bioinformatics 2005, 3: 247-251.
Yang ZR: Biological applications of support vector machines. Brief Bioinform 2004, 5: 328-338.
Wang YF, Chen H, Zhou YH: Prediction and classification of human G-protein coupled receptors based on support vector machines. Genomics Proteomics Bioinformatics 2005, 3: 242-246.
Byvatov E, Schneider G: Support vector machine applications in bioinformatics. Appl Bioinformatics 2003, 2: 67-77.
Vinayagam A, König R, Moormann J, Schubert F, Eils R, Glatting KH, Suhai S: Applying Support Vector Machines for Gene Ontology based gene function prediction. BMC Bioinformatics 2004, 5: 116.
Yang ZR, Chou KC: Bio-support vector machines for computational proteomics. Bioinformatics 2004, 20: 735-741.
Li L, Tang H, Wu Z, Gong J, Gruidl M, Zou J, Tockman M, Clark RA: Data mining techniques for cancer detection using serum proteomic profiling. Artif Intell Med 2004, 32: 71-83.
Li GZ, Yang J, Ye CZ, Geng DY: Degree prediction of malignancy in brain glioma using support vector machines. Comput Biol Med 2006, 36: 313-325.
Glotsos D, Tohka J, Ravazoula P, Cavouras D, Nikiforidis G: Automated diagnosis of brain tumours astrocytomas using probabilistic neural network clustering and support vector machines. Int J Neural Syst 2005, 15: 1-11.
Mattfeldt T, Trijic D, Gottfried HW, Kestler HA: Classification of incidental carcinoma of the prostate using learning vector quantization and support vector machines. Cell Oncol 2004, 26: 45-55.
Papadopoulos A, Fotiadis DI, Likas A: Characterization of clustered microcalcifications in digitized mammograms using neural networks and support vector machines. Artif Intell Med 2005, 34: 141-150.
Comak E, Arslan A, Türkoğlu I: A decision support system based on support vector machines for diagnosis of the heart valve diseases. Comput Biol Med 2007, 37: 21-27.
Yoon Y, Song J, Hong SH, Kim JQ: Analysis of multiple single nucleotide polymorphisms of candidate genes related to coronary heart disease susceptibility by using support vector machines. Clin Chem Lab Med 2003, 41: 529-534.
Pochet NL, Suykens JA: Support vector machines versus logistic regression: improving prospective performance in clinical decision-making. Ultrasound Obstet Gynecol 2006, 27: 607-608.
Clermont G, Angus DC, DiRusso SM, Griffin M, Linde-Zwirble WT: Predicting hospital mortality for patients in the intensive care unit: a comparison of artificial neural networks with logistic regression models. Crit Care Med 2001, 29: 291-296.
Wong LS, Young JD: A comparison of ICU mortality prediction using the APACHE II scoring system and artificial neural networks. Anaesthesia 1999, 54: 1048-1054.
Frize M, Ennett CM, Stevenson M, Trigg HC: Clinical decision support systems for intensive care units: using artificial neural networks. Med Eng Phys 2001, 23: 217-225.
Chen HY, Chen TC, Min DI, Fischer GW, Wu YM: Prediction of tacrolimus blood levels by using the neural network with genetic algorithm in liver transplantation patients. Ther Drug Monit 1999, 21: 50-56.
Lucas PJ, de Bruijn NC, Schurink K, Hoepelman A: A probabilistic and decision-theoretic approach to the management of infectious disease at the ICU. Artif Intell Med 2000, 19: 251-279.
Goldman JM, Cordova MJ: Advanced clinical monitoring: considerations for real-time hemodynamic diagnostics. Proc Annu Symp Comput Appl Med Care 1994, 752-755.
Spencer RG, Lessard CS, Davila F, Etter B: Self-organising discovery, recognition and prediction of haemodynamic patterns in the intensive care unit. Med Biol Eng Comput 1997, 35: 117-123.
Parmanto B, Deneault LG, Denault AY: Detection of hemodynamic changes in clinical monitoring by time-delay neural networks. Int J Med Inform 2001, 63: 91-99.
Lisboa PJ, Taktak AF: The use of artificial neural networks in decision support in cancer: a systematic review. Neural Netw 2006, 19: 408-415.
Lisboa PJ, Wong H, Harris P, Swindell R: A Bayesian neural network approach for modelling censored data with an application to prognosis after surgery for breast cancer. Artif Intell Med 2003, 28: 1-25.
Vapnik VN: The Nature of Statistical Learning Theory. Berlin, Germany: Springer-Verlag; 1995.
Boser BE, Guyon IM, Vapnik VN: A training algorithm for optimal margin classifiers. In Proceedings of the Fifth Annual Workshop on Computational Learning Theory. Pittsburgh, PA: ACM Press; 1992:144-152.
Cortes C, Vapnik V: Support-vector networks. Machine Learning 1995, 20: 273-297.
Schölkopf B, Burges CJC, Vapnik V: Incorporating invariances in support vector learning machines. In Lecture Notes In Computer Science, Proceedings of the 1996 International Conference on Artificial Neural Networks. Volume 1112. London, UK: Springer-Verlag; 1996:47-52.
Guyon I, Elisseeff A: An introduction to variable and feature selection. J Machine Learning Res 2003, 3: 1157-1182.
Aizerman MA, Braverman EM, Rozonoer LI: Theoretical foundations of the potential function method in pattern recognition learning. Automation Remote Control 1964, 25: 821-837.
Smola AJ, Schölkopf B: A tutorial on support vector regression. In NeuroCOLT Technical Report NC-TR-98-030. London, UK: Royal Holloway College, University of London, UK; 1998.
Dempster AP, Laird NM, Rubin DB: Maximum likelihood from incomplete data via the EM algorithm. J Royal Stat Soc Ser B 1977, B 39: 1-38.
Chang CC, Lin CJ: LIBSVM: a library for support vector machines.2001. [http://www.csie.ntu.edu.tw/~cjlin/libsvm]
Effroymson MA: Multiple regression analysis. In Mathematical Methods for Digital Computers. Edited by: Ralston A, Wilf HS. New York, NY: Wiley; 1960:191-203.
Lin CJ, Keerthi S: Asymptotic behaviors of support vector machines with Gaussian kernel. Neural Comput 2003, 15: 1667-1689.
Dennis JE, Torczon V: Derivative-free pattern search methods for multidisciplinary design problems. In Proceedings of the 5th AIAA/USAF/NASA/ISSMO Symposium on Multidisciplinary Analysis and Optimization. American Institute of Aeronautics and Astronautics, Reston, VA; 1994:922-932.
Fukudo M, Yano I, Fukatsu S, Saito H, Uemoto S, Kiuchi T, Tanaka K, Inui K: Forecasting of blood tacrolimus concentrations based on the Bayesian method in adult patients receiving living-donor liver transplantation. Clin Pharmacokinet 2003, 42: 1161-1178.
Willis C, Staatz CE, Tett SE: Bayesian forecasting and prediction of tacrolimus concentrations in pediatric liver and adult renal transplant recipients. Ther Drug Monit 2003, 25: 158-166.
Acknowledgements
The authors thank Tom Fiers and Chris Danneels for their technical support in the data acquisition process.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JD and FDT were responsible for the study concept, design, and overall responsibility. TV performed data acquisition and contributed to the statistical analysis and the drafting of the manuscript. SVL performed data transformation, wrote part of the SVM algorithm, and contributed to the drafting of the manuscript. DB and GVM contributed to the statistical analysis. All authors were responsible for the interpretation of data. All authors, including EH, contributed to the final manuscript. Funding for this study arose in part from project funding by an FWO scholarship and in part from clinical funding by the Ghent University Hospital. SVL and TV contributed equally to this article.
Stijn Van Looy, Thierry Verplancke contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Van Looy, S., Verplancke, T., Benoit, D. et al. A novel approach for prediction of tacrolimus blood concentration in liver transplantation patients in the intensive care unit through support vector regression. Crit Care 11, R83 (2007). https://doi.org/10.1186/cc6081
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1186/cc6081