Abstract
Introduction
Expression of the putative Wnt signalling inhibitor Dickkopf-3 (DKK3) is frequently lost in human cancer tissues because of aberrant 5'-cytosine methylation within the DKK3 gene promoter. Since other Wnt signalling inhibitors have been reported to be targets of epigenetic inactivation in human breast cancer, we questioned if DKK3 expression is also epigenetically silenced during breast carcinogenesis and therefore might contribute to oncogenic Wnt signalling commonly found in this disease.
Methods
DKK3 mRNA expression and DKK3 promoter methylation were determined by RT-PCR, realtime PCR and methylation-specific PCR in breast cell lines (n = 9), normal breast tissues (n = 19) and primary breast carcinomas (n = 150), respectively. In vitro DNA demethylation was performed by incubating breast cell lines with 5-aza-2'-deoxycytidine and trichostatin A. DKK3 protein expression was analysed by immunohistochemistry in breast carcinomas (n = 16) and normal breast tissues (n = 8). Methylation data were statistically correlated with clinical patient characteristics. All statistical evaluations were performed with SPSS 14.0 software.
Results
DKK3 mRNA was downregulated in 71% (five of seven) of breast cancer cell lines and in 68% of primary breast carcinomas (27 of 40) compared with benign cell lines and normal breast tissues, respectively. A DNA demethylating treatment of breast cell lines resulted in strong induction of DKK3 mRNA expression. In tumourous breast tissues, DKK3 mRNA downregulation was significantly associated with DKK3 promoter methylation (p < 0.001). Of the breast carcinomas, 61% (92 of 150) revealed a methylated DKK3 promoter, whereas 39% (58 of 150) retained an unmethylated promoter. Loss of DKK3 expression in association with DKK3 promoter methylation (p = 0.001) was also confirmed at the protein level (p < 0.001). In bivariate analysis, DKK3 promoter methylation was not associated with investigated clinicopathological parameters except patient age (p = 0.007).
Conclusions
DKK3 mRNA expression and consequently DKK3 protein expression become frequently downregulated during human breast cancer development due to aberrant methylation of the DKK3 promoter. Since DKK3 is thought to negatively regulate oncogenic Wnt signalling, DKK3 may be a potential tumour suppressor gene in normal breast tissue.
Similar content being viewed by others
Introduction
The mammalian Dickkopf genes (DKK) encode a class of extracellular signalling molecules that control cell fate during embryonic development and regulate tissue homeostasis in adults [1, 2]. Four DKK gene members have been identified so far. DKK1, DKK2 and DKK4 antagonise canonical Wnt/β-catenin signalling by interaction with LDL-receptor-related proteins (LRP5 and LRP6) [3]. In contrast, DKK3 does not sequester LRPs or Wnt ligands [2, 4, 5]. Its function in antagonising nuclear β-catenin levels, designated as the hallmark of an activated Wnt pathway often found in human tumour tissues [6], has received conflicting reports [7–9]. Most evidence suggest DKK3 exerts a tumour suppressive function by inhibiting a non-canonical Wnt signalling branch referred to as the planar cell polarity (PCP) pathway. The PCP pathway is characterised by the activation of c-Jun kinase (JNK) via recruitment of small GTPases of the Rho/Rac family [10]. It results in changes in cell adhesion, motility and polarity [11] rather than interfering with the networks of proliferation and differentiation, which is mediated by canonical Wnt/β-catenin signalling [6].
In agreement with its putative tumour-suppressive function [9, 12–14]DKK3 is commonly downregulated in human cancers such as lung cancer [15–17], renal clear cell carcinoma [18], pancreatic cancer [19], leukaemia [20], prostate cancer [7, 21], bladder cancer [22], melanoma [23] and gastrointestinal tumours [24]. In many of these diseases transcriptional loss is tightly associated with methylation of the DKK3 promoter [15, 16, 18, 20–22, 24], whereas in other malignancies the cause of downregulation remains to be elucidated or is not related to 5'-cytosine methylation [23]. A study on lung cancer revealed that the rate of DKK3 methylation increased steadily from normal lung tissue, to low-grade and high-grade atypical adenomatous hyperplasia to invasive adenocarcinoma [25], suggesting a potential role of DKK3 methylation in lung cancer progression. In mouse cancer models, DKK3 has proved a promising therapeutic agent capable of repressing tumour progression, for example, in testicular germ cell cancer [14] and prostate cancer [13]. More recently, a breast cancer xenotransplantation model demonstrated that a single adenoviral-mediated intra-tumoural injection of a DKK3 expression vector efficiently discontinued tumour growth, with the induction of apoptosis in these cells [26]. This suggests that DKK3 may have an important tumour-suppressive function that either prevents tumour initiation or attenuates cancer progression. Interestingly, loss of DKK3 expression was first observed in numerous immortalised tumour-derived cell lines [27]. Immortalisation, that is escape from cellular senescence, is an early event in malignant transformation [28], so DKK3 could act as a tumour suppressor gene by mediating the effects of senescence stimuli. In concordance with this hypothesis, DKK3 expression was found to be elevated in organs with predominantly growth-arrested post-mitotic cells, for example in the heart and brain [29] and also in senescent prostate epithelial cells [30].
However, to the authors' knowledge, a comprehensive study on DKK3 gene regulation and its implication in breast cancer has not yet been published. In our study we investigated DKK3 mRNA expression, DKK3 protein expression and DKK3 promoter methylation in breast cell lines as well as in normal and malignant primary breast tissues. Our results demonstrate for the first time that DKK3 expression is frequently downregulated in human breast cancer as a consequence of aberrant DNA methylation within the DKK3 gene promoter.
Materials and methods
Patient material
Nineteen matched tumour and normal tissue samples from breast cancer patients and 131 tissue samples from breast carcinomas were obtained from patients treated by primary surgery for breast cancer at the Departments of Gynecology at the University Hospitals of Aachen, Jena, Regensburg and Düsseldorf, Germany, between 1991 and 2005. The selection of cases was based on availability of tissue, and the sample was recruited in a non-selective, consecutive manner. Female patients presenting with unilateral, invasive breast cancer with no individual breast cancer history were included in the study. Exclusion criteria were: neo-adjuvant chemotherapy before surgery; presentation with secondary breast cancer; or peritumourous carcinoma in situ present in the tumour sample. Patient characteristics are shown in Table 1.
All patients gave informed consent for retention and analysis of their tissue for research purposes and the Institutional Review Boards of the participating centres approved the study.
Tumour histology was determined according to the criteria of the World Health Organization [31], while disease stage was assessed according to International Union Against Cancer [32]. Tumours were graded according to Bloom and Richardson, as modified by Elston and Ellis [33]. After surgery, tumour material was immediately snap-frozen in liquid nitrogen. Sections stained with haematoxylin and eosin were prepared for assessing the percentage of tumour cells; only samples containing more than 70% tumour cells were selected. Samples were dissolved in lysis buffer followed by DNA isolation using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations.
Breast cell lines
The cancerous breast cell lines BT20, Hs578T, MCF7, MDA-MB231, SKBR3, T47D and ZR75-1 and the non-cancerous lines HMEC and MCF12A were obtained from the American Type Culture Collection (ATCC) (Rockville, MA, USA) and cultured under recommended conditions.
RNA expression analysis
RNA isolation, RT-PCR and realtime PCR were performed as previously described [34]. To ensure experiment accuracy, all quantitative measurements were performed in triplicate. Intron-spanning primer sequences and cycling conditions are given in Table 2.
Bisulphite-modification and methylation-specific PCR
About 1 μg of genomic DNA was bisulphite-modified using the EZ DNA Methylation Kit (Zymo Research, Orange, CA, USA) according to the manufacturer's recommendations. Methylation-specific PCR (MSP) was performed as previously described [34]. Primers and cycling conditions are listed in Table 2. Specificity of MSP primers in detecting the DKK3 methylation status were demonstrated using universal unmethylated and universal polymethylated DNA as a template (Epi Tect Control DNA Set; Qiagen, Hilden, Germany).
5-aza-2'-deoxycytidine and trichostatin A treatment
DNA demethylating treatment of breast cancer cell lines was performed with 5-aza-2'-deoxycytidine as described elsewhere [34], modified by the addition of 300 nM of the histone deacetylase inhibitor trichostatin A (Sigma-Aldrich, Deisenheim, Germany) on day three (incubation for 24 hours). Drug concentrations were adjusted in advance to warrant viability and replication of all cell lines.
Immunohistochemistry
Formalin-fixed, paraffin-embedded 2 μm tissue sections were subjected to immunostaining using the Advance Kit (Dako, Hamburg, Germany) following the manufacturer's instructions. Antigen retrieval was performed by pre-treatment with citrate buffer (pH 7) in a microwave oven (20 minutes at 200 W). Sections were incubated for 30 minutes with the primary antibody (AP1523a, Abgent, San Diego, CA, USA; 1:50), washed and incubated for 10 minutes with the secondary antibody (biotinylated polylink; Dako, Hamburg, Germany). Diaminobenzidin (Dako, Hamburg, Germany) was used for antibody detection. An experienced breast cancer pathologist (N.B.) scored the immunohistochemical staining according to the scoring system suggested by Remmele and Stegner [35]. The tissue specimens for immunohistochemical analysis are characterised in Table 3.
Statistical evaluations
Statistical analyses were completed using SPSS 14.0 (SPSS, Chicago, IL, USA). Differences were considered significant when p < 0.05. A non-parametric Mann-Whitney U-test and a Kruskal-Wallis test were applied to examine expression levels among normal tissues, DKK3 unmethylated tumours and DKK3 methylated tumours. A student's t-test was applied on the expression of paired normal and tumour samples. To study statistical associations between clinicopathological factors and DKK3 methylation status contingency tables and a two-sided Fisher's exact test were used.
Results
Differential DKK3mRNA expression in breast cell lines
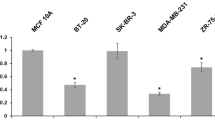
To start our analysis of DKK3 expression in breast cancer patients we analysed mRNA expression in non-malignant and malignant breast cell lines using realtime PCR. Strong DKK3 expression could be detected in non-malignant HMEC and MCF12A cells (Figure 1a) exhibiting expression levels comparable with human placental tissue, which is known to abundantly express DKK3 [22]. Among the malignant cell lines, Hs578T and SKBR3 cells revealed abundant DKK3 mRNA levels comparable with benign breast cells. In five further breast cancer cell lines (BT20, MCF7, MDA-MB231, T47D and ZR75-1) DKK3 expression was substantially reduced.
DKK3 mRNA expression and DKK3 promoter methylation analysis in breast cell lines. (a) DKK3 mRNA expression is differentially expressed in breast cell lines. Non-cancerous cell lines (HMEC and MCF12A) and cancerous breast cell lines (Hs578T, SKBR3, BT20, MCF7, MDA-MB231, T47D and ZR75-1) were analysed by realtime PCR and related to DKK3 mRNA expression in human placental tissue (set to 1). Grey bars = samples expressing abundant DKK3 mRNA; black bars = samples showing strong downregulation of DKK3 expression. (b) Hypermethylation of the DKK3 promoter in breast cell lines. Methylation specific PCR (MSP) was performed with bisulphite-treated DNA from benign and malignant breast cell lines. DNA bands in lanes labelled with U indicate PCR products amplified with primers recognising unmethylated DKK3 promoter sequence. DNA bands in lanes labelled with M represent amplificate generated with methylation-specific primers; water served as 'no template control' (NTC). MSP controls demonstrate the specificity of the DKK3 primers used. Universal poly-methylated bisulphite-converted DNA (UMD) exclusively yields amplification products with primers specific to methylated DKK3 promoter sequence; universal unmethylated bisulphite-converted DNA (UUD) yields exclusive amplification products with primers recognising the unmethylated DKK3 promoter sequence.
Methylation of the DKK3promoter in breast cell lines
Aberrant promoter hypermethylation of tumour suppressor genes during carcinogenesis is an effective mechanism resulting in downregulation and functional inactivation of these genes [36, 37]. Knowing that DKK3 expression was downregulated in most malignant breast cell lines we performed promoter methylation analysis in these cells. By using MSP [38] we found a methylated DKK3 promoter sequence in all cell lines showing reduced DKK3 expression, BT20, MCF7, MDA-MB231, T47D and ZR75-1 (Figure 1b). In contrast to this, all DKK3 expressing cell lines (HMEC, MCF12A, Hs578T and SKBR3) as well as human placental tissue lacked DKK3 promoter methylation in the analysed promoter region.
In vitro demethylation of the DKK3promoter
To prove a direct association of DKK3 promoter methylation with loss of DKK3 mRNA expression we treated seven breast cell lines sequentially with the DNA-methyltransferase inhibitor 5-aza-2'-deoxycytidine and the histone deacetylase inhibitor trichostatin A. Subsequently, we determined DKK3 promoter methylation and DKK3 mRNA expression before and after the drug treatment. MSP analyses after the treatment (Figure 2a) confirmed that promoter demethylation had occurred in all methylated cell lines by the appearance and enhancement of signals indicative of unmethylated DNA sequences. Those cell lines initially bearing a methylated DKK3 promoter showed elevated DKK3 mRNA expression after treatment (BT20, MCF7, MDA-MB231, T47D and ZR75-1; Figure 2b), whereas no DKK3 mRNA gain was achieved in unmethylated MCF12A and only a marginal DKK3 mRNA gain in SKBR3 cells. The induction of DKK3 mRNA transcription after the treatment as determined by realtime PCR ranged from 4.7-fold to 29-fold higher than in originally methylated breast cancer cells.
DKK3 mRNA expression after in vitro DNA demethylation. (a) In vitro demethylation of the DKK3 promoter. Methylation specific PCR of seven breast cell lines was performed with DNA from cells either untreated (-) or treated with 1 μM 5-aza-2'-deoxycytidine (DAC) and 300 nM trichostatin A (TSA) (+). (b) Re-expression of DKK3 mRNA in breast cell lines after treatment with DAC/TSA. All cell lines initially exhibiting low DKK3 mRNA restored expression of DKK3 compared with the untreated control cells. Induction level (fold change) of DKK3 mRNA expression was determined by realtime PCR.
Differential DKK3mRNA expression in primary breast carcinomas
DKK3 mRNA expression in primary breast tissues was then examined by realtime PCR. In the first step, 19 pairs of breast carcinoma tissues and corresponding normal breast tissue were analysed. A significant downregulation of DKK3 mRNA expression in tumour tissue compared with its adjacent normal tissue was detected in 14 (74%) of the 19 pairs (Figure 3a), as defined by an expression fold change of two or more (FC2).
DKK3 mRNA expression analysis in primary breast carcinomas. (a) Realtime PCR of DKK3 mRNA expression in 19 matched pairs (normal vs. tumour). Arrowheads indicate downregulation in the tumour by an expression fold change of more than two (FC2). (b) Relative DKK3 mRNA expression in 40 additional breast cancer specimens. Mean expression of 19 normal breast tissues (N) was set to 1. Based on a FC2, grey bars represent tumour specimens (T) showing normal expression, black bars represent tumour specimens with reduced DKK3 mRNA expression.
To further support this data set we analysed 40 additional breast carcinomas with no corresponding normal breast tissue and referred each individual DKK3 mRNA expression level to the mean DKK3 expression of the previously analysed 19 normal breast tissues (Figure 3b). The frequency of DKK3 mRNA downregulation measured in this tumour cohort (27 of 40, 68% by FC2) is in good agreement to the result achieved with the 19 matched pairs. Therefore downregulation of DKK3 mRNA in breast cancer is considered to affect about 70% of patients.
DKK3promoter methylation in primary breast carcinomas
To address the question of whether DKK3 promoter methylation occurs in primary breast carcinomas, we analysed 150 mammary tumour samples by MSP. Corresponding normal breast tissue was available for 19 tumours. In total, 92 of 150 tumours (61.3%) revealed DKK3 promoter methylation (for example, #7 in Figure 4) whereas in 58 of 150 tumours (38.7%) the DKK3 promoter was unmethylated. In these cases, MSP amplification signals were obtained exclusively in the U-reaction (for example, #5 in Figure 4). Of the normal breast tissues, only a single sample (5.3% of 19 samples) gave a very weak methylation signal (data not shown) in contrast to all other normal tissues that lacked DKK3 promoter methylation. As an additional control that infiltrating immune cells do not contribute to methylation signals in mammary tumours, bisulphite-converted DNA from human peripheral blood lymphocytes were assayed and revealed an unmethylated DKK3 promoter, consistent with results from a previous study [39].
DKK3 promoter methylation analysis in primary breast carcinomas. Methylation specific PCR results from 10 representative matched pairs of primary breast tumour (T) and corresponding normal breast tissue (N) are shown. DNA from human placenta (Plc) as well as from peripheral blood lymphocytes (Lyc) reveals an unmethylated DKK3 promoter. DNA from breast carcinoma cell line BT20 served as positive control. NTC = no template control.
Correlation of DKK3 promoter methylation with loss of DKK3mRNA expression
For all breast carcinoma samples analysed for DKK3 mRNA expression (n = 59) we determined the DKK3 promoter methylation status at the same time. Thus, we were able to directly compare DKK3 methylation and mRNA expression in primary human breast carcinomas. A boxplot (Figure 5) illustrates the distribution and medians of DKK3 RNA expression among normal breast tissues, DKK3 unmethylated tumours and DKK3 methylated tumours. The median DKK3 expression level (exp) of unmethylated tumours (exp = 0.87; FC = 1.2) was comparable with that of normal breast tissues (set to exp = 1). In contrast, DKK3 methylated tumours showed a significant mRNA downregulation (exp = 0.17; FC = 5.9) compared with DKK3 unmethylated tumours and normal breast tissue (global p < 0.001).
Correlation analysis of DKK3 promoter methylation with DKK3 mRNA expression in primary breast carcinomas. Tumours that are unmethylated in the DKK3 promoter express DKK3 mRNA comparable with DKK3 mRNA expression normal breast tissue, whereas in DKK3 methylated tumours DKK3 mRNA expression is significantly reduced. Horizontal lines = group medians; boxes = 25 to 75% quartiles, range, peak and minimum.
Differential DKK3protein expression in primary breast carcinomas
Immunohistochemical staining was used to investigate DKK3 protein expression in normal (n = 8) and malignant (n = 16) breast tissues. DKK3 was strongly expressed in non-malignant luminal and basal epithelial cells, achieving a mean (sd) immunoreactivity score (IRS) [34] of 11.3 (1.4) (Figure 6a) and a median IRS of 12 (range 9 to 12). DKK3 protein was not detectable in stromal cells in the normal breast tissue. Of the breast carcinomas, four of 16 (25%) revealed abundant DKK3 protein expression (IRS = 9 to 12; Figure 6b) in contrast to seven of 16 tumours (44%), which showed partial loss (IRS = 4 to 8; Figure 6c), and five of 16 tumours (31%) with substantial loss of DKK3 protein (IRS = 0 to 3; Figure 6d and Table 3). The mean protein staining intensity in breast carcinomas was determined to have an IRS of 5.9 (3.3) and the median to have an IRS of 6 (range 0 to 12). DKK3 expression levels in the tumour and normal tissue groups were shown to be significantly different (p = 0.002; U-test), and DKK3 protein was also differentially expressed within the eight matched pairs (p < 0.001; t-test). As a continuous variable, a lower IRS in breast carcinoma was significantly associated with the presence of DKK3 promoter methylation (p = 0.001; Fisher's exact test).
DKK3 protein expression in primary breast carcinomas as determined by immunohistochemical staining. (a) Normal mammary tissue without (left) and with application (right) of DKK3 antibody. Abundant DKK3 protein expression is detectable in luminal and basal epithelial cells. (b) Breast carcinoma with unmethylated DKK3 promoter reveals abundant DKK3 protein expression. (c) and (d) Breast carcinomas with a methylated DKK3 promoter exhibit substantial loss of DKK3 protein expression. Original magnifications are given in upper right-hand corner.
Association of DKK3promoter methylation with clinicopathological factors
For descriptive data analysis clinicopathological patient characteristics were correlated with the DKK3 promoter methylation status. In a bivariate analysis, DKK3 promoter methylation was significantly associated with advanced patient age at diagnosis (p = 0.007). Furthermore, DKK3 promoter methylation was not associated with tumour size, lymph node status, histological grade, histological type, and oestrogen receptor or progesterone receptor positivity (data not shown).
Discussion
It was previously reported that expression of the putative Wnt antagonist DKK3 was downregulated in several tumour entities as a consequence of epigenetic DNA modification [15, 16, 18, 20–22, 24]. Our study is the first to analyse DKK3 gene regulation in human breast cancer. Malignant breast cell lines showed strong reduction of DKK3 mRNA in association with DKK3 promoter methylation. Consistently, DKK3 mRNA expression was induced after promoter DNA demethylation in these cells. In primary breast carcinomas, DKK3 mRNA expression was downregulated in 68% of invasive tumours with significant association with methylation of the DKK3 gene promoter (p < 0.001). The total frequency of DKK3 methylation was 61% in breast carcinomas, whereas corresponding normal breast tissues were unaffected by this epimutation. We further showed that a loss of DKK3 protein in breast carcinomas is also associated with DKK3 promoter methylation (p = 0.001) whereas protein expression is abundant in epithelial cells of the normal breast. In summary, our data demonstrate for the first time that promoter methylation-mediated downregulation of DKK3 expression is a frequent and tumour-related epigenetic alteration in the development of human breast cancer.
The implication of aberrant canonical Wnt/β-catenin signalling in the pathogenesis of human cancer has become widely accepted [40]. Its oncogenic effect is mediated by uncontrolled activation of target genes that for example, enhance cell proliferation, such as c-myc and cyclin D1. In breast cancer, several genes encoding inhibitors of canonical Wnt/β-catenin signalling have been reported to be frequently hypermethylated, for example, SFRP1 [34, 41], SFRP2 [42], SFRP5 [43], WIF1 [44] and DKK1 [42]. We suggest that disruption of a non-canonical Wnt signalling branch, the PCP pathway, may also be implicated in human carcinogenesis by pathologically altering the networks of cellular adhesion, motility and cell polarity, because it has been shown that expression of the putative PCP pathway inhibitor DKK3 is commonly downregulated in malignant tissues. As a consequence, loss of DKK3 may promote hyperactivation of the PCP pathway, thereby potentially enhancing tumour aggressiveness.
Recent in vivo experiments support a hypothesis that the loss of DKK3 expression promotes an aggressive cancer phenotype. In a mouse model, DKK3 proved to be a promising therapeutic agent to significantly inhibit tumour growth in testicular germ cell cancer [14]. In an orthotopic prostate cancer model a similar treatment resulted in tumour regression, decreased metastasis and prolonged survival of the host [13]. The most recent findings from a breast cancer study revealed that DKK3 not only attenuates tumour growth in a xenotransplantation mouse model, it also re-sensitised multidrug-resistant MCF7/ADR cells to doxorubicin treatment in a JNK-c-Jun dependent manner [26]. This highlights its potential utility as a gene therapeutic agent in human breast cancer. Our study adds important information to this aspect, because it so far remained unknown if methylation-mediated loss of DKK3 expression also occurred in primary breast cancer, and, if so, how many patients were affected by this epimutation. We have shown that a large proportion (61%) of breast cancer patients have DKK3 promoter methylation in the carcinoma tissue, leading to a functional inactivation of the tumour-protective protein. Therefore, we conclude that a potential gene therapeutic treatment with DKK3 might be of benefit for a large target population of breast cancer patients.
In contrast to other studies, DKK3 promoter methylation in our cohort was not associated with clinicopathological factors indicative of a progressive cancer subtype, such as tumour size, node status or histological grade. The existence of such an association has been demonstrated in prostate cancer [7, 12], in which expression of DKK3 was predominantly lost in high-grade prostatic tumours. Moreover, siRNA-mediated downregulation of DKK3 expression in prostate epithelial cells disrupted acinar morphogenesis [7], which taken with its prevalent expression in growth-arrested cells suggests a functional role of DKK3 in post-mitotic tissue differentiation processes. Whether DKK3 is also involved in maintaining glandular morphology in the normal mammary gland will be elucidated in a further study.
In human breast cancer, hypermethylation of Wnt antagonist genes was reported to be of clinical relevance. Both SFRP1 and SFRP5 methylation were shown to occur frequently and be tumour specific with a strong association to poor clinical patient outcome [34, 43]. An impact of DKK3 promoter methylation on cancer patient survival has been repeatedly found. It was shown to be associated with reduced disease-free survival in acute lymphoblastic leukaemia [20], and also with shorter overall survival in kidney cancer [45] and lung cancer [46]. Due to its functional properties as a potential tumour suppressor in human cancers including breast cancer [26], together with the finding that DKK3 methylation is a significant prognostic factor in three human malignancies, we speculate that DKK3 methylation might also bear prognostic power in breast cancer. This hypothesis is currently being approached in our laboratory in a further study.
In summary, we demonstrate for the first time that the Wnt antagonist gene DKK3 is a frequent target of epigenetic inactivation in human breast cancer, leading to downregulation of DKK3 mRNA and DKK3 protein expression in tumourous tissues. These results suggest a causative implication of DKK3 in the development of human breast cancer. Since DKK3 is believed to negatively regulate Wnt signalling, these results underline the pivotal role of a deregulated Wnt signalling pathway commonly found in this disease.
Conclusion
This study shows that the putative Wnt antagonist DKK3 is frequently downregulated in human breast cancer due to promoter methylation, whereas it is abundantly expressed and unmethylated in normal breast cell epithelium. Since promoter methylation is a primary cause to functionally inactivate tumour suppressor genes, DKK3 may act as a tumour suppressor in the human mammary gland. DKK3 is believed to particularly regulate non-canonical Wnt signalling. Therefore, we conclude that disruption of this Wnt pathway branch may add further tumour growth advantages to those already conferred by canonical Wnt/β-catenin signalling.
Abbreviations
- DKK3:
-
Dickkopf-3
- FC:
-
fold change
- IRS:
-
immunoreactivity score
- JNK:
-
c-Jun-kinase
- LRP:
-
LDL-receptor related protein
- MSP:
-
methylation-specific polymerase chain reaction
- PCP:
-
planar cell polarity pathway
- RT-PCR:
-
reverse transcription polymerase chain reaction
- SD:
-
standard deviation
- siRNA:
-
small interfering ribonucleic acid
- TSA:
-
trichostatin A.
References
Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C: Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998, 391: 357-362. 10.1038/34848.
Krupnik VE, Sharp JD, Jiang C, Robison K, Chickering TW, Amaravadi L, Brown DE, Guyot D, Mays G, Leiby K, Chang B, Duong T, Goodearl AD, Gearing DP, Sokol SY, McCarthy SA: Functional and structural diversity of the human Dickkopf gene family. Gene. 1999, 238: 301-313. 10.1016/S0378-1119(99)00365-0.
He X, Semenov M, Tamai K, Zeng X: LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: arrows point the way. Development. 2004, 131: 1663-1677. 10.1242/dev.01117.
Mao B, Wu W, Li Y, Hoppe D, Stannek P, Glinka A, Niehrs C: LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Nature. 2001, 411: 321-325. 10.1038/35077108.
Mao B, Niehrs C: Kremen2 modulates Dickkopf2 activity during Wnt/LRP6 signaling. Gene. 2003, 302: 179-183. 10.1016/S0378-1119(02)01106-X.
Reya T, Clevers H: Wnt signalling in stem cells and cancer. Nature. 2005, 434: 843-850. 10.1038/nature03319.
Kawano Y, Kitaoka M, Hamada Y, Walker MM, Waxman J, Kypta RM: Regulation of prostate cell growth and morphogenesis by Dickkopf-3. Oncogene. 2006, 25: 6528-6537. 10.1038/sj.onc.1209661.
Yue W, Sun Q, Dacic S, Landreneau RJ, Siegfried JM, Yu J, Zhang L: Downregulation of Dkk3 activates beta-catenin/TCF-4 signaling in lung cancer. Carcinogenesis. 2008, 29: 84-92. 10.1093/carcin/bgm267.
Hoang BH, Kubo T, Healey JH, Yang R, Nathan SS, Kolb EA, Mazza B, Meyers PA, Gorlick R: Dickkopf 3 inhibits invasion and motility of Saos-2 osteosarcoma cells by modulating the Wnt-beta-catenin pathway. Cancer Res. 2004, 64: 2734-2739. 10.1158/0008-5472.CAN-03-1952.
Tada M, Concha ML, Heisenberg CP: Non-canonical Wnt signalling and regulation of gastrulation movements. Semin Cell Dev Biol. 2002, 13: 251-260. 10.1016/S1084-9521(02)00052-6.
Henderson DJ, Phillips HM, Chaudhry B: Vang-like 2 and noncanonical Wnt signaling in outflow tract development. Trends Cardiovasc Med. 2006, 16: 38-45. 10.1016/j.tcm.2005.11.005.
Abarzua F, Sakaguchi M, Takaishi M, Nasu Y, Kurose K, Ebara S, Miyazaki M, Namba M, Kumon H, Huh NH: Adenovirus-mediated overexpression of REIC/Dkk-3 selectively induces apoptosis in human prostate cancer cells through activation of c-Jun-NH2-kinase. Cancer Res. 2005, 65: 9617-9622. 10.1158/0008-5472.CAN-05-0829.
Edamura K, Nasu Y, Takaishi M, Kobayashi T, Abarzua F, Sakaguchi M, Kashiwakura Y, Ebara S, Saika T, Watanabe M, Huh NH, Kumon H: Adenovirus-mediated REIC/Dkk-3 gene transfer inhibits tumor growth and metastasis in an orthotopic prostate cancer model. Cancer Gene Ther. 2007, 14: 765-772. 10.1038/sj.cgt.7701071.
Tanimoto R, Abarzua F, Sakaguchi M, Takaishi M, Nasu Y, Kumon H, Huh NH: REIC/Dkk-3 as a potential gene therapeutic agent against human testicular cancer. Int J Mol Med. 2007, 19: 363-368.
Nozaki I, Tsuji T, Iijima O, Ohmura Y, Andou A, Miyazaki M, Shimizu N, Namba M: Reduced expression of REIC/Dkk-3 gene in non-small cell lung cancer. Int J Oncol. 2001, 19: 117-121.
Kobayashi K, Ouchida M, Tsuji T, Hanafusa H, Miyazaki M, Namba M, Shimizu N, Shimizu K: Reduced expression of the REIC/Dkk-3 gene by promoter-hypermethylation in human tumor cells. Gene. 2002, 282: 151-158. 10.1016/S0378-1119(01)00838-1.
Tsuji T, Nozaki I, Miyazaki M, Sakaguchi M, Pu H, Hamazaki Y, Iijima O, Namba M: Antiproliferative activity of REIC/Dkk-3 and its significant down-regulation in non-small-cell lung carcinomas. Biochem Biophys Res Commun. 2001, 289: 257-263. 10.1006/bbrc.2001.5972.
Kurose K, Sakaguchi M, Nasu Y, Ebara S, Kaku H, Kariyama R, Arao Y, Miyazaki M, Tsushima T, Namba M, Kumon H, Huh NH: Decreased expression of REIC/Dkk-3 in human renal clear cell carcinoma. J Urol. 2004, 171: 1314-1318. 10.1097/01.ju.0000101047.64379.d4.
Hsieh SY, Hsieh PS, Chiu CT, Chen WY: Dickkopf-3/REIC functions as a suppressor gene of tumor growth. Oncogene. 2004, 23: 9183-9189.
Roman-Gomez J, Jimenez-Velasco A, Agirre X, Castillejo JA, Navarro G, Barrios M, Andreu EJ, Prosper F, Heiniger A, Torres A: Transcriptional silencing of the Dickkopfs-3 (Dkk-3) gene by CpG hypermethylation in acute lymphoblastic leukaemia. Br J Cancer. 2004, 91: 707-713.
Lodygin D, Epanchintsev A, Menssen A, Diebold J, Hermeking H: Functional epigenomics identifies genes frequently silenced in prostate cancer. Cancer Res. 2005, 65: 4218-4227. 10.1158/0008-5472.CAN-04-4407.
Urakami S, Shiina H, Enokida H, Kawakami T, Kawamoto K, Hirata H, Tanaka Y, Kikuno N, Nakagawa M, Igawa M, Dahiya R: Combination analysis of hypermethylated Wnt-antagonist family genes as a novel epigenetic biomarker panel for bladder cancer detection. Clin Cancer Res. 2006, 12: 2109-2116. 10.1158/1078-0432.CCR-05-2468.
Kuphal S, Lodermeyer S, Bataille F, Schuierer M, Hoang BH, Bosserhoff AK: Expression of Dickkopf genes is strongly reduced in malignant melanoma. Oncogene. 2006, 25: 5027-5036. 10.1038/sj.onc.1209508.
Sato H, Suzuki H, Toyota M, Nojima M, Maruyama R, Sasaki S, Takagi H, Sogabe Y, Sasaki Y, Idogawa M, Sonoda T, Mori M, Imai K, Tokino T, Shinomura Y: Frequent epigenetic inactivation of DICKKOPF family genes in human gastrointestinal tumors. Carcinogenesis. 2007, 28: 2459-2466. 10.1093/carcin/bgm178.
Licchesi JD, Westra WH, Hooker CM, Machida EO, Baylin SB, Herman JG: Epigenetic alteration of Wnt pathway antagonists in progressive glandular neoplasia of the lung. Carcinogenesis. 2008, 29: 895-904. 10.1093/carcin/bgn017.
Kawasaki K, Watanabe M, Sakaguchi M, Ogasawara Y, Ochiai K, Nasu Y, Doihara H, Kashiwakura Y, Huh NH, Kumon H, Date H: REIC/Dkk-3 overexpression downregulates P-glycoprotein in multidrug-resistant MCF7/ADR cells and induces apoptosis in breast cancer. Cancer Gene Ther. 2008, doi:10.1038/cgt.2008.58.
Tsuji T, Miyazaki M, Sakaguchi M, Inoue Y, Namba M: A REIC gene shows down-regulation in human immortalized cells and human tumor-derived cell lines. Biochem Biophys Res Commun. 2000, 268: 20-24. 10.1006/bbrc.1999.2067.
Namba M, Tsuji T: Early events during neoplastic transformation of human cells in vitro: Genetic and biological aspects of immortalization. Molecular Pathology of Early Cancer. Edited by: Srivastava S, Henson DE, Gazdar A. 1999, Amsterdam: IOS Press, 27-38.
Monaghan AP, Kioschis P, Wu W, Zuniga A, Bock D, Poustka A, Delius H, Niehrs C: Dickkopf genes are co-ordinately expressed in mesodermal lineages. Mech Dev. 1999, 87: 45-56. 10.1016/S0925-4773(99)00138-0.
Tavassoli FA, Devilee P: World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of the Breast and Female Genital Organs. 2003, Lyon: IARC Press
Untergasser G, Koch HB, Menssen A, Hermeking H: Characterization of epithelial senescence by serial analysis of gene expression: identification of genes potentially involved in prostate cancer. Cancer Res. 2002, 62: 6255-6262.
Sobin LH, Wittekind C, eds: TNM classification of malignant tumors. 1997, New York: Wiley Liss, 5
Elston EW, Ellis IO: Method for grading breast cancer. J Clin Pathol. 1993, 46: 189-190. 10.1136/jcp.46.2.189-b.
Veeck J, Niederacher D, An H, Klopocki E, Wiesmann F, Betz B, Galm O, Camara O, Dürst M, Kristiansen G, Huszka C, Knüchel R, Dahl E: Aberrant methylation of the Wnt antagonist SFRP1 in breast cancer is associated with unfavourable prognosis. Oncogene. 2006, 25: 3479-3488. 10.1038/sj.onc.1209386.
Remmele W, Stegner HE: [Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue]. Pathologe. 1987, 8: 138-140.
Das PM, Singal R: DNA methylation and cancer. J Clin Oncol. 2004, 22: 4632-4642. 10.1200/JCO.2004.07.151.
Esteller M: Aberrant DNA methylation as a cancer-inducing mechanism. Annu Rev Pharmacol Toxicol. 2005, 45: 629-656. 10.1146/annurev.pharmtox.45.120403.095832.
Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB: Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996, 93: 9821-9826. 10.1073/pnas.93.18.9821.
Román-Gómez J, Cordeu L, Agirre X, Jiménez-Velasco A, San José-Eneriz E, Garate L, Calasanz MJ, Heiniger A, Torres A, Prosper F: Epigenetic regulation of Wnt-signaling pathway in acute lymphoblastic leukemia. Blood. 2007, 109: 3462-3469. 10.1182/blood-2006-09-047043.
Polakis P: Wnt signaling and cancer. Genes Dev. 2000, 14: 1837-1851.
Lo PK, Mehrotra J, D'Costa A, Fackler MJ, Garrett-Mayer E, Argani P, Sukumar S: Epigenetic suppression of secreted frizzled related protein 1 (SFRP1) expression in human breast cancer. Cancer Biol Ther. 2006, 5: 281-286.
Suzuki H, Toyota M, Caraway H, Gabrielson E, Ohmura T, Fujikane T, Nishikawa N, Sogabe Y, Nojima M, Sonoda T, Mori M, Hirata K, Imai K, Shinomura Y, Baylin SB, Tokino T: Frequent epigenetic inactivation of Wnt antagonist genes in breast cancer. Br J Cancer. 2008, 98: 1147-1156. 10.1038/sj.bjc.6604259.
Veeck J, Geisler C, Noetzel E, Alkaya S, Hartmann A, Knüchel R, Dahl E: Epigenetic inactivation of the Secreted frizzled-related protein-5 (SFRP5) gene in human breast cancer is associated with unfavorable prognosis. Carcinogenesis. 2008, 29: 991-998. 10.1093/carcin/bgn076.
Ai L, Tao Q, Zhong S, Fields CR, Kim WJ, Lee MW, Cui Y, Brown KD, Robertson KD: Inactivation of Wnt inhibitory factor-1 (WIF1) expression by epigenetic silencing is a common event in breast cancer. Carcinogenesis. 2006, 27: 1341-1348. 10.1093/carcin/bgi379.
Urakami S, Shiina H, Enokida H, Hirata H, Kawamoto K, Kawakami T, Kikuno N, Tanaka Y, Majid S, Nakagawa M, Igawa M, Dahiya R: Wnt antagonist family genes as biomarkers for diagnosis, staging, and prognosis of renal cell carcinoma using tumor and serum DNA. Clin Cancer Res. 2006, 12: 6989-6997. 10.1158/1078-0432.CCR-06-1194.
Suzuki M, Shigematsu H, Nakajima T, Kubo R, Motohashi S, Sekine Y, Shibuya K, Iizasa T, Hiroshima K, Nakatani Y, Gazdar AF, Fujisawa T: Synchronous alterations of Wnt and epidermal growth factor receptor signaling pathways through aberrant methylation and mutation in non small cell lung cancer. Clin Cancer Res. 2007, 13: 6087-6092. 10.1158/1078-0432.CCR-07-0591.
Acknowledgements
The expert technical assistance of Sevim Alkaya, Sonja von Serényi and Inge Losen is greatly appreciated. We thank Dr Dieter Niederacher (Heinrich-Heine University, Düsseldorf, Germany) and Professor Matthias Dürst (Friedrich-Schiller University, Jena, Germany) for kindly providing patient samples. This work is a research project within the German Human Genome Project and has been supported by a Bundesministerium für Bildung und Forschung grant 01KW0401 to Edgar Dahl.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
Edgar Dahl has declared that he has submitted a patent application on the use of DKK3 promoter methylation. The other authors have no competing interests.
Authors' contributions
JV carried out the gene expression analyses, immunohistochemical studies, methylation experiments and statistical evaluations, participated in the conception and design of the study, and wrote the manuscript. NB participated in the immunohistochemical analysis, performed data interpretation and critically revised the manuscript. AH provided clinical samples and clinicopathological data, performed data interpretation, supported in statistical analyses and critically revised the manuscript. GK provided clinical samples and clinicopathological data, performed data interpretation and critically revised the manuscript. UH provided clinical samples and clinicopathological data, participated in data interpretation and critically revised the manuscript. RK participated in the design and co-ordination of the study and critically revised the manuscript. ED planned and co-ordinated the study, and critically revised the manuscript. All authors have given final approval of the text to be published.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Veeck, J., Bektas, N., Hartmann, A. et al. Wnt signalling in human breast cancer: expression of the putative Wnt inhibitor Dickkopf-3 (DKK3) is frequently suppressed by promoter hypermethylation in mammary tumours. Breast Cancer Res 10, R82 (2008). https://doi.org/10.1186/bcr2151
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1186/bcr2151