Abstract
Introduction
There is inconsistent association between urate transporters SLC22A11 (organic anion transporter 4 (OAT4)) and SLC22A12 (urate transporter 1 (URAT1)) and risk of gout. New Zealand (NZ) Māori and Pacific Island people have higher serum urate and more severe gout than European people. The aim of this study was to test genetic variation across the SLC22A11/SLC22A12 locus for association with risk of gout in NZ sample sets.
Methods
A total of 12 single nucleotide polymorphism (SNP) variants in four haplotype blocks were genotyped using TaqMan® and Sequenom MassArray in 1003 gout cases and 1156 controls. All cases had gout according to the 1977 American Rheumatism Association criteria. Association analysis of single markers and haplotypes was performed using PLINK and Stata.
Results
A haplotype block 1 SNP (rs17299124) (upstream of SLC22A11) was associated with gout in less admixed Polynesian sample sets, but not European Caucasian (odds ratio; OR = 3.38, P = 6.1 × 10-4; OR = 0.91, P = 0.40, respectively) sample sets. A protective block 1 haplotype caused the rs17299124 association (OR = 0.28, P = 6.0 × 10-4). Within haplotype block 2 (SLC22A11) we could not replicate previous reports of association of rs2078267 with gout in European Caucasian (OR = 0.98, P = 0.82) sample sets, however this SNP was associated with gout in Polynesian (OR = 1.51, P = 0.022) sample sets. Within haplotype block 3 (including SLC22A12) analysis of haplotypes revealed a haplotype with trans-ancestral protective effects (OR = 0.80, P = 0.004), and a second haplotype conferring protection in less admixed Polynesian sample sets (OR = 0.63, P = 0.028) but risk in European Caucasian samples (OR = 1.33, P = 0.039).
Conclusions
Our analysis provides evidence for multiple ancestral-specific effects across the SLC22A11/SLC22A12 locus that presumably influence the activity of OAT4 and URAT1 and risk of gout. Further fine mapping of the association signal is needed using trans-ancestral re-sequence data.
Similar content being viewed by others
Introduction
Hyperuricemia is a primary risk factor for gout [1], leading to the formation of monosodium urate crystals and gout in some people. Genome-wide association studies (GWAS) have identified 28 loci that account for approximately 6% of the variance in serum urate (SU) levels in European Caucasians [2]. Loci with the strongest effect encode proteins involved in secretion and renal filtration of uric acid. Two of these, SLC2A9 and ABCG2, have a very strong effect in gout (risk allele odds ratio (OR) >2.0) in multiple ancestral groups [2–5], whilst a third locus SLC17A1, has a weaker effect (OR <1.5) [2, 6, 7]. The SLC22A11 (OAT4) and SLC22A12 (URAT1) genes are also associated with SU at a genome-wide level of significance [2]. However, evidence for association with gout is ambiguous.
SLC22A11 (OAT4) and SLC22A12 (URAT1) encode renal urate transporters located together on chromosome 11. SLC22A12 is expressed across multiple tissue types and developmental stages, whilst SLC22A11 expression is restricted to the kidneys and placenta [8]. URAT1 is a high-affinity apical urate transporter [9], whilst OAT4 transports multiple organic anions and has low affinity for urate [10]. Both URAT1 and OAT4 may play a role in gout secondary to diuretic use due to competitive transport of uric acid and diuretics [9, 10].
Multiple single nucleotide polymorphisms (SNPs) at the SLC22A11/SLC22A12 locus have been associated with SU in European Caucasians, with two independent effects reported within the locus [2]. These SNPs are spread across a 1.3-Mb region and constitute four distinct linkage disequilibrium (LD) blocks, two of which contain SLC22A11 and SLC22A12[2]. This suggests a complex pattern of association with SU in European Caucasians, at least. In non-European populations, studies specifically on the SLC22A12 gene have revealed associations with SU in Chinese, Korean and Japanese sample sets that are consistent with the those reported in European Caucasians (rs505802 and SNPs in LD) [11–14] whilst additional associations are evident in Chinese (rs475688 and rs7932775, in low LD with rs505802) [14, 15].
Variants in the haplotype block that includes SLC22A12 have previously been tested for association with gout in European Caucasian with no association of rs505802 (OR = 0.99) [16] or rs478607 (OR = 0.97) [2]. There is conflicting evidence for association of common SLC22A12 variants with gout in Chinese sample sets [15, 17]. Rare variants in the SLC22A12 locus have been associated with SU and gout in African-Americans [18] and gout in Japanese [19] and Micronesians [20]. At SLC22A11, Köttgen et al.[2] reported association of the minor (C) allele of rs2078267 with prevalent gout in a meta-analysis of European Caucasian cases nested within population-based cohorts (OR = 1.16, P = 4.4 × 10-6). However, Stark et al. [16] reported no evidence for association of rs17300741 (OR = 1.02, P = 0.80; in very strong LD with rs2078267 (r2 = 0.96)) with gout. To our knowledge, no previous studies have tested for association of SLC22A11 with SU and gout in non-European populations.
There is ambiguity regarding association of SLC22A11 and SLC22A12 with gout - previous reports have studied different population groups, with various methods for determination of gout; clinical ascertainment using 1977 American Rheumatism Association (ARA) criteria [15, 17], self-report of physician-diagnosed gout, use of gout medications [2, 18], a combination of self-report and examination of medical records [16]. Here our aim was to clarify the association with gout according to the ARA classification criteria at the SLC22A11-SLC22A12 locus in multiple ancestral groups drawn from the New Zealand (NZ) population, including Māori and Pacific Island. These groups have a prevalence of gout double that of European Caucasians [21], with earlier onset and more severe gout and a higher prevalence of co-morbidities [6].
Methods
Study participants
All gout cases (Additional file 1: Table S1; n = 1,003) had a confirmed diagnosis of gout as defined by the 1977 ARA preliminary classification criteria for acute gout [22] and were recruited from community-based settings, primary and secondary care. Controls (n = 1,156) self-reported having no history of arthritis and were convenience-sampled from workplaces and community focal points in the Auckland region of NZ. Co-morbid conditions (type 2 diabetes, renal disease, hypertension, dyslipidemia and heart disease) were self-reported. Ethical approval was given by the NZ Multi-Region Ethics Committee and all participants provided written informed consent for the collection of samples and subsequent analysis.
Participants were divided into four sample sets according to self-reported grandparental ancestry; NZ European Caucasian (420 cases, 638 controls), Eastern Polynesian (EP) - NZ Māori and Cook Island Māori (315 cases, 349 controls), Western Polynesian (WP) - Tongan, Samoan and Niuean (249 cases, 144 controls), and a small mixed eastern and western ancestry group (EP/WP) (19 cases, 25 controls) (Additional file 1: Table S1). These groupings were based on previous evidence for genetic heterogeneity between Eastern and Western Polynesia (see [5] and references therein). The EP sample set (predominantly NZ Māori) was further split into two groups according to Polynesian ancestry estimates obtained using 67 genomic control markers [6]. Individuals of higher EP ancestry formed the EP(High) sample set (256 cases, 187 controls) and those of lower ancestry formed the EP(Low) sample set (59 cases, 162 controls).
SNP selection
Data on 1,000 genomes [23] for Caucasian (CEU) and Han Chinese (CHB), as the most closely related and available population to the NZ Māori and Pacific populations, was used to define haplotype blocks based on all the SNPs previously associated with SU in GWAS studies within the 1.3-Mb region of chromosome 11 (Additional file 2: Figure S1). Four distinct haplotype blocks were identified (block 1: approximately 26Kb upstream, Block 2: SLC22A11, Block 3: including SLC22A12, and block 4: approximately 840Kb downstream) (Additional file 1: Table S2) and this information was then used to select variants for genotyping (Additional file 2: Figure S2). SNPs were selected largely on the basis of previous literature reports of association with SU. For block 1 and block 4 both the SNP most associated with SU (rs17299124; β = −3.75 μmolL-1, P = 8.6 × 10-16 and rs642803; β = −2.56 μmolL-1, P = 4.5 × 10-14, respectively) and the most associated tag-SNP (in high LD with all other associated SNPs (rs475414; β = −2.38 μmolL-1, P = 3.1 × 10-12 and rs12289836; β = −2.62 μmolL-1, P = 6.6 × 10-14, respectively) were chosen [2, 18, 24]. For block 2 (SLC22A11) one tag-SNP common to CEU and CHB was identified (rs693591) and combined with the previously studied rs17300741 and rs2078267[2, 24]. One tag-SNP for block 3 (SLC22A12) was selected (rs476037) and included with the previously reported SNPs rs3825018 (in complete LD with rs505802), rs475688 and rs7932775[2, 24]. A fifth SNP (rs478607) from block 3 was also added as representative of a second (weaker) effect potentially independent of rs2078267 (SLC22A12) ([2]; β = −2.80 μmolL-1, P = 4.4 × 10-11) (Additional file 2: Figure S3, S4). Three of the twelve chosen SNPs have a predicted biological function according to FuncPred [25]; rs3825018 is situated in a potential transcription factor binding site and may enhance or silence splicing, rs7932775 may also enhance or silence splicing and rs476037 is situated at a potential miRNA binding site.
Genotyping
SNPs were genotyped using either TaqMan® assays (Applied Biosystems, Foster City, CA, USA) in a Lightcycler® 480 Real-Time PCR System (Roche, Indianapolis, IN, USA) (rs693591 (assay ID C_925815_10), rs17300741 (C_3234214_10), rs3825018 (C_27162391_10)), rs475688 (C_925797_10), rs476037 (C_3108662_20), rs7932775 (C_3108664_10), or the MassArray system (Sequenom iPLEXassay, San Diego, CA, USA): rs17299124, rs642803, rs475414, rs12289836, rs478607 and rs2078267.
Association analysis
Allelic ORs were obtained from a sex-adjusted logistic regression in PLINK [26]. These were relevant to the allele in-phase with the rs3825018 G allele as determined using 1,000 genomes LD data. To assess experiment-wide significance a Bonferroni correction was applied to P-values. A conservative correction factor of 48 was applied to the single SNP analysis (P <0.00104; 12 SNPs meta-analyzed by inverse-variance weighting over four sample sets - EP(High), WP, EP/WP), all Polynesian (including EP(Low)), European Caucasian and EP(Low), and all five sample sets combined) and a factor of 12 for the haplotype analysis (P <0.0042; meta-analysis of three haplotype blocks over four sample sets). Interaction terms for co-morbid phenotypes and ancestry were included in the logistic regression model where appropriate.
Ancestrally defined sample sets were combined by an inverse-variance weighted fixed-effect method, using STATA 8.0 software [27]. A Q-statistic was calculated to determine the heterogeneity between cohorts and for SNPs showing heterogeneity (PHet <0.05) the fixed-effect model was replaced with a random-effect model. Power was calculated for a nominal P = 0.05 (Additional file 2: Figure S5) [28]. All sample sets were adequately powered (>80%) to detect moderate effects (OR = 1.5) at minor allele frequency >0.1.
Haplotype association analysis
Haplotype imputation by expectation maximization was performed in PLINK to produce most-likely haplotype calls (haplotypes with a frequency <0.01 were excluded). These data were then used to perform a sex-adjusted logistic regression in PLINK, producing ORs and P- values. Inverse-variance weighted fixed-effect analysis in STATA 8.0 was performed to combine the independent datasets (as described for the allelic association analysis).
Locus-specific gout genetic risk score
A weighted genetic risk score was calculated using the most associated SNP in each of block one to three. The gout risk score calculation was ((rs17299124(C) × β) + (rs2078267(C) × β) + (rs3825018(G) × β)), where each SNP ID followed by an allele in brackets denotes the allele count per individual, and the β after each SNP denotes the natural log of the ancestry-specific OR for that allele. Individuals who did not have complete genotype information (9.22%) were excluded.
The mean genetic risk score was calculated for each sample set and a two-tailed t-test performed to identify any significant differences between case and control values. A further sex-adjusted logistic regression was performed to test association with gout. All risk-score analysis was conducted in STATA 8.0.
Results
Allelic associations
The European Caucasian sample set revealed nominally significant (P <0.05) associations with gout at two block-3 SNPs: rs475688 and rs7932775 (SLC22A12; OR = 1.26, P = 0.043; OR = 1.32, P = 0.033, respectively) (Table 1). The EP(High) sample set revealed four nominally significant associations within block 1 (rs17299124; OR = 2.77, P = 0.019), block 2 (SLC22A11) (rs693591; OR = 1.41, P = 0.031), and block 3 (SLC22A12) (rs3825018; OR = 1.46, P = 0.028, and rs476037; OR = 1.56, P = 0.022). The EP(Low) sample set showed no significant associations whereas the WP sample set revealed association with the block-1 SNP rs17299124 (OR = 5.65, P = 0.011) and the block-3 SNP rs7932775 (OR = 1.49, P = 0.019). The mixed Eastern and Western Polynesian sample set revealed a nominally significant association at rs693591 (OR = 4.55, P = 0.025) (Table 1).
Within block 1, observing that the effect of rs1729914 was stronger in the less admixed Polynesian sample sets (EP(High), WP, EP/WP) (Table 1), we combined these sample sets by inverse-variance weighting for rs17299124 and tested for association with gout in these sample sets, revealing experiment-wide evidence for association (Table 2; OR = 3.38, P = 6.1 × 10-4). The heterogeneity P- value of 0.010 (in the combined analysis of all ancestries) suggested an ancestral specific effect. Based on this, all SNPs were combined by inverse-variance weighting in four groups - less admixed Polynesian (EP(High), WP, EP/WP), all Polynesian (including EP(Low)), European Caucasian and EP(Low), and the five sample sets combined. Of the other SNPs, only block-3 rs3825018 approached experiment-wide significance in all ancestries combined (OR = 1.27, P = 0.002). Because no nominal association with block 4 was seen in any sample set the relevant SNPs (rs12289836 and rs642803) were not included in further analyses.
Block-1 to −3 SNPs rs17299124, rs2078267 and rs3825018 were tested for association with SU levels in controls (Additional file 1: Table S3). The only nominally significant associations observed (P <0.05) were with rs2078267 in less admixed Polynesian and total Polynesian (β = 0.044 mmolL-1, P = 0.007 and β = 0.030 mmolL-1, P = 0.022 respectively). The minor allele increased SU, consistent with the increased risk mediated for gout by the minor allele in these sample sets (Table 2). These associations were, however, not significant after correcting P- values for multiple testing (n = 12).
Interaction terms were included in the sex-adjusted logistic regression analyses in order to test for interaction between genotype (of the most associated SNP in blocks 1 to 3), gout and body mass index, type 2 diabetes, dyslipidemia, heart disease and hypertension in the European Caucasian, less admixed Polynesian and total Polynesian samples (Additional file 1: Table S4). There were no significant interactions observed (after correcting for the number of tests done, n = 45). We also tested for interaction with ancestry with significant interaction observed, after correcting for the number of tests done (n = 3), with ancestry for block-1 SNP rs17299124 (Pc = 9.9 × 10-6) and block-2 SNP rs2078267 (Pc = 2.6 × 10-5). The interaction with ancestry at rs17299124 was consistent with the PHet = 0.01 in the inverse-variance weighted analysis with the rs2078267 association also specific to Polynesian (Table 2). The PInteraction at rs3825018 was 0.033.
Haplotype association analysis
We initially combined by inverse-variance weighting haplotypes in the same combinations of sample sets as used in Tables 2 and 3. At block 1 there was a haplotype (rs475414 (allele T) - rs17299124 (allele A)) that exhibited significant heterogeneity (PHet = 0.007) in the combined analysis of all ancestries. Consistent with the rs17299124 data the effect, which was protective, was restricted to the less admixed Polynesian sample sets (OR = 0.28, P = 6.0 × 10-4). This protective T-A haplotype was rarer in the less admixed Polynesian sample set (Table 4 control frequency 0.044-0.052) compared with EP(Low) 0.203 and European Caucasian 0.245.
At block 2 no haplotype association was observed. At block 3 the only other experiment- wide significant association was observed, a protective haplotype in the combined sample set (A-C-T-G-A; OR = 0.80, P = 0.004). This haplotype was more common in European Caucasian (control frequency of 0.717) than Polynesian (control frequency range of 0.303-0.545). The block-3 G-T-C-G-G haplotype showed evidence for heterogeneity (PHet = 0.014) in the combined analysis of all ancestries – closer examination showed that this owed to a risk effect in European Caucasian and more admixed Polynesian (EP(Low)) (OR = 1.41, P = 0.007) and a protective effect in less admixed Polynesian (OR = 0.63, P = 0.028) (Tables 3 and 4). The frequency of this haplotype varied considerably, even within the Polynesian sample sets (Table 4 control frequency: WP 0.034, EP(High) 0.139, EP(Low) 0.131, European Caucasian 0.133).
Gout genetic risk-score association
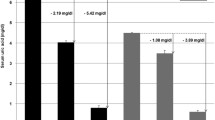
A weighted genetic risk score was calculated to determine the cumulative effect of the approximately 360-Kb (block 1 to block 3) region on gout risk, using the most associated marker in each ancestral block. Genetic risk scores ranged from 0 to 18.03 with no European Caucasian individual risk score being >7 (range 0 to 6.21). In each sample set the average case risk-score was significantly larger than the average control risk-score (Table 5). There was a significant association between gout and the genetic risk score in EP(High) (OR = 1.22, P = 0.002) and WP (OR = 1.14, P = 0.041) but not in European Caucasian (OR = 1.01, P = 0.80), EP(Low) (OR = 1.03, P = 0.77) or the small EP/WP sample set (OR = 1.32, P = 0.21).
Discussion
We analyzed three haplotype blocks - a region upstream of SLC22A11 containing no known genes (block 1), SLC22A11 (block 2) and SLC22A12 (block 3). Collectively our data indicate a complex pattern of association that is ancestry-dependent, with risk appearing to be driven by protective haplotypes that are at a higher frequency in European Caucasian than in people of Polynesian ancestry. The genetic risk-score analysis (Table 5) emphasized the relative important of the locus on the risk of gout in less admixed Polynesian (% variance explained 1.4 to 1.9) compared to the more admixed Polynesian and European Caucasian sample sets (<0.1% variance explained). Common variation in this locus is likely to be part of the explanation for why Polynesian people exhibit higher SU and a higher prevalence of gout [6, 21].
The block-1 region has previously never been studied for association with gout. The single SNP data of rs17299124 showed the strongest effect in the less admixed Polynesian sample sets (OR = 3.38) with non-significant effects closer to OR = 1.00 in the more admixed Eastern Polynesian and European Caucasian sample sets (OR = 1.20 and 0.91, respectively). This SNP is therefore a novel candidate risk variant to test for association with gout in South East Asian populations (ancestrally related to Polynesian). Haplotype analysis indicated that this effect in less admixed Polynesians is driven by a genetic variant that protects from gout. The lack of any known gene within the region (Ensembl release 70, January 2013) suggests that the effect seen at rs17299124 may operate by influencing the expression of a nearby gene, perhaps SLC22A11. Previously rs2186571 has been associated with SU levels in the Pacific Micronesian population of Kosrae [20]. This SNP, which maps several hundred kilobases upstream of SLC22A11, further implicates this region in the etiology of gout. There are two inversions and a deletion spanning this region [29], possibly able to cause a difference in the regulation of SLC22A11 or SLC22A12 expression.
At block 2 (SLC22A11) rs2078267 has previously been convincingly associated with gout in European Caucasian (OR = 0.88, P = 2.3 × 10-5; [2]) but the NZ European Caucasian sample set is inconsistent with these findings (OR = 0.98, P = 0.82). While low power may be a factor, we expected to observe an effect size consistent with that reported by Köttgen et al.[2] (OR = 1.14 to the C allele). It is possible that differences in phenotype among gout cases are important at SLC22A11. In the Köttgen et al. study the association was restricted to the prevalent gout cases (OR = 0.86, P = 4.4 × 10-6) and not observed in the incident cases (OR = 0.96, P = 0.55). Ascertainment in the prevalent group was dominated by a combination of self-report or use of urate-lowering medication and colchicine (prescribed for alleviating pain from acute gout), whereas the incident group was ascertained, as were the NZ gout cases (OR = 0.98, P = 0.82), by use of the ARA classification criteria for gout, a superior ascertainment method [30] (although the NZ cases were ascertained by clinical examination whereas the incident ones by a self-administered questionnaire [31]). It is therefore counter-intuitive that the association of SLC22A11 should be restricted to gout cases ascertained by less stringent criteria. Understanding this intriguing paradox may reveal new information on the role of SLC22A11 (OAT4) in the control of SU and in the risk of gout. There are physiological data showing that OAT4 mediates hydrochlorothiazide-induced hyperuricemia [10] and also evidence for interaction of rs2078267 with diuretic use in determining the risk of gout [32]. It is possible that non-additive interaction with diuretics could be a factor in the inconsistencies. In the Polynesian analysis there was nominal evidence for association with gout at rs2078267, with the minor allele conferring risk (OR = 1.51, P = 0.022), consistent with the urate-increasing effect of this allele in European Caucasians [2]. However this association was not significant when corrected for multiple testing. Given the lower allele frequency of this variant in Polynesians (<0.10 in the less admixed samples), a larger sample size is required for more robust conclusions about the possible role of SLC22A11 in gout in Polynesian people.
At SLC22A12 the clearest findings came from analysis of haplotype association. There was a gout-protective variant associated in the combined analysis (A-C-T-G-A; OR = 0.80, P = 0.004), with a consistent direction of effect in Polynesians and European Caucasians. There was also a haplotype with ancestral specific effects; G-T-C-G-G was protective in the less admixed Polynesian sample sets, yet conferred risk in the more admixed EP and European Caucasian sample sets. Our data therefore suggest that multiple common variants within the SLC22A12 locus contribute to the risk of gout, in a population-dependent manner. In the European Caucasian sample set there were nominal P- values for rs475688 and rs7932775 of 0.043 (OR = 1.26) and 0.033 (OR = 1.32), respectively. This is, to our knowledge, the first report of nominally significant association of SLC22A12 with gout in European Caucasians. However, given the likely weak effect, confirmation of this will require larger gout sample sets.
The block-3 SNP rs475688, in moderate LD (r2 = 0.80) with rs3825018 in European Caucasians but low LD in CHB and Polynesians (r2 = 0.33 and 0.36, respectively) (Additional file 2: Figures S2 and S3), showed weak evidence for association with gout in the NZ European Caucasian sample set (OR = 1.26, P = 0.04) consistent with the minor (T) allele increasing SU levels in Caucasians (β = 3.28 μmolL-1, P = 1.3 × 10-17) [2]. This variant, however, has been associated with gout and SU in Han Chinese and Solomon Island (Melanesian) sample sets with the T allele conferring protection against gout (OR = 0.54, P = 1 × 10-4) and decreasing serum urate levels (β = −11.90 μmolL-1, P = 0.02) [15], a direction of association opposite to that observed in NZ European Caucasians. This opposite effect between Polynesians and European Caucasians at rs475688 is consistent with the opposing direction of association seen with the block-3 G-T-C-G-G haplotype (rs475688 is the second marker) between less admixed Polynesians, and European Caucasians and the more admixed Eastern Polynesians. It will be informative to test this haplotype for association with SU and gout in Asian and other Pacific populations.
Conclusions
We present several important findings: 1) novel association of rs17299124 upstream of SLC22A11 with gout in Polynesians and not Caucasians, which is driven by a protective effect; 2) evidence for association of the SLC22A11 SNP rs2078267 with gout in Polynesians with the previously reported association in European Caucasians not replicated; and 3) the SLC22A12 analysis was the first report of nominally significant association with gout in European Caucasians, with the haplotype analysis suggesting multiple alleles conferring risk at SLC22A12 in an ancestry-specific manner.
Abbreviations
- ARA:
-
American Rheumatism Association
- CEU:
-
Centre d’Etude du Polymorphisme Humain from Utah
- CHB:
-
Han Chinese from Beijing
- EP:
-
Eastern Polynesian
- GWAS:
-
Genome-wide association studies
- LD:
-
Linkage disequilibrium
- NZ:
-
New Zealand
- OAT4:
-
Organic anion transporter 4
- OR:
-
Odds ratio
- SNP:
-
Single nucleotide polymorphism
- SU:
-
Serum urate
- URAT1:
-
Urate transporter 1
- WP:
-
Western Polynesian
- NZ:
-
New Zealand.
References
Campion EW, Glynn RJ, DeLabry LO: Asymptomatic hyperuricemia. Risks and consequences in the Normative Aging Study. Am J Med. 1987, 82: 421-426. 10.1016/0002-9343(87)90441-4.
Köttgen A, Albrecht E, Teumer A, Vitart V, Krumsiek J, Hundertmark C, Pistis G, Ruggiero D, O’Seaghdha CM, Haller T, Yang Q, Tanaka T, Johnson AD, Kutalik Z, Smith AV, Shi J, Struchalin M, Middelberg RP, Brown MJ, Gaffo AL, Pirastu N, Li G, Hayward C, Zemunik T, Huffman J, Yengo L, Zhao JH, Demirkan A, Feitosa MF, Liu X: Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet. 2013, 45: 145-154.
Hollis-Moffatt JE, Xu X, Dalbeth N, Merriman ME, Topless R, Waddell C, Gow PJ, Harrison AA, Highton J, Jones PB, Stamp LK, Merriman TR: Role of the urate transporter SLC2A9 gene in susceptibility to gout in New Zealand Maori, Pacific Island, and Caucasian case-control sample sets. Arthritis Rheum. 2009, 60: 3485-3492. 10.1002/art.24938.
Matsuo H, Takada T, Ichida K, Nakamura T, Nakayama A, Ikebuchi Y, Ito K, Kusanagi Y, Chiba T, Tadokoro S, Takada Y, Oikawa Y, Inoue H, Suzuki K, Okada R, Nishiyama J, Domoto H, Watanabe S, Fujita M, Morimoto Y, Naito M, Nishio K, Hishida A, Wakai K, Asai Y, Niwa K, Kamakura K, Nonoyama S, Sakurai Y, Hosoya T: Common defects of ABCG2, a high-capacity urate exporter, cause gout: a function-based genetic analysis in a Japanese population. Sci Transl Med. 2009, 1: 5ra11-
Phipps-Green AJ H-MJ, Dalbeth N, Merriman ME, Topless R, Gow PJ, Harrison AA, Highton J, Jones PB, Stamp LK, Merriman TR: A strong role for the ABCG2 gene in susceptibility to gout in New Zealand Pacific Island and Caucasian, but not Maori, case and control sample sets. Hum Mol Genet. 2010, 19: 4813-4819. 10.1093/hmg/ddq412.
Hollis-Moffatt JE, Phipps-Green AJ, Chapman B, Jones GT, van Rij A, Gow PJ, Harrison AA, Highton J, Jones PB, Montgomery GW, Stamp LK, Dalbeth N, Merriman TR: The renal urate transporter SLC17A1 locus: confirmation of association with gout. Arthritis Res Ther. 2012, 14: R92-10.1186/ar3816.
Urano W, Taniguchi A, Anzai N, Inoue E, Kanai Y, Yamanaka M, Endou H, Kamatani N, Yamanaka H: Sodium-dependent phosphate cotransporter type 1 sequence polymorphisms in male patients with gout. Ann Rheum Dis. 2010, 69: 1232-1234. 10.1136/ard.2008.106856.
Bleasby KCJ, Roberts CJ, Cheng C, Bailey WJ, Sina JF, Kulkarni AV, Hafey MJ, Evers R, Johnson JM, Ulrich RG, Slatter JG: Expression profiles of 50 xenobiotic transporter genes in humans and pre-clinical species: a resource for investigations into drug disposition. Xenobiotica. 2006, 36: 963-988. 10.1080/00498250600861751.
Enomoto AKH, Chairoungdua A, Shigeta Y, Jutabha P, Cha SH, Hosoyamada M, Takeda M, Sekine T, Igarashi T, Matsuo H, Kikuchi Y, Oda T, Ichida K, Hosoya T, Shimokata K, Niwa T, Kanai Y, Endou H: Molecular identification of a renal urate anion exchanger that regulates blood urate levels. Nature. 2002, 417: 447-452.
Hagos YSD, Ugele B, Burckhardt G, Bahn A: Human renal organic anion transporter 4 operates as an asymmetric urate transporter. J Am Soc Nephrol. 2007, 18: 430-439. 10.1681/ASN.2006040415.
Jang WCNY, Park SM, Ahn YC, Park SH, Choe JY, Shin IH, Kim SK: T6092C polymorphism of SLC22A12 gene is associated with serum uric acid. Clin Chim Acta. 2008, 398: 140-144. 10.1016/j.cca.2008.09.008.
Jang WCNY, Ahn YC, Park SM, Yoon IK, Choe JY, Park SH, Her M, Kim SK: G109T polymorphism of SLC22A12 gene is associated with serum uric acid level, but not with metabolic syndrome. Rheumatol Int. 2012, 32: 2257-2263. 10.1007/s00296-011-1952-5.
Kamatani Y, Matsuda K, Okada Y, Kubo M, Hosono N, Daigo Y, Nakamura Y, Kamatani N: Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat Genet. 2010, 4: 210-215.
Li C, Han L, Levin AM, Song H, Yan S, Wang Y, Wang Y, Meng D, Lv S, Ji Y, Xu X, Liu X, Wang Y, Zhou L, Miao Z, Mi QS: Multiple single nucleotide polymorphisms in the human urate transporter 1 (hURAT1) gene are associated with hyperuricaemia in Han Chinese. J Med Genet. 2010, 47: 204-210. 10.1136/jmg.2009.068619.
Tu HPCC, Lee CH, Tovosia S, Ko AM, Wang SJ, Ou TT, Lin GT, Chiang SL, Chiang HC, Chen PH, Chang SJ, Lai HM, Ko YC: The SLC22A12 gene is associated with gout in Han Chinese and Solomon Islanders. Ann Rheum Dis. 2010, 69: 1252-1254. 10.1136/ard.2009.114504.
Stark KRW, Grassl M, Erdmann J, Schunkert H, Illig T, Hengstenberg C: Common polymorphisms influencing serum uric acid levels contribute to susceptibility to gout, but not to coronary artery disease. PLoS One. 2009, 4: e7729-10.1371/journal.pone.0007729.
Guan M, Zhang J, Chen Y, Liu W, Kong N, Zou H: High-resolution melting analysis for the rapid detection of an intronic single nucleotide polymorphism in SLC22A12 in male patients with primary gout in China. Scand J Rheumatol. 2009, 38: 276-281. 10.1080/03009740802572483.
Tin AWO, Kao WH, Liu CT, Lu X, Nalls MA, Shriner D, Semmo M, Akylbekova EL, Wyatt SB, Hwang SJ, Yang Q, Zonderman AB, Adeyemo AA, Palmer C, Meng Y, Reilly M, Shlipak MG, Siscovick D, Evans MK, Rotimi CN, Flessner MF, Köttgen M, Cupples LA, Fox CS, Köttgen A, CARe and CHARGE Consortia: Genome-wide Association Study for Serum Urate Concentrations and Gout among African Americans Identifies Genomic Risk Loci and a Novel URAT1 Loss-of-Function Allele. Hum Mol Genet. 2011, 20: 4056-4068. 10.1093/hmg/ddr307.
Taniguchi AUW, Yamanaka M, Yamanaka H, Hosoyamada M, Endou H, Kamatani N: A common mutation in an organic anion transporter gene, SLC22A12, is a suppressing factor for the development of gout. Arthritis Rheum. 2005, 52: 2576-2577. 10.1002/art.21242.
Kenny EE, Kim M, Gusev A, Lowe JK, Salit J, Smith JG, Kovvali S, Kang HM, Newton-Cheh C, Daly MJ, Stoffel M, Altshuler DM, Friedman JM, Eskin E, Breslow JL, Pe’er I: Increased power of mixed models facilitates association mapping of 10 loci for metabolic traits in an isolated population. Hum Mol Genet. 2011, 20: 827-839. 10.1093/hmg/ddq510.
Winnard D, Wright C, Taylor WJ, Jackson G, Te Karu L, Gow PJ, Arroll B, Thornley S, Gribben B, Dalbeth N: National prevalence of gout derived from administrative health data in Aotearoa New Zealand. Rheumatology (Oxford). 2012, 51: 901-909. 10.1093/rheumatology/ker361.
Wallace SL, Robinson H, Masi AT, Decker JL, McCarty DJ, Yu TF: Preliminary criteria for the classification of the acute arthritis of primary gout. Arthritis Rheum. 1977, 20: 895-900. 10.1002/art.1780200320.
1000 genomes: a deep catalog of human genetic variation. [http://www.1000genomes.org]
Yang Q, Köttgen A, Dehghan A, Smith AV, Glazer NL, Chen MH, Chasman DI, Aspelund T, Eiriksdottir G, Harris TB, Launer L, Nalls M, Hernandez D, Arking DE, Boerwinkle E, Grove ML, Li M, Linda Kao WH, Chonchol M, Haritunians T, Li G, Lumley T, Psaty BM, Shlipak M, Hwang SJ, Larson MG, O’Donnell CJ, Upadhyay A, van Duijn CM, Hofman A: Multiple genetic loci influence serum urate levels and their relationship with gout and cardiovascular disease risk factors. Circ Cardiovasc Genet. 2010, 3: 523-530. 10.1161/CIRCGENETICS.109.934455.
SNP function prediction. [http://snpinfo.niehs.nih.gov/snpinfo/snpfunc.htm]
Purcell S, Neale B, Todd , Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007, 81: 559-575. 10.1086/519795.
Stata | Data analysis and statistical software. [http://www.stata.com/]
Johnson GC, Esposito L, Barratt BJ, Smith AN, Heward J, Di Genova G, Ueda H, Cordell HJ, Eaves IA, Dudbridge F, Twells RC, Payne F, Hughes W, Nutland S, Stevens H, Carr P, Tuomilehto-Wolf E, Tuomilehto J, Gough SC, Clayton DG, Todd JA: Haplotype tagging for the identification of common disease genes. Nat Genet. 2001, 29: 233-237. 10.1038/ng1001-233.
Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, Taillon BE, Chen Z, Tanzer A, Saunders AC, Chi J, Yang F, Carter NP, Hurles ME, Weissman SM, Harkins TT, Gerstein MB, Egholm M, Snyder M: Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007, 318: 420-426. 10.1126/science.1149504.
Taylor W: Diagnosis of gout: considering clinical and research settings. Curr Rheum Rev. 2011, 7: 97-105. 10.2174/157339711795304988.
Choi HK, Atkinson K, Karlson EW, Willett W, Curhan G: Alcohol intake and risk of incident gout in men: a prospective study. Lancet. 2004, 363: 1277-1281. 10.1016/S0140-6736(04)16000-5.
McAdams-DeMarco MA, Maynard JW, Baer AN, Kao LW, Kottgen A, Coresh J: A urate gene-by-diuretic interaction and gout risk in participants with hypertension: results from the ARIC study. Ann Rheum Dis. 2013, 72: 701-706. 10.1136/annrheumdis-2011-201186.
Acknowledgements
This work was supported by the Health Research Council of New Zealand, Arthritis New Zealand, New Zealand Lottery Health, Genetics Otago and the University of Otago. The authors would like to thank Chris Franklin, Jill Drake, Roddi Laurence and Gabrielle Sexton for assistance in recruitment.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
TJF, JEH-M, and TRM helped to design the study, oversee its execution, and prepare the manuscript. AP-G, MEM, RT, LKS and ND helped to provide clinical recruitment and prepare the manuscript. BC and GWM helped to collect data and prepare the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
13075_2013_4084_MOESM1_ESM.docx
Additional file 1: Table S1: Demographic and clinical characteristics of study participants. Table S2: Haplotype block summary. Table S3: Association of block-1 to −3 single nucleotide polymorphisms (SNPs) with serum urate in control individuals. Table S4: Interaction analysis between genotype, gout and co-morbid phenotypes. (DOCX 114 KB)
13075_2013_4084_MOESM2_ESM.docx
Additional file 2: Figure S1: Intermarker linkage disequilibrium of single nucleotide polymorphisms (SNPs) previously associated with serum urate. Figure S2: Common linkage disequilbrium block haplotypes in European Caucasian and Han Chinese. Figure S3: Intermarker linkage disequilibrium between the 12 genoytped SNPs in European Caucasian and Han Chinese. Figure S4: Intermarker linkage disequilibrium between the 12 genoytped SNPs in New Zealand sample sets. Figure S5: Power calculations. (DOCX 3 MB)
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Flynn, T.J., Phipps-Green, A., Hollis-Moffatt, J.E. et al. Association analysis of the SLC22A11 (organic anion transporter 4) and SLC22A12 (urate transporter 1) urate transporter locus with gout in New Zealand case-control sample sets reveals multiple ancestral-specific effects. Arthritis Res Ther 15, R220 (2013). https://doi.org/10.1186/ar4417
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/ar4417