Abstract
The transcription factor SOX9 is important in maintaining the chondrocyte phenotype. To identify novel genes regulated by SOX9 we investigated changes in gene expression by microarray analysis following retroviral transduction with SOX9 of a human chondrocytic cell line (SW1353). From the results the expression of a group of genes (SRPX, S100A1, APOD, RGC32, CRTL1, MYBPH, CRLF1 and SPINT1) was evaluated further in human articular chondrocytes (HACs). First, the same genes were investigated in primary cultures of HACs following SOX9 transduction, and four were found to be similarly regulated (SRPX, APOD, CRTL1 and S100A1). Second, during dedifferentiation of HACs by passage in monolayer cell culture, during which the expression of SOX9 progressively decreased, four of the genes (S100A1, RGC32, CRTL1 and SPINT1) also decreased in their expression. Third, in samples of osteoarthritic (OA) cartilage, which had decreased SOX9 expression compared with age-matched controls, there was decreased expression of SRPX, APOD, RGC32, CRTL1 and SPINT1. The results showed that a group of genes identified as being upregulated by SOX9 in the initial SW1353 screen were also regulated in expression in healthy and OA cartilage. Other genes initially identified were differently expressed in isolated OA chondrocytes and their expression was unrelated to changes in SOX9. The results thus identified some genes whose expression appeared to be linked to SOX9 expression in isolated chondrocytes and were also altered during cartilage degeneration in osteoarthritis.
Similar content being viewed by others
Introduction
The chondrocytes within articular cartilage are responsible for the maintenance of the specialized extracellular matrix (ECM) of the tissue and for its biomechanical properties. The chondrocyte phenotype is characterized by the expression of specific genes, such as collagen type II and the transcription factor SOX9 [1]. Collagen type II is an abundant component in the cartilage ECM and is essential for its integrity. Damage to collagen type II and loss of other cartilage ECM components occur during degenerative joint diseases such as osteoarthritis (OA), which result in severe disability and present a major health problem in the ageing population [2]. This may arise from complex pathogenic mechanisms, which result in decreased matrix synthesis and upregulated pathways of tissue degradation [3]. Characteristic of cartilage in OA are changes in the expression of ECM genes and the downregulation of the key chondrogenic transcription factor SOX9 [4, 5].
A large number of cartilage matrix genes have been shown to come under the transcriptional control of SOX9. They include COL2A1, COL9A1, COL11A2, aggrecan and cartilage link protein (CRTL1) genes [6–9], all of which play important roles in articular cartilage structure and function. Furthermore, SOX9 is expressed in presumptive cartilage during embryo development, and mutations in the human SOX9 gene, which result in haploinsufficiency of SOX9, cause campomelic dysplasia with skeletal malformation and dwarfism [10]. Moreover, mice chimaeras containing both wild-type and SOX9-null cells develop normally, but there is no contribution by the SOX9-null cells towards cartilage formation [11].
The expression of SOX9 declines rapidly in chondrocytes that are isolated and cultured in monolayer [12], and this is accompanied by a reduction in the expression of cartilage matrix genes such as COL2A1 [13]. Overexpression of SOX9 in human chondrocytes passaged in culture increases COL2A1 expression and increases their capacity to reform a cartilage ECM when placed in chondrogenic culture [14–16]. In view of the importance of SOX9 in the development and maintenance of the chondrocyte phenotype, its down regulation in OA is clearly likely to contribute to cartilage pathology. We investigated SOX9 transduction of a human chondrocytic cell line to identify genes that are differentially expressed in the presence by SOX9. We then investigated the expression of these genes in both cDNA samples representative of normal or OA cartilage and in primary human articular chondrocytes during culture and dedifferentiation to establish whether they were similarly regulated in vitro and in cartilage pathology.
Materials and methods
Tissue collection
Osteoarthritic cartilage was obtained from patients undergoing total knee arthroplasty for clinically and radiologically diagnosed OA [17]. Patients were excluded if there was any history of inflammatory arthropathies, or infection within the knee. Normal articular cartilage was obtained from patients undergoing above-knee amputation who had no history of joint disease. All tissue was obtained with fully informed consent and ethical approval. For tissue culture, cartilage from intact regions of joints with clinical confirmation of degenerative OA was harvested and subject to sequential trypsin/collagenase digestion to isolate chondrocytes as previously described [14]. For gene-expression studies, paired full depth samples were taken from each joint (8 normal joints and 15 OA joints), with one sample being harvested from a major loaded area on the medial femoral condyle (MFC), and one from the less loaded lateral posterior condyle (LPC), placed in RNAlater and transferred to the laboratory on ice.
Culture and retroviral transduction of cells
Monolayer cultures of SW1353 cells were kept in Dulbecco's modified Eagles medium (DMEM) supplemented with 10% foetal bovine serum (FBS), 100 units/ml penicillin and 100 units/ml streptomycin (all from Cambrex, Wokingham, UK) at 37°C, 5% CO2. For retroviral transductions, 40% confluent cultures were infected in standard culture medium with an RKAT retrovirus containing a bicistronically expressed cDNA encoding human FLAG tagged SOX9 and green fluorescent protein (GFP), at a titre of 5 × 106 [14]. After three repeated transductions, more than 90% of the cells were transduced and the cells were designated SOX9-SW1353. Cells transduced with a GFP-only retrovirus were used as controls and designated GFP-SW1353. SOX9 protein was assessed in the cells by immunoblotting using an anti-SOX9 goat polyclonal antibody (H-90, Santa Cruz Biotechnology, Calne UK). Human articular chondrocytes were isolated from cartilage on OA knee joints and maintained in culture in medium (as above) [14]. Cells were harvested for gene-expression analysis within the first week of culture (P0) and after 1 and 2 passages (P1 and P2). HACs were transduced with SOX9 or GFP-only retrovirus at passage 4 after first increasing their proliferation rate by adding platelet derived growth factor BB, transforming growth factor β1 and fibroblast growth factor 2 to the culture medium [14]. Gene expression in these cells was analysed at passage 6–8.
Microarray analysis
Glass spotted microarrays (Human known gene SGC oligo set array number 1) were obtained from the Human Genome Mapping Project. Each glass slide contains 9600 spotted oligonucleotides printed in duplicate, approximately 600 bp in length corresponding to the 3' region of each gene's mRNA. Probes were created from RNA, which was isolated from confluent monolayer cultures of GFP-SW1353 or SOX9-SW1353 using Tri Reagent (Sigma, Poole UK). 50 μg of total RNA was added to 2 μg of oligo d(T)16 (Invitrogen, Paisley, UK) and incubated at 70°C for 10 minutes before snap cooling on ice for 2 minutes. RNA was reverse transcribed to produce a cDNA probe in a labelling mix containing 1× first strand synthesis buffer, 500 μM DTT, 500 μM dATP, 500 μM dTTP, 500 μM dGTP, 100 μM dCTP, 400 units of superscript II reverse transcriptase (Invitrogen) and either 100 μM Cy3–dCTP or 100 μM Cy5–dCTP (Amersham, Uxbridge, UK). Labelling reactions were incubated for 2 hours at 42°C before adding EDTA to 1 mM to stop the reaction. RNA in the samples was degraded by adding sodium hydroxide to 25 mM and heating at 70°C for 10 minutes. Samples were neutralised by addition of hydrochloric acid, and labelled cDNA was purified using a PCR clean up kit (Qiagen, Crawley UK). The purified probe was eluted in 50 μl of nuclease free water and combined with 10 μg of human Cot-1 DNA, 6 μg oligo d(A)10–20, and 3 μg oligo d(T)16 (all from Invitrogen), and the volume reduced to 18 μl by vacuum centrifugation. The probe was then combined with 18 μl of a 2× hybridisation solution (final concentration: 25% formamide, 5 × SSC and 0.1% SDS), boiled for 3 minutes and hybridised overnight at 42°C under a glass cover slip. The arrays were then washed for 3 minutes each in 2× SSC, 0.1× SSC/0.1% SDS, and 0.1× SSC. Raw intensities at 635 nm and 532 nm were obtained for analysis from four independently probed arrays using the same starting RNA sample using a GenePix 4000A confocal microarray scanner. This data was imported into MaxDView software [18] and each pair of red/green measurements were subjected to intensity-dependant normalization. This removed intensity-dependent bias introduced by the use of the two different fluorophores as probe labels using the Loess (Lowess) method [19] and converted the data to a log ratio with the mean set to zero following normalisation. Quadruplicate log ratios were averaged and standard deviations were determined. In addition, t-tests were carried out comparing the four replicates to zero to determine potentially significantly regulated genes. Data was filtered to display only those genes with P < 0.05 and greater than twofold change in expression. Raw data from each individual channel of each array was also subjected to principle components analysis (PCA) and hierarchical clustering following normalisation of data (log2, mean set to 0 and standard deviation set to 1) using Partek software. All pre-normalised data has been submitted to MIAMExpress [20] at the European Bioinformatics Institute to allow public access (ArrayExpress Accession number E-MEXP-826).
RNA extraction and cDNA synthesis
Cell culture
Total RNA was prepared from monolayer SW1353 and HAC cultures using Tri Reagent. cDNA was synthesised from 1 μg of total RNA using M-MLV reverse transcriptase and primed with random hexamers oligonucleotides (Promega, Southampton, UK) in a 25 μl reaction.
Tissue extraction
Total RNA from cartilage was obtained by homogenization with Braun mikrodismembrator followed by Trizol extraction and chloroform/ethanol purification. Total RNA was then isolated using RNeasy minicolumns and reagents, according to the manufacturers instructions (Qiagen, Crawley, Surrey, UK) including on-column digestion of residual DNA using a RNase-Free DNAse kit (Qiagen) [21, 22]. cDNA was synthesised from 10–100 ng of total RNA using global amplification methodology [23].
Real time PCR analysis
Real time PCR was used to determine the expression of chondrocyte genes identified as being regulated by SOX9 in the microarray experiments. Amplification by PCR was carried out in 25 μl reaction volumes on a MJ Research Opticon 2 using reagents from a SYBR Green Core Kit (Eurogentec, Seraing, Belgium) with gene-specific primers designed using Applied Biosystems Primer Express software. Relative expression levels were normalised to GAPDH and calculated using the 2-ΔCCt method [24]. Primer sequences for GAPDH, COL1A1, COL2A1, aggrecan, SOX6, and SOX9 for identification of the effect of SOX9 transduction in SW1353 have been described previously [15]. Primer sequences for the other genes of interest were designed with a 3' bias to allow accurate quantification of the globally amplified cartilage cDNA libraries (Table 1).
Database analysis of conserved SOX9 binding regions
Candidate gene alignments were visualised using Vista Browser [25]. Conserved SOX9 consensus binding sites, defined by the Transfac database, were identified by comparing the human genome with that of mouse or dog using rVista [26]. SPINT1 and GAPDH genes were analysed in their entirety as well as up to 7 kb upstream of the transcription start site. Due to the very large intron size of the APOD, RGC32 and SRPX genes, only 7 kb upstream of the transcription start site and the regions within the first intron of these genes were included in this analysis. Conserved sites identified in both species comparisons were accepted as potential SOX9 binding regions.
Statistical analysis
Unpaired t-tests were used to compare the effect of SOX9 transduction on cultured cells. Statistical analyses to identify the effect of both disease state and site within the joint on gene expression were performed using mixed effects linear regression models following transformation of the data to allow normal distribution.
Results
Changes in gene expression in SOX9transduced SW1353 cells
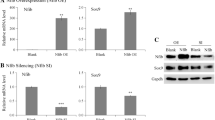
Retroviral transduction with SOX9 was carried out on a human chondrocytic cell line (SW1353), which had previously been shown to have responses to growth factors and cytokines similar to primary chondrocytes [27] and provided RNA in amounts appropriate for microarray analysis. The SW1353 cells were transduced at ~90% efficiency with a SOX9-GFP bicistronic retroviral vector (SOX9-SW1353), and controls were transduced with a SOX9-free GFP-retrovirus (GFP-SW1353) (Figure 1a). The SW1353 cells were of interest for this study as their normal expression of SOX9 was much lower than human chondrocytes in cartilage (relative to GAPDH), whereas the level of SOX9 expression following transduction was increased by 18-fold (Figure 1c) and approached the level found in cartilage. There was no discernable change in morphology following SOX9 transduction. Immunoblotting confirmed that SOX9-SW1353 synthesised increased levels of the SOX9 protein compared with controls (Figure 1b). The cells also showed increased gene expression of SOX6 (up to 14-fold) and COL2A1 (up to 13-fold), but aggrecan expression was low and was unchanged by SOX9. The SW1353 cells expressed high levels of COL1A1 and this was reduced 6-fold by SOX9 transduction. The stimulation of both SOX6 and COL2A1 by SOX9 confirmed that SW1353 cells were responsive to this factor, unlike other non-chondrocytic cells, such as dermal fibroblasts, which failed to upregulate cartilage matrix genes in response to SOX9 transduction [28].
Retroviral expression of SOX9 in SW1353 chondrosarcoma cells. (a) Phase contrast micrograph demonstrating the morphology of SW1353 chondrosarcoma cells transduced with a retrovirus containing GFP or bicistronic SOX9-GFP. Scale Bar = 50 μm. (b) Cell lysates from GFP- or SOX9-SW1353 cells were analysed by western blotting using an anti-SOX9 antibody. (c) Real time PCR analysis of cDNA derived from green-fluorescent protein (GFP; black bars) or bicistronic SOX9-GFP (grey bars) transduced SW1353 chondrosarcoma cells.
Microarray analysis of SOX9-transduced SW1353 cells
Dual hybridisations were performed in quadruplicate (including duplicated orientations of dye to sample) using probes produced from single RNA samples from SOX9-SW1353 or GFP-SW1353 cells. Extensive filtering of the normalised data was carried out as described in the materials and methods section. From the original 9,600 different genes on each array, 22 were found to be upregulated and 9 were downregulated by SOX9. From these, eight of the most strongly regulated genes were selected for further analysis (Table 2).
Real time PCR analysis of SOX9-regulated genes
Gene-expression analysis by quantitative real time (qRT) PCR was used to confirm the gene changes identified by the microarray analysis. Statistically significant upregulation was observed for SRPX (1.8-fold), S100A1 (7.9-fold), APOD (4.2-fold) RGC32 (2.3-fold) and CRTL1 (2 fold) with analyses from separately cultured SW1353 cells. Regulation of the expression of SPINT1, CRLF1 and MYBPH could not be confirmed.
Having previously shown that retroviral transduction with SOX9 of passaged human OA chondrocytes re-activated their potential to form cartilage matrix [15], we investigated the expression of the novel genes identified in SW1353 cells in human articular chondrocytes that had been expanded in monolayer culture and transduced with SOX9-retrovirus. The results showed significant upregulation (P < 0.05) of S100A1 (26.8-fold), CRTL1 (3.0-fold) and SRPX (1.7-fold) following SOX9 transduction. Interestingly, SPINT1 was also significantly upregulated in the chondrocytes (2.1-fold), which differed from the finding in the SW1353 cells. APOD was expressed at very low level in transduced and control human OA chondrocytes in monolayer culture, and although its expression appeared to be increased slightly following SOX9 transduction, no statistical analysis was possible. MYBPH expression was again unaffected by SOX9. Therefore, there were examples of genes that displayed similar responses to SOX9 transduction in both SW1353 cells and primary chondrocytes, but also genes for which there were clear regulatory differences between the cell types.
Primary chondrocyte culture with decrease in SOX9 expression
To determine whether the expression of genes identified in this study were altered by non-viral-mediated changes in SOX9 expression, we investigated in vitro cultures of freshly isolated human articular chondrocytes (Figure 2). These cells were from OA cartilage, and had a lower expression of SOX9 in culture than in tissue, but still higher (relative to GAPDH) than in SW1353 cells. During monolayer culture of the OA chondrocytes there was a further 8–10-fold decrease in SOX9 expression, and we examined whether this was accompanied by any change in expression of the newly identified genes (Figure 2). A number of the genes including S100A1, RGC32, CRTL1 and SPINT1 were down regulated under these conditions, and therefore correlated with the reduction in SOX9. SRPX, in contrast, did not correlate with SOX9 in the monolayer cultured HAC, and its expression increased with passage. The expression of another gene, CRLF1, was unchanged during the fall in SOX9. The expression of MYBPH and APOD (one of the genes most strongly upregulated by SOX9 in SW1353 cells) were very low in these primary human articular chondrocytes, and significant regulation could not be identified.
Regulation of candidate genes during chondrocyte dedifferentiation. Real time PCR analysis of candidate gene expression in cDNA from human articular chondrocytes at passage (P) 0, 1 or 2. Mean fold-change values (where P0 = 1) with standard errors are presented from chondrocytes cultures obtained from 3 donors. * indicates significant difference in expression compared with passage 0 levels P < 0.05 by paired students t-test.
Expression of SOX9-regulated genes in normal and osteoarthritic cartilage
In a previous study [5] we showed that osteoarthritic cartilage consistently showed reduced expression of SOX9 compared with healthy age-matched control tissue. It was therefore of great interest to understand whether the newly identified genes were also altered in expression in OA cartilage. We therefore probed globally amplified cDNA samples from normal and OA femoral knee cartilage [5] for their expression (Figure 3). Furthermore, the tissues samples analysed were paired cartilage samples from high-load-bearing (MFC) and low-load-bearing regions (LPC) of the same joints. SOX9 gene expression was reduced (P < 0.0001) in the osteoarthritic samples compared with the age-matched controls, and there was no difference between differently loaded sites. Of the genes investigated, five were expressed at lower level in osteoarthritic cartilage (CRTL1 (P < 0.01), SRPX (P < 0.0001), SPINT1 (P < 0.0001), RGC32 (P < 0.001) and APOD (P < 0.0001)). One gene (CRLF1) was significantly upregulated in OA tissue compared with normal tissue (P < 0.01), and S100A1 showed a wide range of expression and no significant difference between OA and controls; however, analysis of the results from all the cartilage samples showed that its expression was correlated with SOX9. Most genes investigated showed similar expression in both the more highly loaded MFC and the lower loaded LPC sites. The exceptions to this were APOD, which was further reduced (P < 0.01) in expression in the more loaded and damaged cartilage, while both S100A1 (p < 0.03) and CRLF1 (p < 0.02) were expressed at higher levels in the more loaded tissue. The analysis of cartilage oligomeric matrix protein (COMP) gene expression showed that it was unaffected in OA (or location in the joint), and demonstrated that there was no generalised downregulation of all gene expression in OA chondrocytes.
Comparison of the expression levels of candidate genes in normal and osteoarthritic cartilage. Real time PCR analysis of candidate gene expression in globally amplified cDNA representative of mRNA levels from normal (n = 8) or osteoarthritic (n = 15) human articular cartilage samples. Cartilage for the analysis was derived from either the medial or lateral femoral condyles. NM = normal medial, NL = normal lateral, OM = osteoarthritic medial and OL = osteoarthritic lateral. Symbols above bars indicate statistically significant regulation of that gene caused by:* disease (P < 0.05 mixed effects regression model) or ◆ joint location (P < 0.05 mixed effects regression model).
Genomic analysis of potential SOX9 binding sites in candidate genes
The candidate genes SPINT1, SRPX, APOD, RGC32, CRTL1 and S100A1 were among those whose expression followed that of SOX9 in most of the experimental systems that we examined. Of these genes, CRTL1 and S100A1 have previously been shown to possess SOX9 binding sequences within their promoter regions [9, 29]. Potential SOX9 binding sites in non-coding, conserved regions of the genome in and around the other four gene loci were studied using rVista. This tool identifies conserved transcription factor binding sites in sequences based on homologies of such sites between different species, and in these genes it identified binding site conservation in human, mouse and dog sequences (Table 3). The analysis demonstrated conserved SOX9 binding regions in all four candidate genes. As a control, analysis of the house-keeping gene GAPDH revealed no potential SOX9 binding sites common to all three genomic sequences.
Discussion
The transcription factor SOX9 has been shown to control the transcription of a number of important cartilage matrix genes. It is able to interact with a conserved cartilage-specific enhancer element in the COL2A1 gene and can bind to promoter and enhancer regions in a number of other cartilage matrix genes [6–9]. This work has now identified a number of genes whose expression was changed in SW1353 cells by increasing SOX9 expression by retroviral transduction and may similarly contain conserved SOX9 response elements
The human SW1353 cells have previously been used to elucidate cytokine regulation of ECM-degrading proteases as model chondrocytes [27, 30] and their SOX9 expression was shown to be increased by fibroblast growth factors 1, 2 and 9 and decreased by IL1β and TNFα [31]. They have also been used to identify cyclic AMP response element binding protein and p300 as novel partners of SOX9 that bind at cartilage-specific promoter sites [32]. Thus the SW1353 cells have some features of chondrocytes, but as with other chondrocytic cells in monolayer culture they expressed low levels of both cartilage ECM genes and SOX9, 6 and 5 [33]. Their transduction of cytokine signals has also been reported to differ from that seen in primary articular chondrocytes [33]. In this study SOX9-transduction increased the expression of target genes (such as COL2A1), although others appeared unaffected (such as aggrecan). The SW1353 cells therefore appear to lack some chondrocyte properties, but their response to SOX9 transduction was clearly more chondrocyte-like than dermal fibroblasts, which showed no regulation cartilage matrix genes in response to the overexpression of SOX9 [28].
From the initial microarray analysis we followed up gene-expression changes by qRT-PCR analysis in SOX9-SW1353 cells to confirm their regulation. Investigation of changes in the expression of this panel of genes in primary human chondrocytes following SOX9 transduction showed that some genes showed evidence of similar control to SW1353 cells, although some showed no comparable response. To extend these observations we investigated the expression in articular chondrocytes under conditions where the expression of SOX9 was changed by both natural and pathological factors. The expression was investigated in primary human chondrocytes cultured and passaged in monolayer, under which conditions the expression of SOX9 progressively becomes reduced. It was only after culture of the OA chondrocytes that the expression of SOX9 became reduced to the level found in the SW1353 cells before transduction. The change in expression during this fall in endogenous SOX9 expression showed S100A1, RGC32, CRTL1 and SPINT1 to decrease, which were therefore correlated with SOX9, as in SW1353 cells.
The identification of these SOX9-regulated genes led us to probe a human normal and OA cartilage library of globally amplified cDNA representing mRNA levels in chondrocytes in cartilage taken from load bearing or non-load-bearing regions from age-matched normal and OA human knees. SOX9 has been shown to be downregulated in osteoarthritis, and this may contribute to the pathological process by causing a reduction in the expression of ECM genes [4, 5]. We found that many of the genes whose expression was altered by SOX9 in SW1353 cells and/or isolated primary chondrocytes displayed altered expression levels in OA cartilage (CRTL1 (P < 0.01), SRPX (P < 0.0001), SPINT1 (P < 0.0001), RGC32 (P < 0.001) and APOD (P < 0.0001)) compared with age-matched controls. It is worth noting that even a gene such as COL2A1, which is known to have SOX9 regulatory elements, has been demonstrated to poorly correlate with the expression of SOX9 in control and osteoarthritic cartilage [4], suggesting that in OA its expression is more dominantly controlled by other factors. It was therefore more interesting to identify genes such as CRTL1, RGC32, S100A1 and APOD, which had a pattern of expression closely correlating with SOX9 expression levels in SW1353 cells, in primary chondrocytes, and also in OA cartilage. SRPX generally correlated with SOX9, except during chondrocyte dedifferentiation, which may indicate that other factors predominantly influence it during this process.
APOD, which was expressed at relatively low levels in SW1353 cells, was expressed more strongly in cartilage, and the expression was reduced in OA, which was consistent with the decrease in SOX9. APOD encodes apolipoprotein D, which is a protein component of low density lipoprotein in human plasma [34], and is reported to be a transit protein in the skin [35]. It may therefore have some function in cartilage ECM. The finding that APOD is downregulated in OA agrees with two previous microarray studies comparing normal and OA tissue [36, 37]. The present results showed further that APOD expression was not only downregulated in OA, but was also most strongly downregulated in the highly loaded, more physically damaged cartilage. APOD expression thus correlated with cartilage damage, whereas matrix genes, such as CTRL1 and SOX9, were similarly changed in OA in both low-loaded and high-loaded cartilage sites.
S100A1 encodes an intercellular calcium-binding protein, which can control myocardial contractility [38] and has recently been identified as an important SOX9 regulated gene that controls the terminal differentiation of chondrocytes [29]. S100A1 has previously been reported to be downregulated in osteoarthritis [36]. In the OA and control cartilage samples investigated here, S100A1 had lower mean expression in OA, but the difference was not statistically significant. However, its expression was found to be significantly correlated with SOX9 when the results from all cartilage samples were analysed (data not shown).
SRPX expression was increased by SOX9 transduction in SOX9-SW1353 and in primary human articular chondrocytes, and its expression was greatly reduced in OA cartilage. It therefore correlated well with SOX9 expression, although during chondrocyte dedifferentiation its expression increased more than sevenfold by passage 2 and was clearly unrelated to SOX9. This perhaps emphasises that any loss of chondrocyte phenotype in OA cartilage does not occur through a mechanism closely related to the loss of phenotype that occurs in these cells in monolayer culture. SRPX has a recognised role in ocular biology and disease. The SRPX gene encodes a putative membrane protein expressed abundantly in the retina, and was discovered as a candidate gene responsible for X-linked retinitis pigmentosa [39]. SOX9 has a potential regulatory role in the development of the retina, and may regulate the synthesis of collagen type II in the vitreous of the eye [40]. Furthermore, disrupted SOX9 expression in the 'odd sex' transgenic mouse, which results in sex reversal, also causes an eye phenotype with microphthalmia with cataracts [41]. The expression of SRPX may therefore be regulated by SOX9 during ocular development and may also have a role in cartilage biology.
Despite being unable to confirm any regulation by SOX9 in SW1353 by real time PCR, and with its expression unaffected in primary chondrocytes by the transition to monolayer culture, it was interesting that CRLF1 was significantly upregulated in osteoarthritic cartilage. CRLF1 protein is a member of the cytokine type I receptor family, and when expressed as a heterodimer with the cardiotrophin-like cytokine (CLC) can activate the membrane bound ciliary neurotrophic factor receptor-α (CNTFRα), which causes an interaction between gp130 and leukaemia inhibitory factor receptor, leading to cell signalling [42, 43]. Further work characterising the expression of the genes encoding CNTFRα and CLC in cartilage is required as does the possibility that upregulation of CRLF1 expression could have a use as a marker of OA.
This study identified genes whose expression in chondrocytes was consistently correlated with changes in SOX9 expression. The results suggested that the expression of these genes may be regulated by SOX9, and as SOX9 is essential for chondrocyte phenotype, the novel genes with unknown function may help control the differentiated state of chondrocytes within cartilage. The correlation of expression with SOX9 linked these genes to changes in cartilage in OA. As OA is characterized by degenerative changes in cartilage it will be important to establish how the changes in the expression of SOX9-regulated genes contribute to the progressive loss of chondrocyte function and the compromise in cartilage integrity that occurs in OA.
Conclusion
We have identified genes in a human chondrosarcoma cell line whose expression is altered by the overexpression of the chondrogenic transcription factor SOX9. Some of these genes were similarly regulated in primary human chondrocytes in response to changes in SOX9 induced by overexpression or by dedifferentiation in culture. The expression of some of these genes was also correlated with SOX9 expression in intact human articular cartilage, and was therefore suppressed in OA cartilage compared with age-matched control cartilage.
Abbreviations
- CLC:
-
= cardiotrophin-like cytokine
- CNTFR:
-
= ciliary neurotrophic factor receptor
- DMEM:
-
= Dulbecco's modified Eagle's medium
- ECM:
-
= extracellular matrix
- FBS:
-
= foetal bovine serum
- GFP:
-
= green fluorescent protein
- LPC:
-
= lateral posterior condyle
- MFC:
-
= medial femoral condyle
- OA:
-
= osteoarthritis.
References
Zhao Q, Eberspaecher H, Lefebvre V, De Crombrugghe B: Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev Dyn. 1997, 209: 377-386. 10.1002/(SICI)1097-0177(199708)209:4<377::AID-AJA5>3.0.CO;2-F.
Buckwalter JA, Saltzman C, Brown T: The impact of osteoarthritis: implications for research. Clin Orthop Relat Res. 2004, S6-15. 10.1097/01.blo.0000143938.30681.9d.
Aigner T, McKenna L: Molecular pathology and pathobiology of osteoarthritic cartilage. Cell Mol Life Sci. 2002, 59: 5-18. 10.1007/s00018-002-8400-3.
Aigner T, Gebhard PM, Schmid E, Bau B, Harley V, Poschl E: SOX9 expression does not correlate with type II collagen expression in adult articular chondrocytes. Matrix Biol. 2003, 22: 363-372. 10.1016/S0945-053X(03)00049-0.
Brew CJ, Andrew JG, Boot-Handford R, Hardingham TE: Late osteoarthritic cartilage shows down regulation of SOX9 and aggrecan expression but little evidence of chondrocyte hypertrophy. Trans Orthop Res Soc. 2004, 50: 938-
Sekiya I, Tsuji K, Koopman P, Watanabe H, Yamada Y, Shinomiya K, Nifuji A, Noda M: SOX9 enhances aggrecan gene promoter/enhancer activity and is up-regulated by retinoic acid in a cartilage-derived cell line, TC6. J Biol Chem. 2000, 275: 10738-10744. 10.1074/jbc.275.15.10738.
Zhang P, Jimenez SA, Stokes DG: Regulation of human COL9A1 gene expression. Activation of the proximal promoter region by SOX9. J Biol Chem. 2003, 278: 117-123. 10.1074/jbc.M208049200.
Bridgewater LC, Lefebvre V, de Crombrugghe B: Chondrocyte-specific enhancer elements in the Col11a2 gene resemble the Col2a1 tissue-specific enhancer. J Biol Chem. 1998, 273: 14998-15006. 10.1074/jbc.273.24.14998.
Kou I, Ikegawa S: SOX9-dependent and -independent transcriptional regulation of human cartilage link protein. J Biol Chem. 2004, 279: 50942-50948. 10.1074/jbc.M406786200.
Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Bricarelli FD, Keutel J, Hustert E, et al: Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell. 1994, 79: 1111-1120. 10.1016/0092-8674(94)90041-8.
Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B: Sox9 is required for cartilage formation. Nat Genet. 1999, 22: 85-89. 10.1038/8792.
Stokes DG, Liu G, Dharmavaram R, Hawkins D, Piera-Velazquez S, Jimenez SA: Regulation of type-II collagen gene expression during human chondrocyte de-differentiation and recovery of chondrocyte-specific phenotype in culture involves Sry-type high-mobility-group box (SOX) transcription factors. Biochem J. 2001, 360: 461-470. 10.1042/0264-6021:3600461.
Hardingham T, Tew S, Murdoch A: Tissue engineering: chondrocytes and cartilage. Arthritis Res. 2002, 4 (Suppl 3): S63-68. 10.1186/ar561.
Li Y, Tew SR, Russell AM, Gonzalez K, Hardingham TE, Hawkins RE: Transduction of human articular chondrocytes with adenoviral, retroviral and lentiviral vectors and the effects of enhanced expression of SOX9. Tissue Eng. 2004, 10: 575-584. 10.1089/107632704323061933.
Tew SR, Li Y, Pothacharoen P, Tweats LM, Hawkins RE, Hardingham TE: Retroviral transduction with SOX9 enhances re-expression of the chondrocyte phenotype in passaged osteoarthritic human articular chondrocytes. Osteoarthritis Cartilage. 2005, 13: 80-89. 10.1016/j.joca.2004.10.011.
Lefebvre V, Huang W, Harley VR, Goodfellow PN, de Crombrugghe B: SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol Cell Biol. 1997, 17: 2336-2346.
Kellgren JH, Lawrence JS: Radiological assessment of osteo-arthrosis. Ann Rheum Dis. 1957, 16: 494-502.
Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP: Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002, 30: e15-10.1093/nar/30.4.e15.
MIAMExpress. [http://www.ebi.ac.uk/miamexpress/]
Reno C, Marchuk L, Sciore P, Frank CB, Hart DA: Rapid isolation of total RNA from small samples of hypocellular, dense connective tissues. Biotechniques. 1997, 22: 1082-1086.
Flannery CR, Little CB, Caterson B, Hughes CE: Effects of culture conditions and exposure to catabolic stimulators (IL-1 and retinoic acid) on the expression of matrix metalloproteinases (MMPs) and disintegrin metalloproteinases (ADAMs) by articular cartilage chondrocytes. Matrix Biol. 1999, 18: 225-237. 10.1016/S0945-053X(99)00024-4.
Al-Taher A, Bashein A, Nolan T, Hollingsworth M, Brady G: Global cDNA amplification combined with real-time RT-PCR: accurate quantification of multiple human potassium channel genes at the single cell level. Yeast. 2000, 17: 201-210. 10.1002/1097-0061(20000930)17:3<201::AID-YEA30>3.0.CO;2-R.
Livak KJ, Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001, 25: 402-408. 10.1006/meth.2001.1262.
Vista browser. [http://pipeline.lbl.gov/cgi-bin/gateway2]
Loots GG, Ovcharenko I, Pachter L, Dubchak I, Rubin EM: rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res. 2002, 12: 832-839. 10.1101/gr.225502. Article published online before print in April 2002.
Liacini A, Sylvester J, Li WQ, Zafarullah M: Mithramycin downregulates proinflammatory cytokine-induced matrix metalloproteinase gene expression in articular chondrocytes. Arthritis Res Ther. 2005, 7: R777-783. 10.1186/ar1735.
Ikeda T, Kamekura S, Mabuchi A, Kou I, Seki S, Takato T, Nakamura K, Kawaguchi H, Ikegawa S, Chung UI: The combination of SOX5, SOX6, and SOX9 (the SOX trio) provides signals sufficient for induction of permanent cartilage. Arthritis Rheum. 2004, 50: 3561-3573. 10.1002/art.20611.
Saito T, Ikeda T, Nakamura K, Chung UI, Kawaguchi H: S100A1 and S100B, transcriptional targets of SOX trio, inhibit terminal differentiation of chondrocytes. EMBO Rep. 2007
Shi J, Schmitt-Talbot E, DiMattia DA, Dullea RG: The differential effects of IL-1 and TNF-alpha on proinflammatory cytokine and matrix metalloproteinase expression in human chondrosarcoma cells. Inflamm Res. 2004, 53: 377-389. 10.1007/s00011-004-1271-3.
Schaefer JF, Millham ML, de Crombrugghe B, Buckbinder L: FGF signaling antagonizes cytokine-mediated repression of Sox9 in SW1353 chondrosarcoma cells. Osteoarthritis Cartilage. 2003, 11: 233-241. 10.1016/S1063-4584(02)00354-0.
Tsuda M, Takahashi S, Takahashi Y, Asahara H: Transcriptional co-activators CREB-binding protein and p300 regulate chondrocyte-specific gene expression via association with Sox9. J Biol Chem. 2003, 278: 27224-27229. 10.1074/jbc.M303471200.
Gebauer M, Saas J, Sohler F, Haag J, Soder S, Pieper M, Bartnik E, Beninga J, Zimmer R, Aigner T: Comparison of the chondrosarcoma cell line SW1353 with primary human adult articular chondrocytes with regard to their gene expression profile and reactivity to IL-1beta. Osteoarthritis Cartilage. 2005, 13: 697-708. 10.1016/j.joca.2005.04.004.
Fielding PE, Fielding CJ: A cholesteryl ester transfer complex in human plasma. Proc Natl Acad Sci USA. 1980, 77: 3327-3330. 10.1073/pnas.77.6.3327.
Zeng C, Spielman AI, Vowels BR, Leyden JJ, Biemann K, Preti G: A human axillary odorant is carried by apolipoprotein D. Proc Natl Acad Sci USA. 1996, 93: 6626-6630. 10.1073/pnas.93.13.6626.
Tardif G, Hum D, Pelletier JP, Boileau C, Ranger P, Martel-Pelletier J: Differential gene expression and regulation of the bone morphogenetic protein antagonists follistatin and gremlin in normal and osteoarthritic human chondrocytes and synovial fibroblasts. Arthritis Rheum. 2004, 50: 2521-2530. 10.1002/art.20441.
Gebauer M, Saas J, Haag J, Dietz U, Takigawa M, Bartnik E, Aigner T: Repression of anti-proliferative factor Tob1 in osteoarthritic cartilage. Arthritis Res Ther. 2005, 7: R274-284. 10.1186/ar1479.
Most P, Bernotat J, Ehlermann P, Pleger ST, Reppel M, Borries M, Niroomand F, Pieske B, Janssen PM, Eschenhagen T, et al: S100A1: a regulator of myocardial contractility. Proc Natl Acad Sci USA. 2001, 98: 13889-13894. 10.1073/pnas.241393598.
Meindl A, Carvalho MR, Herrmann K, Lorenz B, Achatz H, Apfelstedt-Sylla E, Wittwer B, Ross M, Meitinger T: A gene (SRPX) encoding a sushi-repeat-containing protein is deleted in patients with X-linked retinitis pigmentosa. Hum Mol Genet. 1995, 4: 2339-2346. 10.1093/hmg/4.12.2339.
Ihanamaki T, Saamanen AM, Suominen J, Pelliniemi LJ, Harley V, Vuorio E, Salminen H: Expression of Sox9 and type IIA procollagen during ocular development and aging in transgenic Del1 mice with a mutation in the type II collagen gene. Eur J Ophthalmol. 2002, 12: 450-458.
Qin Y, Kong LK, Poirier C, Truong C, Overbeek PA, Bishop CE: Long-range activation of Sox9 in Odd Sex (Ods) mice. Hum Mol Genet. 2004, 13: 1213-1218. 10.1093/hmg/ddh141.
Elson GC, Lelievre E, Guillet C, Chevalier S, Plun-Favreau H, Froger J, Suard I, de Coignac AB, Delneste Y, Bonnefoy JY, et al: CLF associates with CLC to form a functional heteromeric ligand for the CNTF receptor complex. Nat Neurosci. 2000, 3: 867-872. 10.1038/78765.
Lelievre E, Plun-Favreau H, Chevalier S, Froger J, Guillet C, Elson GC, Gauchat JF, Gascan H: Signaling pathways recruited by the cardiotrophin-like cytokine/cytokine-like factor-1 composite cytokine: specific requirement of the membrane-bound form of ciliary neurotrophic factor receptor alpha component. J Biol Chem. 2001, 276: 22476-22484. 10.1074/jbc.M101681200.
Acknowledgements
The authors wish to thank Andrew Hayes and Leo Zeef for technical and analytical assistance with the microarray study and to the Human Genome Mapping Project for kindly providing the arrays. This work was funded by Biotechnology and Biological Sciences Research Council, Medical Research Council and Engineering and Physical Sciences Research Council, The Wellcome Trust (Research Leave Fellowship GR067462MA to PDC) and the Arthritis Research Campaign (Clinical Research Training Fellowship to CJB).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SRT conceived, designed and performed the experimental work associated with the microarray and was responsible for the initial versions of this manuscript. CJB collected the normal and OA cartilage and produced the cDNA libraries from femoral cartilage. CMR undertook the laboratory work associated with real time PCR analysis of the normal and OA cartilage libraries. PDC performed the statistical analyses, designed and validated the PCR primers, and supervised the project. TEH supervised and oversaw the completion of the studies as well as the writing of this manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Tew, S.R., Clegg, P.D., Brew, C.J. et al. SOX9 transduction of a human chondrocytic cell line identifies novel genes regulated in primary human chondrocytes and in osteoarthritis. Arthritis Res Ther 9, R107 (2007). https://doi.org/10.1186/ar2311
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1186/ar2311