Abstract
A major obstacle to identifying improved treatments for pediatric low-grade brain tumors (gliomas) is the inability to reproducibly generate human xenografts. To surmount this barrier, we leveraged human induced pluripotent stem cell (hiPSC) engineering to generate low-grade gliomas (LGGs) harboring the two most common pediatric pilocytic astrocytoma-associated molecular alterations, NF1 loss and KIAA1549:BRAF fusion. Herein, we identified that hiPSC-derived neuroglial progenitor populations (neural progenitors, glial restricted progenitors and oligodendrocyte progenitors), but not terminally differentiated astrocytes, give rise to tumors retaining LGG histologic features for at least 6 months in vivo. Additionally, we demonstrated that hiPSC-LGG xenograft formation requires the absence of CD4 T cell-mediated induction of astrocytic Cxcl10 expression. Genetic Cxcl10 ablation is both necessary and sufficient for human LGG xenograft development, which additionally enables the successful long-term growth of patient-derived pediatric LGGs in vivo. Lastly, MEK inhibitor (PD0325901) treatment increased hiPSC-LGG cell apoptosis and reduced proliferation both in vitro and in vivo. Collectively, this study establishes a tractable experimental humanized platform to elucidate the pathogenesis of and potential therapeutic opportunities for childhood brain tumors.
Similar content being viewed by others
Introduction
Low-grade gliomas (LGGs; World Health Organization grade 1 and 2 astrocytomas) are the most frequently occurring cancers of the central nervous system (CNS) in children [41], accounting for one third of all pediatric brain tumors [44]. The most common pediatric LGG is the grade 1 pilocytic astrocytoma (PA), which arises sporadically or in the context of the neurofibromatosis type 1 (NF1) tumor predisposition syndrome. In contrast to gliomas in adults, pediatric LGGs are typically slow-growing neoplasms located in the cerebellum, brainstem, and optic pathway [61], with an overall 10-year survival rate of > 90% [19]. Importantly, PAs in children rarely progress to high-grade malignancy and infrequently result in death. As such, pediatric LGGs represent a chronic condition that causes significant life-long morbidity [5], including vision loss [16], neurologic deficits, and endocrine complications [18, 54].
While preclinical mouse and pig models of NF1-associated optic pathway glioma (NF1-OPG [7, 29, 70]) and BRAF-driven [20] sporadic pediatric LGGs have helped elucidate the pathobiology of these tumors and served as platforms for therapeutic drug testing [22, 34], they only partially capture the essential properties of their human counterparts. Unfortunately, efforts to develop patient-derived pediatric LGG xenograft (PDX) models have been hindered by multiple factors, including their intrinsic slow growth rates [28], propensity to undergo premature senescence [30, 49], low clonogenic frequency [58], and heavy dependence on a supportive microenvironment [47]. In addition, the few existing human pediatric LGG xenograft models harbor genetic mutations (TP53/CDKN1A or CDKN2A/RB1 alterations) uncharacteristic of these childhood gliomas, especially PAs, which were specifically introduced to permit pediatric LGG cells to escape cellular senescence [55, 62]. In the current report, we leverage human induced pluripotent stem cell (hiPSC) engineering to generate humanized NF1-associated and sporadic KIAA1549:BRAF-driven pediatric LGG models, identify potential cells of origin for these tumors, and discover a CD4+ T cell-astrocyte Cxcl10 axis critical for LGG xenograft formation. Disruption of Cxcl10 expression established a new murine platform for in vivo patient-derived pediatric LGG xenograft modeling and therapeutic evaluation.
Results
NF1-null and KIAA1549:BRAF-expressing human induced pluripotent stem cell (hiPSC)-derived neural progenitor cells (iNPCs) exhibit increased cell proliferation
To model NF1 loss of heterozygosity in pediatric LGGs arising in the NF1 brain tumor predisposition syndrome, we engineered two different hiPSCs lines, each with a distinct homozygous germline NF1 patient-derived NF1 mutation (c.2041C > T−/− and c.6513T > A−/−; Additional file 1: Fig. S1a). Although the cell of origin for human pediatric LGGs is currently unknown, murine Nf1 LGGs of the optic nerve/chiasm (optic pathway gliomas; OPGs) arise from neural stem or neuroglial (progenitor) cells [36]. As such, we first differentiated NF1-null (c.2041C > T−/− and c.6513T > A−/−) and control (isogenic iPSCs that have undergone identical CRISPR/Cas9 processing as the NF1-null iPSC lines, but without the introduction of a mutation) hiPSCs into multipotent human neural stem cells (iNPCs) capable of generating both neuronal and glial lineage cells (Additional file 1: Fig. S1a). Consistent with the established role of the NF1 protein, neurofibromin, as a negative RAS regulator [43], these NF1-null iNPCs had increased RAS activity (2041C > T−/−, 9.2-fold; 6513T > A−/−, 13.9-fold; 2041C > T+/−, 2.2-fold−; 6513T > A+/−, 1.8-fold relative to CTL iNPCs; Additional file 1: Fig. S1b), but similar cAMP levels (2041C > T−/−, 51% reduction; 6513T > A−/−, 57% reduction; 2041C > T+/−, 49% reduction; 6513T > A+/−, 61% reduction; Additional file 1: Fig. S1c), relative to NF1-mutant iNPCs heterozygous for the same germline NF1 mutations, as we previously reported in mice [4]. In addition, to model sporadic pediatric LGGs resulting from genomic rearrangements involving the BRAF kinase gene [32, 67], we generated KIAA1549:BRAF-expressing iNPCs. Both NF1-null and KIAA1549:BRAF-expressing iNPCs had increased proliferation, as measured by BrdU incorporation (2041C > T−/−, 7.9-fold; 6513T > A−/−, 8.2-fold; 2041C > T+/−, 3.3-fold−; 6513T > A+/−, 3.5-fold; KIAA1549:BRAF, 7.5-fold) and direct cell counting (2041C > T−/−, 4.2-fold; 6513T > A−/−, 4.6-fold; 2041C > T+/−, 1.8-fold−; 6513T > A+/−, 2-fold; KIAA1549:BRAF, 3.1-fold), relative to control iNPCs (Additional file 1: Fig. S1d, e). Moreover, KIAA1549:BRAF-expressing iNPCs demonstrated increased phosphorylation of ERK1/2 relative to their isogenic controls, indicative of increased MAPK pathway activation (Additional file 1: Fig. S1f).
iNPCs form LGGs in Rag1 −/− mice
To determine whether NF1-null iNPCs generate LGGs in immunocompromised mice, 1 × 104 to 5 × 105 CTL or NF1-null iNPCs were implanted into the brainstems of Rag1−/− mice at 0–3 days of age (PN0-3). We specifically chose the brainstem for several reasons: (1) it is the second most common brain location for pediatric LGGs arising in children with NF1 [21, 42], (2) we have previously used brainstem injections for murine Nf1-OPG stem cell tumor modeling [47], and (3) injections into the mouse optic nerve, the most common site for NF1-pediatric LGGs, create major tissue damage and induce a reactive immune microenvironment. Using established neuropathological criteria for pediatric LGGs [10], we defined a LGG as (1) a mass-occupying lesion with architectural distortion by standard H&E staining (and by MRI in a subset of cases), with (2) increased proliferation (Ki67 labeling index > 1%) and (3) immunopositivity for glial immunohistochemical markers used in the routine diagnosis of human low-grade gliomas (e.g., GFAP and OLIG2). Based on these criteria and leveraging this hiPSC/murine platform, both NF1-null iNPC lines (2041C > T−/−, 6513T > A−/−) formed LGGs 1 month post-injection (mpi) in approximately 50% of mice injected with 1 × 105 iNPCs and in > 85% of animals injected with 5 × 105 iNPCs (Additional file 1: Fig. S2a). Similarly, to model sporadic pediatric LGGs, we orthotopically transplanted 5 × 105 KIAA1549:BRAF-expressing iNPCs into the cerebella of Rag1−/− mice, the most common location for sporadic PA with this molecular alteration. Over 85% of all Rag1−/− mice injected with KIAA1549:BRAF-expressing iNPCs formed LGGs at 1mpi (Fig. 1a, b). LGG sections were reviewed by an expert human neuropathologist (F.J.R.).
Rag1−/− mice develop NF1-null and KIAA1549:BRAF-expressing low-grade gliomas (LGGs) a Injection of NF1-null iNPCs (2041C > T−/−, 6513T > A−/−) into the brainstems or KIAA1549:BRAF-expressing iNPCs into the cerebella of Rag1−/− mice result in the formation of LGGs detectable by MRI (denoted by red dotted lines). b Summary of injected Rag1−/− mice harboring iNPC LGGs. c LGGs are hypercellular (H&E staining) and immunopositive for Ku80 (human-specific antibody), Ki67, glial (GFAP, OLIG2) and neuronal (synaptophysin) marker expression, by 1 month post-injection (mpi). The tumors had both glial only (top panels) and mixed embryonal/glial (lower panels) histological characteristics. Scale bars: left, Ku80/ H&E panels, 1 mm; other panels, 100 µm. d LGGs contain CD133+, ABCG1+, PDPN+, BLBP+ and GFAP+ cells (white arrowheads). No Ku80+ LGG tumor cells had SOX10 or p16 expression. Scale bar, 100 µm
NF1-associated and KIAA1549:BRAF-driven lesions were also detectable by magnetic resonance imaging (4.7-Tesla MRI, Fig. 1a) and exhibited many of the histopathologic features of human pediatric LGGs. These tumors were composed of human cells (Ku80+ cells), were hypercellular, mostly parenchymal with exophytic components, either anterior or lateral to the midbrain/brainstem tissue (NF1-null iNPC tumors) or anterior to the cerebellum (KIAA1549:BRAF-expressing iNPC tumors), and well-circumscribed (Fig. 1c). The tumors contained GFAP- and OLIG2-immunopositive cells (Fig. 1c), as seen in pediatric LGGs. All of the iNPC-lesions contained both glial areas, as determined by H&E, GFAP and OLIG2 (glial) immunopositivity, and embryonal-like hypercellular areas with neuronal (synaptophysin+) components, some of which contained neuroepithelial rosettes. Since the resulting tumors did not exhibit some histologic features routinely observed in pilocytic astrocytomas (eosinophilic granular bodies, Rosenthal fibers), they were classified as LGGs, which is the routine diagnostic approach in clinical practice. Moreover, the Ku80+ cells in these LGGs also expressed CD133 and ABCG1 (Fig. 1d), markers of glioma stem cells [15, 59], but were immunonegative for SOX10 and p16 expression, which can be observed in some patient pediatric LGGs (Fig. 1d). iNPC LGGs were negative for the OCT4 and NANOG hiPSC pluripotency markers, as well as for SMA and AFP endoderm and mesoderm markers, but were immunopositive for the Nestin neural stem cell marker (Additional file 1: Fig. S2b). Additionally, KIAA1549:BRAF-expressing LGGs demonstrated increased ERK1/2 activity relative to the surrounding non-neoplastic brain tissue (Additional file 1: Fig. S2c). Importantly, neither heterozygous NF1-mutant nor control iNPCs formed LGGs in Rag1−/− mice (Additional file 1: Fig. S2d), consistent with the requirement for NF1 loss of heterozygosity in NF1-LGG tumorigenesis.
RNA sequencing or methylation studies were not used to compare the humanized iNPC-LGGs with resected patient LGGs: iNPC-LGGs are composed entirely of human neoplastic cells and cannot be directly compared with human LGG biospecimens, in which 30–50% of the cells are monocytes, neurons, and T cells [13, 23, 50, 51, 57]. In addition, while methylation has proven valuable for separating classes of brain tumors, it is not as accurate in distinguishing subclasses of pediatric LGGs and glioneuronal tumors. This is due in part to the variable contributions of non-neoplastic elements, with the diagnostic standard involving histology, immunohistochemistry, and genetic analysis for the specific drivers involved (e.g., BRAF, NF1) [12, 52]. For these reasons, we employed two additional complementary approaches to compare hiPSC-LGGs to their spontaneously arising clinical counterparts. First, leveraging human bulk RNA sequencing data [46], we identified PDPN as a gene differentially expressed both in NF1-associated and sporadic pilocytic astrocytomas relative to control non-neoplastic brain tissue (Additional file 1: Fig. S2e). Second, we found that FABP7, which we previously showed to be overexpressed in mouse optic gliomas relative to control optic nerve by bulk RNA sequencing [15], was increased both in NF1-associated and sporadic pediatric LGGs relative to control non-neoplastic human brain (Additional file 1: Fig. S2e). Similar to their spontaneously arising pediatric LGG counterparts, both PDPN and BLBP (FABP7 protein) were expressed in the humanized NF1-null and KIAA1549:BRAF-associated LGGs in situ (Fig. 1d).
As expected for pediatric low-grade tumors, and mirroring clinical observations, mice with hiPSC-derived LGGs did not exhibit increased mortality nor obvious abnormal neurologic findings. To determine whether these lesions reflected the chronic non-malignant nature of pediatric LGGs, we assessed tumors at 1, 3 and 6 mpi (Fig. 2). Using power calculations with a two-sided, two portions test, where power was set at 80% and significance at 0.05, we estimated that a minimum of 4, 4, and 5 mice would be required to detect tumor formation at 1 mpi, 3 mpi and 6 mpi, respectively. With appropriately powered cohorts, mice injected with NF1-null and KIAA1549:BRAF-expressing iNPCs formed LGGs that were histologically similar at 1, 3 and 6 mpi (Fig. 2a). While these lesions grew in size over time, occupying on average 10–36% of the brainstem or 10–15% of the cerebellum, respectively (Fig. 2b), they had similar proliferative indices (Ki67+ cells) at 1, 3 and 6 mpi (Fig. 2c). The glioma tumor areas exhibited low proliferative indices (4–6% Ki67+ cells), within the upper range of proliferation rates seen in pediatric PAs [64], whereas the embryonal-like areas exhibited slightly higher proliferative rates (8–15% Ki67+ cells) (Fig. 2a, c). There was no evidence of increased apoptosis (0.38–0.41% TUNEL+ cells; Fig. 2d) or increased cellular senescence (0.3–0.4% β-galactosidase+ cells; Additional file 1: Fig. S2f) in vivo. Taken together, these findings demonstrate that iNPCs generate both NF1-associated and sporadic LGGs in vivo.
Rag1−/− mice develop persistent LGGs a Percentage of Rag1−/− mice harboring LGGs at 1, 3 and 6 mpi. The total number of injected mice is denoted in the parentheses. G, glial only; G/E, mixed glial/embryonal. b Representative H&E staining of NF1-null and KIAA1549:BRAF-expressing LGGs 1, 3 and 6 mpi. The lesions are outlined by the dotted lines. The percentage of the area occupied by the LGG is indicated within each panel. Scale bars, 200 µm. c Representative Ki67 immunohistochemistry images (top panel) and percentages of Ki67+ cells in Rag1−/− mice harboring NF1-null or KIAA1549:BRAF-expressing iNPC glial or mixed/glial embryonal tumors at 1, 3 and 6 mpi. Scale bars, 200 µm d No change in TUNEL+ cells (apoptotic cells) in NF1-null and KIAA1549:BRAF-expressing iNPC-derived LGGs were observed 1 (0.41%; 0.38%; 0.38%), 3 (0.36%, 0.37%; 0.34%), and 6 months (0.38%, 0.37%; 0.38%) after injection. Scale bars, 100 µm. Data are represented as means ± SEM, one-way ANOVA with Bonferroni post-test correction, Individual p values are indicated within each graph. ns not significant
hiPSC-derived glial restricted progenitors (iGRPs) and oligodendrocyte progenitors (iOPCs) form LGGs in Rag1 −/− mice
Because iNPCs are a multipotent population that generates neuronal, glial and oligodendroglial progenitor-like cells in vitro and within the LGGs in vivo (Fig. 1d), we leveraged this hiPSC platform to examine the potential of derivative restricted progenitors to serve as cells of origin for LGGs. To this end, we differentiated control, NF1-null, and KIAA1549:BRAF-expressing iNPCs into glial restricted progenitors (iGRPs: CD133+, SOX2+, O4+, GFAP+ and S100β+ cells), oligodendrocyte progenitor cells (iOPCs: O4+, MBP+, GFAP+ and NG2neg cells), and astrocytes (GFAP+, S100β+, EAAT1+ and EAAT2+ cells) (Fig. 3a) for injection into the brainstems of Rag1−/− mice (Additional file 1: Figs. S3 and S4). 1 month following the injection of 5 × 105 NF1-null or KIAA1549:BRAF-expressing cells, transplanted astrocytes did not form tumors (Fig. 3b), suggesting that terminally-differentiated preneoplastic astrocytes are unlikely to be the cells of origin for pediatric LGGs, similar to that observed for murine Nf1 optic gliomas [36]. In contrast, LGGs formed in 100% of Rag1−/− mice injected with 5 × 105 NF1-null (n = 13) and KIAA1549:BRAF-expressing (n = 5) iGRPs or 5 × 105 NF1-null (n = 14) and KIAA1549:BRAF-expressing (n = 5) iOPCs (Fig. 3b, d, Additional file 1: Fig. S4).
iGRPs form LGGs in Rag1−/− mice a Differentiation of human iNPCs into iOPCs, iGRPs and astrocytes. b Rag1−/− mice harbor LGGs at 1mpi following CTL, 2041C > T−/− and 6513T > A−/− NF1-null iGRP and iOPC brainstem injections, as well as KIAA1549:BRAF-expressing iGRP and iOPC cerebellum injections. No LGGs were observed following NF1-null or KIAA1549:BRAF-expressing hiPSC-astrocyte injections. G, glial only; G/E, mixed embryonal/glial. c Analysis of 2041C > T−/− NF1-null differentiated cells revealed CD133+, SOX2+, ABCG1+, O4+, GFAP+ and S100β + iGRPs (top panel), GFAP+, S100b+, EAAT+ and EAAT2+ astrocytes (middle panel) and O4+, MBP+, GFAP+ and NG2neg iOPCs ( lower panel). d Representative low-magnification H&E images of 2041C > T−/− and 6513 T > A−/− NF1-null and KIAA1549:BRAF-expressing iGRP and iOPC LGGs. Scale bars, 1 mm. e H&E, Ku80, Ki67, GFAP, OLIG2, synaptophysin, BLBP, CD133, ABCG1, PDPN, SOX10 and P16 immunostaining images of representative 2041C > T−/− NF1-null iGRP- (top) and iOPC- (bottom) LGGs at 1mpi. Scale bars, 100 µm. (f) Summary of relative immunopositivity scoring for GFAP, OLIG2, synaptophysin and Ki67 in LGG-bearing 1mo mice. −, 0%; +, < 25%; ++, 50%; +++, > 75% immunopositive cells. Data are represented as means ± SEM, one-way ANOVA with Bonferroni post-test correction, p values are not significant
While both iGRPs and iOPCs formed LGGs, they each exhibited unique histopathologic features seen in human PAs: iGRP LGGs were compact and hypercellular, forming as either purely parenchymal masses or parenchymal masses with exophytic components. A small subset of iGRP LGGs contained glial/embryonal tumor areas that were moderately immunopositive for synaptophysin (Fig. 3d, e, Additional file 1: Fig. S4). In contrast, iOPC LGGs had a looser, less compact architecture with numerous microcysts, and were located exophytically between the midbrain/brainstem and the hippocampus. In contrast to the strongly GFAP-immunoreactive iGRP LGGs, iOPC LGGs were more intensely OLIG2-immunoreactive and mostly negative for GFAP and synaptophysin expression (Fig. 3d, e, Additional file 1: Fig. S4). Both iGRP and iOPC-LGGs exhibited low proliferative indexes (4.1% and 3.2% Ki67+ cells, respectively) (Fig. 3e), similar to most childhood brainstem pediatric LGGs.
iNPC-LGG formation requires CD4+ T cell depletion
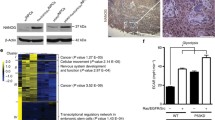
Similar to human patient brain tumor xenografts, iNPC lineage cells did not form LGGs following injection into wild type mice (Fig. 4a). To identify mouse strains that permit LGG formation, NF1-null iNPCs were injected into the brainstems of a series of immunodefective mouse strains (listed in Fig. 4a). Whereas LGGs readily formed in NOD/SCID, CD4-deficient and CD4/CD8-deficient mice at 1mpi, no tumors developed in CD8-deficient mice or strains deficient in the expression of microglia or T cell chemokine receptors (Cx3cr1, Ccr2) [45], components required for murine Nf1 optic glioma formation and growth (Additional file 1: Fig. S5a). To identify potential responsible molecular etiologies underlying CD4+ T cell deficiency-mediated LGG formation, we performed transcriptomal analysis on whole brainstems of wild type and Rag1−/− mice (Fig. 4b, c; Additional file 1: Fig. S5b). We initially identified eight downregulated transcripts and one upregulated transcript in Rag1−/− relative to wild type mouse brainstems (Fig. 4d). While three downregulated transcripts were validated in independently-acquired Rag1−/− brainstem samples (Chil3, Cd59, Cxcl10; Fig. 4e; Additional file 1: Fig. S5c), only Cxcl10 expression was decreased in all immunodefective mouse strains that permitted LGG formation.
CD4+ T cells control iNPC LGG formation in a Cxcl10-dependent manner a Mouse strains harboring NF1-null iNPC-derived LGGs at 1 mpi. b Schematic detailing the experimental design. c Heat map analysis of RNA sequencing performed on whole brainstem tissues from naïve wild type (WT) and Rag1−/− mice reveals segregation of transcript expression. d List of > 3-fold differentially expressed transcripts from the brainstems of Rag1−/− mice relative to WT controls. e Relative expression (R.E.) of Cxcl10 transcripts in WT and immunodeficient (CD8-deficient, Rag1−/−, NOD/SCID, CD4/8-deficient and CD4-deficient) mice. n = 5; data are represented as means ± SEM; one-way ANOVA with Bonferroni post-test correction. Individual p values are indicated above each bar. ns, not significant. f Relative Cxcl10 expression is reduced in Rag1−/− astrocytes compared to WT controls. n = 3; data are represented as means ± SEM, two-tailed student’s t-test; p = 0.0089. g Schematic of the experimental design, and histogram demonstrating that Cxcl10 protein levels are increased by 6.5- and 24.4-fold in Rag1−/− astrocytes treated with CD8+ and CD4+ T cell conditioned media (TCM), respectively, relative to untreated Rag1−/− astrocytes. n = 6; one-way ANOVA with Bonferroni post-test correction; individual p values are indicated above each bar
Since Cxcl10, a cytokine belonging to the CXC chemokine family, is predominantly expressed by microglia and astrocytes [68], we next analyzed these cell types in uninjected wild type and immunodeficient mouse brainstems. Whereas microglia density and morphology was relatively unaltered in all uninjected mouse brains analyzed (Additional file 1: Fig. S6a), there were fewer GFAP+, EAAT2+ and ALDH1L1+ astrocytes in the brainstems of all immunodefective mouse strains that permitted LGG formation (Additional file 1: Fig. S6b). This finding suggested that LGG formation may require a deficit in astrocytes. Consistent with an astrocyte defect, astrocytes isolated from Rag1−/− mice had reduced Cxcl10 expression relative to wild type controls (86.3% reduction, Fig. 4f). Importantly, incubation of Rag1−/− astrocytes with activated wild type mouse T cell-conditioned medium (TCM) induced Cxcl10 protein production. The greatest increase in Cxcl10 protein production was observed after induction of Rag1−/− astrocytes with TCM from CD4+ (24.4-fold), relative to CD8+ (6.5-fold), T cells (Fig. 4g). Taken together, these results indicate that reduced astrocytic Cxcl10 expression in glioma-bearing mouse strains likely reflects an absence of CD4 T cells, which induce Cxcl10 expression in astrocytes to hinder LGG growth in vivo.
Cxcl10 inhibits pediatric LGG formation
To determine whether Cxcl10 inhibits cell cycle progression or induces progenitor cell differentiation, NF1-null iNPCs were either engineered to ectopically express Cxcl10 or incubated with increasing concentrations of recombinant murine Cxcl10 protein. Both treatments induced a decrease in cell number (ectopic Cxcl10 expression, 20%; 25 pg/mL Cxcl10, 9%; 100 pg/mL Cxcl10, 13% decrease; Fig. 5a) and an increase in programmed cell death (cleaved caspase-3+ cells; ectopic Cxcl10 expression, 9.4-fold; 25 pg/mL Cxcl10, 4.6-fold; 100 pg/mL Cxcl10, 20.5-fold increase; Fig. 5b). Additionally, both treatments increased GFAP+ astrocytic differentiation of the pluripotent iNPCs (ectopic Cxcl10 expression, 8.3-fold; 25 pg/mL Cxcl10, 8.2-fold; 100 pg/mL Cxcl10, 20.5-fold increase; Fig. 5b). In this respect, 95–100% of the differentiated GFAP+ cells were cleaved caspase-3-positive, and 83.3–88.8% of the total number of cells undergoing apoptosis were astrocytes (Fig. 5b).Together, these results demonstrate that Cxcl10 induces astrocytic differentiation, as well as cell death, in vitro.
Cxcl10 absence is both necessary and sufficient for LGG formation a NF1-null iNPC cell numbers decrease (direct cell count, top; 9–20% decrease) and the percent of cleaved caspase-3+ cells increases (bottom; 8.3–20.5-fold) following ectopic Cxcl10 expression or incubation with 25 or 100 pg/mL of recombinant Cxcl10 protein. b Representative immunocytochemistry and quantification, demonstrating that ectopic Cxcl10 expression or treatment with increasing concentrations of recombinant Cxcl10 peptide induce an increase in GFAP+ astrocytic differentiation (Cxcl10: 8.3-fold, 25 pg/mL Cxcl10: 8.2-fold and 100 pg/mL Cxcl10: 20.5-fold increase), while 95–100% of the differentiated GFAP+ cells are undergoing apoptosis (cleaved caspase-3+) and 83.3–88.8% of the total number of cells undergoing apoptosis (cleaved caspase-3+) are GFAP+. c Top panel: Ectopic expression of murine Cxcl10 in 2041C > T−/− NF1-null iNPCs and iGRPs (GFP expression, Western blot). α-tubulin was used as an internal protein loading control. Bottom panel: Ectopic murine Cxcl10 expression inhibits LGG formation in Rag1−/− mice at 1mpi (H&E images). Scale bar, 1 mm. d Representative images of H&E, GFAP, OLIG2, synaptophysin and Ki67 staining of 2041C > T−/− NF1-null iNPC- and iGRP- derived LGGs in Cxcl10−/− mice at 1mpi. The number of LGG-bearing mice is indicated within the H&E panels. Scale bars: left H&E panel, 1 mm; other panels, 100 µm. e Bar graphs denoting the percent of Ki67+ cells in the lesions. Data are represented as means ± SEM, a and b one-way ANOVA with Dunnett’s multiple comparisons test, e two-tailed student’s t-test. Individual p values are indicated within each graph. ns not significant
To demonstrate that stromal Cxcl10 is sufficient to inhibit LGG formation in vivo, NF1-null iNPCs and iGRPs engineered to ectopically express murine Cxcl10 were injected into the brainstems of Rag1−/− mice. In contrast to vector-infected controls, no LGGs formed in mice following the injection of NF1-null iNPCs and iGRPs expressing murine Cxcl10 (Fig. 5c). Conversely, to establish that Cxcl10 is necessary to inhibit LGG formation, we transplanted NF1-null iNPCs and iGRPs into the brainstems of Cxcl10−/− mice. At 1 mpi, 92% and 85% of the injected mice developed iNPC- and iGRP-derived LGGs, respectively (Fig. 5d). These lesions were hypercellular, exophytic, and immunopositive for GFAP and OLIG2 expression, with 4.9% and 4.9% Ki67+ cells, respectively (Fig. 5e). Taken together, these data demonstrate that Cxcl10 is both necessary and sufficient to suppress LGG cell growth both in vitro and in vivo.
Human pediatric LGG cell lines develop LGGs in Cxcl10 −/− mice
To extend these findings to human PDX modeling, we leveraged two primary human PA cell lines from one patient with an NF1-PA (JHH-NF-PA; NF1 loss) and another with a sporadic PA (Res186). While these lines grow briefly in larval zebrafish (6 days), they do not form tumors in athymic nude mice over the course of 12 months [65]. Similar to the experiments using hiPSC neuroglial lineage cells, 5 × 105 PA cell lines were injected into brainstems of Rag1−/− and Cxcl10−/− mice. Whereas human pediatric PA cells did not form gliomas in wild type mice, both Rag1−/− and Cxcl10−/− mice developed LGGs at 1mpi and 6mpi. These LGGs were hypercellular with microcystic components, parenchymal with exophytic components, immunopositive for OLIG2, but mostly negative for GFAP, expression (Fig. 6). These lesions exhibited proliferative indices between 4.2 and 4.7% at 1mpi and 3.4–3.5% at 6 mpi. Together, these results establish Cxcl10−/− mice as a tractable in vivo platform for human pediatric LGG modeling.
Pediatric LGG cells form lesions in Rag1−/− and Cxcl10−/− mice a and b Representative images of H&E, Ki67, GFAP and OLIG2 staining of pediatric LGG tumors in Rag1−/− (top) and Cxcl10−/− (bottom) mice at 1mpi and 6mpi using two pediatric LGG lines: JHH-NF-PA (a) and Res186 (b). The number of LGG-bearing mice is indicated within the H&E panels. Bar graphs denote the % Ki67+ cells in the lesions. Data are represented as means ± SEM. Scale bars: left H&E panel, 1 mm; other panels, 100 µm
hiPSC-LGG growth is reduced following MEK inhibition
Finally, to provide a proof-of-principle demonstration that this humanized LGG platform could be used for preclinical drug evaluation, we assayed the ability of a MEK inhibitor (PD0325901) to block hiPSC-LGG growth beginning at 1 mpi in vivo. The treatment lasted a total of four weeks, a time frame we previously reported to inhibit Nf1 mouse low-grade optic glioma growth [34]. While Ku80+ iNPC-LGGs were still detectable in mice following PD0325901 administration, there was increased tumor cell apoptosis (TUNEL+ cells), as well as reduced tumor cell proliferation (Ki67+ cells), relative to vehicle-treated LGGs (Fig. 7). In vitro PD0325901 treatment of NF1 null iNPCs, iGRPs and iOPCs also resulted in decreased cell proliferation and increased apoptosis (Additional file 1: Fig. S7), thus establishing this experimental platform for preclinical therapeutic studies.
PD0325901 treatment induces apoptosis and decreases cell proliferation in iNPC-LGGs a and b TUNEL+ cells are increased (apoptotic cells; green; a), while Ki67+ cells are decreased (proliferating cells; green; b), in NF1−/− iNPC-derived LGGs following PD0325901 treatment. Human tumor cells are Ku80+ (red; b). The % area occupied by the iNPC-LGGs is indicated within the images. Data are represented as means ± SEM, two-tailed student’s t-test; Individual p values are indicated within each graph. Scale bars, 50 µm
Discussion
Building upon prior studies using hiPSC-derived 2D and 3D organoid cultures to study high-grade gliomas [1, 35, 39, 53] and medulloblastoma [63], we developed a humanized xenograft platform to model sporadic and NF1-associated pediatric LGGs and elucidate the pathogenesis, cellular origins, and signaling pathway dependencies. Beyond the value of this system to PDX pediatric LGG future preclinical experimentation, this study raises several important points germane to human brain tumor pathobiology.
First, leveraging hiPSC engineering for humanized tumor modeling represents an efficient system applicable to other low-grade nervous system tumor xenografts, which have been extremely challenging to establish in mice [28, 30, 49]. Relevant to pediatric LGGs, like NF1-PA and BRAF-driven PAs, the derivative tumor cells undergo oncogene-induced senescence [49] and display a senescence-associated secretory phenotype (SASP) in vitro unless provided with fibroblast conditioned medium and ROCK inhibition [65] or senolytic inhibitors [11]. The fragility of these PA tumor cell cultures likely reflects their profound stromal (tumor microenvironment) dependence, as well as their limited intrinsic self-renewal capacity [59], as demonstrated using genetically engineered mouse models and murine pediatric LGG explant systems. In these studies, Nf1 optic glioma growth in mice requires T cell and microglia support through the elaboration of critical cytokines and growth factors, as Nf1 optic glioma stem cells cannot form glioma-like lesions in mice lacking these stromal cells [22, 47]. Similarly, KIAA1549:BRAF-expressing cerebellar stem cells cannot form tumors in mice lacking the T cell and microglia Ccr2 chemokine receptor [14]. Moreover, the SASP represents a cellular state characterized by the secretion of inflammatory cytokines and immune modulators, which may counter the pro-tumoral support provided by T cells and monocytes in the tumor microenvironment.
Second, the hiPSC-LGG explant system provides a tractable platform to define the putative cells of origin for histologically distinct tumors, as well as histologically similar tumors arising in different locations. This is particularly important with respect to the cellular ontogeny of brain cancers. In this regard, previous mouse modeling experiments have demonstrated that multiple cell types, including differentiated cells (astrocytes [6], neurons [17]), can give rise to high-grade gliomas. Using other murine modeling approaches, NPCs [37, 45, 60], or their derivative progenitor cells [2, 8], have been shown to be the putative cells of origin, while restricted progenitors of the NPC lineage give rise to oligodendrogliomas [48] and proneural glioblastomas [40]. Moreover, additional cellular constraints, including the specific neuroglial progenitor population [60] and the particular brain location (third ventricle versus lateral ventricle germinal zone [36]) are critical determinants that dictate glioma formation and latency. In this report, we leveraged specific human neuroglial lineage cells to determine that not all lineages faithfully recapitulate LGG histological features. Whereas astrocytes do not give rise to these tumors, iGRPs, iOPCs and iNPCs with NF1 loss or KIAA1549:BRAF expression form LGGs within a month. It is intriguing to note that iGRPs give rise to more compact tumors with stronger GFAP than OLIG2 immunoreactivity, resembling optic pathway and brainstem gliomas, while the iOPC-derived lesions have looser stroma with microcystic changes and a higher density of OLIG2+ than GFAP+ cells, similar to many cerebellar human PAs. In contrast, Olig2+, NG2neg glial progenitors form Nf1 optic gliomas with delayed latency [60], which may reflect innate differences between neuroglial cell populations in humans and rodents and/or different potential cells of origin [38, 56].
Third, the finding that humanized pediatric LGGs develop in a mouse strain not lacking immune cells opens a new avenue for the in vivo study of low-grade PDXs. Whereas some orthotopically injected high-grade PDXs grow in wild type mice [25], the majority of PDX studies leverage immunocompromised (usually athymic and NOD/SCID) mice, due to their inherent inability to reject engrafted human cells. However, even in these immune-impaired animals, not all tumors are able to grow [25, 31], possibly due to the lack of a trophic environment provided by a complete immune system. Since T cells present in human PAs [13] and Nf1 murine low-grade gliomas [22] establish a supportive microenvironment for low-grade glioma growth in vivo, it is important to use host systems with immune systems with limited immunologic impairment, such as Cxcl10−/− mice. Additional studies are in progress to define immune function in Cxcl10-deficient mice.
The relevance of Cxcl10 production to xenograft establishment and tumorigenesis is further strengthened by prior reports demonstrating that elevated CXCL10 in bronchoalveolar lavage fluid is associated with acute lung transplant rejection [26] and that viral CXCL10 gene therapy improves cervical cancer xenograft responses to radiotherapy [69]. In addition, increased Cxcl10 expression is inversely correlated with tumor growth and is associated with cardiac [24] allograft rejections in the PNS, such that enhanced systemic Cxcl10 expression is a prognostic biomarker of graft-versus-host disease [33]. Moreover, reduced Cxcl10 levels in HGG spheroids caused by long-term in vitro culture correlates with higher engraftment rate in immunocompetent rats [27]. Finally, CXCL10 is induced as part of the SASP [66], which limits primary human LGG cell growth in vitro through increased senescence [11, 49]. Understanding the mechanisms underlying CXCL10-mediated hiPSC- and patient-derived xenograft growth inhibition will be critical to the development of second-generation PDX models of pediatric LGGs.
Conclusions
It should be appreciated that this xenograft model system has some inherent limitations. Rag1−/− mice have impaired immune systems and the resulting LGGs lack some of the non-neoplastic cell types (e.g., T cells) important for murine LGG growth [3, 22, 46, 47]. Similarly, the impact of Cxcl10 loss on immune system function is unknown. Ongoing studies are focused on co-introducing hiPSC-derived monocyte populations, as well as defining the impact of Cxcl10 loss on non-neoplastic cell function. While additional work will be required to create preclinical models that fully recapitulate all of the elements of pediatric LGGs, we have generated an experimentally tractable in vivo platform for humanized orthotopic pediatric LGG modeling. Further refinement of this model has great potential to galvanize the study of a variety of different types of brain and nerve tumors, including previously unexplored low-grade subtypes, as well as advance our understanding of the molecular and cellular origins of these common brain tumors in children.
Materials and methods
Study approval
All experiments were performed under active and approved Animal Studies Committee protocols at Washington University.
Animals
Mice were maintained on a 12 light/ dark cycle in a barrier facility, had ad libitum access to food and water, were exclusively used for the purposes of this study, and had not undergone any additional procedures. Mating cages housed one male and two females of the same genotype, and males were separated from dams at the time of delivery. Injections were performed on entire litters of 0–3-day-old mice of the following strains: C57BL/6J, Rag1−/− (B6.129S7-Rag1tm1Mom/J; strain 002216, Jackson laboratories), Cxcl10−/− (B6.129S4-Cxcl10tm1Adl/J; strain 006087, Jackson laboratories), CD4-deficient (B6.129S2-Cd4tm1Mak/J; strain 002663, Jackson laboratories), CD8-deficient (B6.129S2-Cd8atm1Mak/J; strain 002665, Jackson laboratories), CD4/CD8-deficient (intercrossed strains 002663 and 002665), NOD/SCID (NOD.Cg-Prkdcscid/J; strain 001,303, Jackson laboratories), Ccl5−/− (B6.129P2-Ccl5tm1Hso/J; strain 005090; Jackson laboratories) and Cx3cr1−/−; Ccr2−/− [47] mice. Injected pups were allowed to recover and were subsequently immediately returned to their maternal cage until weaning. Mice of both sexes were randomly assigned to all experimental groups without bias, and the investigators were blinded until final data analysis during all of the experiments.
Human induced pluripotent stem cell culture and differentiation
NF1 patient homozygous and heterozygous germline NF1 gene (Transcript ID NM_000267) mutations (c.2041C > T; c.6513T > A) were CRISPR/Cas9-engineered into a single commercially available male control human iPSC line (BJFF.6) by the Washington University Genome Engineering and iPSC Core Facility (GEiC). Homozygous mutations were confirmed by NGS sequencing [9], and two different clones were expanded for each of the NF1−/− and control lines for all subsequent differentiation procedures. Human induced pluripotent stem cells (hiPSCs) were grown on Matrigel (Corning)-coated culture flasks, and were fed daily with mTeSR Plus medium (STEMCELL Technologies). hiPSCs were passaged as needed with ReLeSR medium (STEMCELL technologies) following the manufacturer’s instructions. For neural progenitor cell (iNPC) differentiation, hiPSCs were transferred to poly-l-ornithine (Sigma-Aldrich)/Laminin (Fisher)-coated tissue culture flasks and incubated for 3 days in NPC basic media [50% DMEM/F12, 50% Neurobasal medium, 1 × N-2 supplement, 1 × B-27 supplement, 2 mM GlutaMax (all Thermo Fisher Scientific)] supplemented with 10 ng/mL human LIF, 4 μM CHIR99021, 3 μM SB431542, 2 μM Dorsomorphin and 0.1 μM Compound E (all STEMCELL Technologies). Subsequently, cells were incubated for 5 days in NPC basic medium supplemented with 10 ng/mL human LIF, 4 μM CHIR99021, 3 μM SB431542, and 0.1 μM Compound E. Finally, iNPCs were incubated and maintained in NPC basic medium supplemented with 10 ng/mL human LIF, 3 μM CHIR99021 and 2 μM SB431542. The medium was refreshed daily, and iNPCs passaged as needed with Accutase (STEMCELL Technologies) following the manufacturer’s instructions. For astrocyte differentiation, iNPCs were transferred onto Primaria-coated plates and maintained in Astrocyte growth medium (ThermoFisher Scientific). Astrocytes were fed 3 times a week and passaged as needed with 0.05% Trypsin (Fisher) following the manufacturer’s instructions. For glial restricted progenitor (iGRP) differentiation, iNPCs were dissociated with Accutase (STEMCELL Technologies) following the manufacturer’s instructions, and floating cells transferred to low-attachment culture flasks to allow for gliosphere formation. iGRPs were incubated for 2 weeks in the following medium: Basal GRP medium [DMEM/F12 supplemented with Sodium bicarbonate (Sigma-Aldrich), 1 × B-27 Supplement (Thermo Fisher Scientific), 1 × N-2 Supplement (Thermo Fisher Scientific), 1% penicillin–streptomycin (Thermo Fisher), 1% L-glutamine (Thermo Fisher Scientific), and 1% nonessential amino acids (Thermo Fisher Scientific)] supplemented with 10 ng/mL NT-3 (Peprotech), 10 μM forskolin (Tocris), 60 ng/mL 3,3′,5-triiodo-l-thyronine (T3; Sigma-Aldrich), 20 μg/mL ascorbic acid (Sigma-Aldrich) and 25 μg/mL insulin (Sigma-Aldrich). The supernatant was refreshed 3 times a week by gentle aspiration. For subsequent glial differentiation, gliospheres were incubated for 3 weeks in basal GRP medium supplemented with 20 ng/mL PDGF-AA (Peprotech), 10 ng/mL IGF-1 (Fisher), 10 ng/mL NT-3 (Peprotech), 10 μM forskolin (Tocris), 60 ng/mL T3 (Sigma-Aldrich), and 10 μg/mL insulin (Sigma-Aldrich). The supernatant was refreshed 3 times a week by gentle aspiration. For oligodendrocyte progenitor cell (OPC) differentiation, embryoid bodies (EBs) were generated directly from iPSCs by seeding 60,000 iPSCs at the bottoms of ultra-low cell attachment U-bottom 96 well plates, and incubating them for 5 days in NIM (DMEM/F12, 1% NEAA, 1 × N-2 supplement). Subsequently, the EBs were transferred onto poly-l-ornithine/Laminin-coated 6-well plates and incubated for 11 days in NIM supplemented with 20 ng/mL bFGF (Peprotech) and 2 µg/mL heparin (STEMCELL technologies), 3 days in NIM supplemented with 100 nM retinoic acid (RA; Sigma-Aldrich), 7 days in NIM supplemented with 100 nM RA, 1 µM Purmorphamine (Pur; STEMCELL Technologies) and 1 × B-27, and then 11 days incubation in NIM supplemented with 10 ng/mL bFGF, 1 µM Pur and 1 × B-27. For OPC specification and maturation, the spheres were then transferred to low-attachment culture flasks and incubated for 120 days in glial induction medium [DMEM/F12, 1 × N1 (Sigma-Aldrich), 1 × B27, 60 ng/mL T3, 100 ng/mL Biotin (Sigma-Aldrich) and 1 µM cAMP (Peprotech)] supplemented with 10 ng/mL PDGF-AA, 10 ng/mL IGF-1 and 10 ng/mL NT3.
Intracranial injections
Postnatal day 0–4 animals were anaesthetized and intracranially injected in accordance with active Animal Studies Committee protocols at Washington University. 1 × 104, 5 × 104, 1 × 105 or 5 × 105 cells resuspended in 2 µL ice-cold PBS were injected 0.7 mm to the right of the midline into either the midbrain (0.5 mm posterior to Lambda; 2 mm deep), or the cerebellum (2 mm posterior to Lambda; 1 mm deep) of neonatal mice using a Hamilton syringe. Animals were aged to 1, 3 or 6 months post-injection prior to tissue harvesting and analysis.
PD0325901 treatments
Twenty 4-week-old Rag1−/− mice of both sexes harboring NF1-null iNPC-LGGs 1 month post-injection were intraperitoneally injected either with 10% DMSO in saline or with 5 mg/kg/day PD0325901 (Sigma-Aldrich), 6 days a week, for a total of four weeks [34]. Treated mice were collected for histopathologic analysis. For in vitro experiments, iNPCs, iGRPs and iOPCs were treated with 10 nM PD0325901 for 24 h prior to immunocytochemical analysis.
Magnetic resonance imaging (MRI)
Injected mice were transcardially perfused with Ringer’s solution and 4% paraformaldehyde (PFA). The entire brain was removed from the animals and post-fixed in 4% PFA overnight prior to rehydration in phosphate buffered saline (PBS) for a minimum of 7 days. Rehydrated brain samples underwent MRI. Each mouse brain was then packed into a 2 mL plastic vial, supported with fiberglass and immersed in Fluorinert (FC-3283; 3 M Company, St. Paul, MN). MRI experiments were performed using a 4.7-T small-animal MR scanner built around an Agilent/Varian (Santa Clara, CA) DirectDriveTM console and an Oxford Instruments (Oxford, United Kingdom) horizontal-bore superconducting magnet. The plastic vial containing the mouse brain was loaded into a laboratory-built solenoid RF coil (1-cm diameter; 2-cm length). MR images were collected with a 3D gradient echo (GE3D) sequence: TR 5.0 ms, TE 2.2 ms, flip angle 30°, matrix size 128 × 128 × 128, FOV 16 × 16 × 16 mm3, 0.19 mm isotropic resolution, 4 averages, and 5.5 min data acquisition time. The images were loaded into MatLab (MathWorks®, Natick, MA) and converted into NIfTI (.nii) format for tumor inspection and segmentation with ITK-SNAP (www.itksnap.org).
Tissue fixation and immunohistochemistry
Injected mice were transcardially perfused at 1, 3 or 6 months post-injection, initially with Ringer’s solution, and then followed by ice-cold 4% paraformaldehyde. Whole mouse brains were harvested, post-fixed in 4% PFA and dehydrated in 70% ethanol. Dehydrated samples were then paraffin-embedded and serially sectioned (5 µm). Hematoxylin and eosin (H&E), as well as antibody immunohistochemical staining, were performed using the primary antibodies, Vectastain ABC kit (Vector Laboratories) and appropriate biotinylated secondary antibodies provided in Additional file 1: Table S1. Immunostaining was performed using the primary antibodies listed in Additional file 1: Table S1 with appropriate Alexa Fluor-conjugated secondary antibodies. Bis-benzamide (Hoechst) was used as a nuclear counterstain.
Immunocytochemistry
Immunocytochemistry was performed on hiPSCs, iNPCs, iGRPs, iOPCs and astrocytes using the primary antibodies described in Additional file 1: Table S1. Briefly, adherent cells were washed, fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 in PBS, before blocking in 10% goat serum and incubation overnight at 4 °C in primary antibodies diluted in 2% goat serum at the manufacturer’s suggested concentrations. Appropriate secondary Alexa-Fluor-conjugated secondary antibodies diluted to 1:200 were employed and bis-benzamide (Hoechst) was used as a nuclear counterstain. Cells were mounted with Immu-mount (Fisher) and imaged on a Leica DMi1 fluorescent microscope using the Leica LAS X software, as per manufacturer’s instructions.
BrdU proliferation, RAS activity, cAMP and Cxcl10 ELISA
RAS activity (ThermoFisher), cAMP (Fisher) detection, BrdU proliferation assays (Roche) and direct cell counting were performed according to the manufacturer’s instructions. Cxcl10 (Ray Biotech) ELISA assays were performed according to the manufacturer’s instructions. Each assay was performed using a minimum of three independently generated biological replicates.
Lentivirus production, cell infection and CXCL10 peptide treatment
Cxcl10 cDNA (Sino Biological) and lentiviral packaging vectors, or KIAA1549:BRAF adenoviral DNA, were transfected in HEK293T cells using Fugene HD (Promega) following the manufacturer’s instructions. Viral supernatants were collected 48 h post-transfection, filtered and used to directly infect iNPCs and iGRPs for 24 h. Cxcl10-GFP expression was confirmed by Western blotting as previously described (Anastasaki et al., 2015). KIAA1549:BRAF-infected cells were puromycin-selected for 2 weeks prior to further expansion. Infected cells were used for intracranial injections in Rag1−/− neonatal mice. For cell proliferation, cell death and differentiation analyses, iNPCs were treated with 25 or 100 pg/mL of murine recombinant Cxcl10 (PeproTech) for 24 h. Each experiment was performed a minimum of three times on independently generated samples.
RNA extraction, quantitative real-time PCR and RNA sequencing and analysis
RNA was extracted using a QIAGEN RNeasy mini-kit from snap-frozen brainstem tissues of 1-month-old adult mice following the manufacturer’s instructions (QIAGEN). For quantitative real-time PCR (qPCR) studies, total RNA was reverse-transcribed using the High-Capacity cDNA Reverse Transcription Kit protocol following the manufacturer’s instructions (Thermo Fisher Scientific). qPCR was performed on a Bio-Rad CFX thermocycler using pre-designed TaqMan Gene Expression Assays (Cxcl10, Chil3, CD59a, Gfap; Additional file 1: Table S2) and a commercially available Taqman mastermix (Thermo Fisher Scientific) following the manufacturer’s instructions. Relative transcript expression was calculated using the ΔΔCT analysis method and normalized to Gapdh as an internal control following the manufacturer’s instructions (ThermoFisher). For RNA sequencing, total RNA from three C57BL/6J and three Rag1−/− brainstem samples was submitted to Washington University Genome Technology Access Center (GTAC). Samples were prepared according to the library kit manufacturer’s protocol, indexed, pooled, and sequenced on an Illumina HiSeq. Basecalls and demultiplexing were performed with Illumina bcl2fastq software and a custom python demultiplexing program with a maximum of one mismatch in the indexing read. The analysis was generated using Partek Flow software, version 8.0. RNA sequencing reads were aligned to the mm10—RefSeq Transcripts 83 assembly with STAR version 2.5.3a. Gene counts and isoform expression were derived from the annotation model output. Sequencing performance was assessed for the total number of aligned reads, total number of uniquely aligned reads, and features detected. Normalization size factors were calculated for all gene counts by CPM to adjust for differences in library size. Gene specific analysis was then performed using the lognormal with shrinkage model (limma-trend method) to analyze for differences between Rag1−/− and control C57 mice. The results were filtered for only those genes with p values ≤ 0.05 and log fold-changes ≥ ± 2. Principal component analysis was conducted in Partek Flow using normalized gene counts. RNA sequencing data have been deposited in the GEO portal (GSE174624).
T cell isolation and culture
4–6-week-old C57BL/6J and Rag1−/− mouse spleens were homogenized into single cell suspensions by digestion in PBS containing 0.1% BSA and 0.6% sodium citrate. The homogenates were subsequently washed and incubated with 120 Kunitz units of DNase I for 15 min following red blood cell lysis (eBioscience). Cells were then filtered through a 30 µM cell strainer to obtain a single cell suspension. CD4+ and CD8+ T cells were isolated using CD8a (Miltenyi Biotech) or CD4 (Miltenyi Biotech) T cell isolation kits, respectively. T cells were maintained at 2.5 × 106 cells ml−1 in RPMI-1640 medium supplemented with 10% FBS and 1% penicillin/ streptomycin. T cells were activated by 1.25 µg ml−1 anti-mouse CD3 (Fisher Scientific) and 2 µg ml−1 anti-mouse CD28 (Fisher Scientific) antibody treatment for 48 h. T cell conditioned media (TCM) was collected both from non-activated and activated T cells following 22 µM filtration for subsequent chemokine assay, ELISA and co-culture experiments.
Astrocyte isolation and culture
4- to 6-week-old C57BL/6 J and Rag1−/− mice were transcardially perfused with DPBS and whole brains collected following the removal of the cerebellum and olfactory bulbs. A single cell suspension was generated using the Miltenyi Biotech adult brain dissociation kit following the manufacturer’s instructions. The resulting cells were seeded in Poly-l-Lysine-coated (Millipore Sigma) T75 flasks and incubated in Minimal Essential Medium supplemented with 1 mM l-glutamine, 1 mM sodium pyruvate, 0.6% D-(+)-glucose, 100 μg ml−1 penicillin/ streptomycin and 10% FBS. A complete media change was performed after 24 h and every third day after that for 12–14 days, in order to obtain mixed glial cultures composed of microglia on an astrocyte monolayer. To release the microglia, flasks were mechanically shaken at 200 rpm for 5 h at 37 °C, and the microglia-containing supernatant discarded. Astrocyte-enriched cultures containing > 95% GFAP-positive astrocyte monolayers were passaged with 0.1% trypsin (Invitrogen) for subsequent experiments. Conditioned media from naïve or TCM-treated astrocytes was collected following 22 µM filtration.
Quantification and statistical analysis
All statistical tests were performed using GraphPad Prism 5 software. 2-tailed Student’s t-tests, or one-way analysis of variance (ANOVA) with Bonferroni post-test correction using GraphPad Prism 5 software. Statistical significance was set at p < 0.05, and individual p values are indicated within each graphical figure. A minimum of 3 independently generated biological replicates was employed for each of the analyses. Numbers (n) are noted for each individual analysis.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files. RNA sequencing data have been deposited in the GEO portal (GSE174624).
References
Abdullah KG, Bird CE, Buehler JD, Gattie LC, Savani MR, Sternisha AC, Xiao Y, Levitt MM, Hicks WH, Li W et al (2022) Establishment of patient-derived organoid models of lower-grade glioma. Neuro Oncol 24:612–623. https://doi.org/10.1093/neuonc/noab273
Alcantara Llaguno S, Chen J, Kwon CH, Jackson EL, Li Y, Burns DK, Alvarez-Buylla A, Parada LF (2009) Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell 15:45–56. https://doi.org/10.1016/j.ccr.2008.12.006
Anastasaki C, Mo J, Chen JK, Chatterjee J, Pan Y, Scheaffer SM, Cobb O, Monje M, Le LQ, Gutmann DH (2022) Neuronal hyperexcitability drives central and peripheral nervous system tumor progression in models of neurofibromatosis-1. Nat Commun 13:2785. https://doi.org/10.1038/s41467-022-30466-6
Anastasaki C, Woo AS, Messiaen LM, Gutmann DH (2015) Elucidating the impact of neurofibromatosis-1 germline mutations on neurofibromin function and dopamine-based learning. Hum Mol Genet 24:3518–3528. https://doi.org/10.1093/hmg/ddv103
Armstrong GT, Liu Q, Yasui Y, Huang S, Ness KK, Leisenring W, Hudson MM, Donaldson SS, King AA, Stovall M et al (2009) Long-term outcomes among adult survivors of childhood central nervous system malignancies in the childhood cancer survivor study. J Natl Cancer Inst 101:946–958. https://doi.org/10.1093/jnci/djp148
Bachoo RM, Maher EA, Ligon KL, Sharpless NE, Chan SS, You MJ, Tang Y, DeFrances J, Stover E, Weissleder R et al (2002) Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell 1:269–277. https://doi.org/10.1016/s1535-6108(02)00046-6
Bajenaru ML, Hernandez MR, Perry A, Zhu Y, Parada LF, Garbow JR, Gutmann DH (2003) Optic nerve glioma in mice requires astrocyte Nf1 gene inactivation and Nf1 brain heterozygosity. Cancer Res 63:8573–8577
Becher OJ, Hambardzumyan D, Walker TR, Helmy K, Nazarian J, Albrecht S, Hiner RL, Gall S, Huse JT, Jabado N et al (2010) Preclinical evaluation of radiation and perifosine in a genetically and histologically accurate model of brainstem glioma. Cancer Res 70:2548–2557. https://doi.org/10.1158/0008-5472.CAN-09-2503
Bell CC, Magor GW, Gillinder KR, Perkins AC (2014) A high-throughput screening strategy for detecting CRISPR-Cas9 induced mutations using next-generation sequencing. BMC Genom 15:1002. https://doi.org/10.1186/1471-2164-15-1002
Board WCoTE World Health Organization Classification of Tumours of the Central Nervous System. City
Buhl JL, Selt F, Hielscher T, Guiho R, Ecker J, Sahm F, Ridinger J, Riehl D, Usta D, Ismer B et al (2019) The senescence-associated secretory phenotype mediates oncogene-induced senescence in pediatric pilocytic astrocytoma. Clin Cancer Res 25:1851–1866. https://doi.org/10.1158/1078-0432.CCR-18-1965
Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, Koelsche C, Sahm F, Chavez L, Reuss DE et al (2018) DNA methylation-based classification of central nervous system tumours. Nature 555:469–474. https://doi.org/10.1038/nature26000
Chen J, Sinha N, Cobb O, Liu C, Ersen A, Phillips JJ, Tihan T, Gutmann DH, Dahiya S (2021) Immune cell analysis of pilocytic astrocytomas reveals sexually dimorphic brain region-specific differences in T-cell content. Neurooncol Adv 3:vdab068. https://doi.org/10.1093/noajnl/vdab068
Chen R, Keoni C, Waker CA, Lober RM, Chen YH, Gutmann DH (2019) KIAA1549-BRAF expression establishes a permissive tumor microenvironment through NFkappaB-mediated CCL2 production. Neoplasia 21:52–60. https://doi.org/10.1016/j.neo.2018.11.007
Chen YH, McGowan LD, Cimino PJ, Dahiya S, Leonard JR, Lee DY, Gutmann DH (2015) Mouse low-grade gliomas contain cancer stem cells with unique molecular and functional properties. Cell Rep 10:1899–1912. https://doi.org/10.1016/j.celrep.2015.02.041
Fisher MJ, Loguidice M, Gutmann DH, Listernick R, Ferner RE, Ullrich NJ, Packer RJ, Tabori U, Hoffman RO, Ardern-Holmes SL et al (2012) Visual outcomes in children with neurofibromatosis type 1-associated optic pathway glioma following chemotherapy: a multicenter retrospective analysis. Neuro Oncol 14:790–797. https://doi.org/10.1093/neuonc/nos076
Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, Marumoto T, Singer O, Ellisman MH, Verma IM (2012) Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science 338:1080–1084. https://doi.org/10.1126/science.1226929
Gan HW, Phipps K, Aquilina K, Gaze MN, Hayward R, Spoudeas HA (2015) Neuroendocrine morbidity after pediatric optic gliomas: a longitudinal analysis of 166 children over 30 years. J Clin Endocrinol Metab 100:3787–3799. https://doi.org/10.1210/jc.2015-2028
Gnekow AK, Falkenstein F, von Hornstein S, Zwiener I, Berkefeld S, Bison B, Warmuth-Metz M, Driever PH, Soerensen N, Kortmann RD et al (2012) Long-term follow-up of the multicenter, multidisciplinary treatment study HIT-LGG-1996 for low-grade glioma in children and adolescents of the German speaking society of pediatric oncology and hematology. Neuro Oncol 14:1265–1284. https://doi.org/10.1093/neuonc/nos202
Gronych J, Korshunov A, Bageritz J, Milde T, Jugold M, Hambardzumyan D, Remke M, Hartmann C, Witt H, Jones DT et al (2011) An activated mutant BRAF kinase domain is sufficient to induce pilocytic astrocytoma in mice. J Clin Investig 121:1344–1348. https://doi.org/10.1172/JCI44656
Guillamo JS, Creange A, Kalifa C, Grill J, Rodriguez D, Doz F, Barbarot S, Zerah M, Sanson M, Bastuji-Garin S et al (2003) Prognostic factors of CNS tumours in neurofibromatosis 1 (NF1): a retrospective study of 104 patients. Brain 126:152–160. https://doi.org/10.1093/brain/awg016
Guo X, Pan Y, Xiong M, Sanapala S, Anastasaki C, Cobb O, Dahiya S, Gutmann DH (2020) Midkine activation of CD8(+) T cells establishes a neuron-immune-cancer axis responsible for low-grade glioma growth. Nat Commun 11:2177. https://doi.org/10.1038/s41467-020-15770-3
Gutmann DH, McLellan MD, Hussain I, Wallis JW, Fulton LL, Fulton RS, Magrini V, Demeter R, Wylie T, Kandoth C et al (2013) Somatic neurofibromatosis type 1 (NF1) inactivation characterizes NF1-associated pilocytic astrocytoma. Genome Res 23:431–439. https://doi.org/10.1101/gr.142604.112
Hancock WW, Gao W, Csizmadia V, Faia KL, Shemmeri N, Luster AD (2001) Donor-derived IP-10 initiates development of acute allograft rejection. J Exp Med 193:975–980. https://doi.org/10.1084/jem.193.8.975
Hoffmann N, Fernandez V, Pereira RC, Rancati S, Pelizzoli R, De Pietri Tonelli D (2020) A xenotransplant model of human brain tumors in wild-type mice. iScience 23:100813. https://doi.org/10.1016/j.isci.2019.100813
Husain S, Resende MR, Rajwans N, Zamel R, Pilewski JM, Crespo MM, Singer LG, McCurry KR, Kolls JK, Keshavjee S et al (2014) Elevated CXCL10 (IP-10) in bronchoalveolar lavage fluid is associated with acute cellular rejection after human lung transplantation. Transplantation 97:90–97. https://doi.org/10.1097/TP.0b013e3182a6ee0a
Huszthy PC, Sakariassen PO, Espedal H, Brokstad KA, Bjerkvig R, Miletic H (2015) Engraftment of human glioblastoma cells in immunocompetent rats through acquired immunosuppression. PLoS ONE 10:e0136089. https://doi.org/10.1371/journal.pone.0136089
Hutt-Cabezas M, Karajannis MA, Zagzag D, Shah S, Horkayne-Szakaly I, Rushing EJ, Cameron JD, Jain D, Eberhart CG, Raabe EH et al (2013) Activation of mTORC1/mTORC2 signaling in pediatric low-grade glioma and pilocytic astrocytoma reveals mTOR as a therapeutic target. Neuro Oncol 15:1604–1614. https://doi.org/10.1093/neuonc/not132
Isakson SH, Rizzardi AE, Coutts AW, Carlson DF, Kirstein MN, Fisher J, Vitte J, Williams KB, Pluhar GE, Dahiya S et al (2018) Genetically engineered minipigs model the major clinical features of human neurofibromatosis type 1. Commun Biol 1:158. https://doi.org/10.1038/s42003-018-0163-y
Jacob K, Quang-Khuong DA, Jones DT, Witt H, Lambert S, Albrecht S, Witt O, Vezina C, Shirinian M, Faury D et al (2011) Genetic aberrations leading to MAPK pathway activation mediate oncogene-induced senescence in sporadic pilocytic astrocytomas. Clin Cancer Res 17:4650–4660. https://doi.org/10.1158/1078-0432.CCR-11-0127
Jimenez-Valerio G, Martinez-Lozano M, Bassani N, Vidal A, Ochoa-de-Olza M, Suarez C, Garcia-Del-Muro X, Carles J, Vinals F, Graupera M et al (2016) Resistance to antiangiogenic therapies by metabolic symbiosis in renal cell carcinoma PDX models and patients. Cell Rep 15:1134–1143. https://doi.org/10.1016/j.celrep.2016.04.015
Jones DT, Kocialkowski S, Liu L, Pearson DM, Backlund LM, Ichimura K, Collins VP (2008) Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res 68:8673–8677. https://doi.org/10.1158/0008-5472.CAN-08-2097
Kariminia A, Holtan SG, Ivison S, Rozmus J, Hebert MJ, Martin PJ, Lee SJ, Wolff D, Subrt P, Abdossamadi S et al (2016) Heterogeneity of chronic graft-versus-host disease biomarkers: association with CXCL10 and CXCR3+ NK cells. Blood 127:3082–3091. https://doi.org/10.1182/blood-2015-09-668251
Kaul A, Toonen JA, Cimino PJ, Gianino SM, Gutmann DH (2015) Akt- or MEK-mediated mTOR inhibition suppresses Nf1 optic glioma growth. Neuro Oncol 17:843–853. https://doi.org/10.1093/neuonc/nou329
Koga T, Chaim IA, Benitez JA, Markmiller S, Parisian AD, Hevner RF, Turner KM, Hessenauer FM, D’Antonio M, Nguyen ND et al (2020) Longitudinal assessment of tumor development using cancer avatars derived from genetically engineered pluripotent stem cells. Nat Commun 11:550. https://doi.org/10.1038/s41467-020-14312-1
Lee DY, Gianino SM, Gutmann DH (2012) Innate neural stem cell heterogeneity determines the patterning of glioma formation in children. Cancer Cell 22:131–138. https://doi.org/10.1016/j.ccr.2012.05.036
Lee JH, Lee JE, Kahng JY, Kim SH, Park JS, Yoon SJ, Um JY, Kim WK, Lee JK, Park J et al (2018) Human glioblastoma arises from subventricular zone cells with low-level driver mutations. Nature 560:243–247. https://doi.org/10.1038/s41586-018-0389-3
Lin CY, Erkek S, Tong Y, Yin L, Federation AJ, Zapatka M, Haldipur P, Kawauchi D, Risch T, Warnatz HJ et al (2016) Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature 530:57–62. https://doi.org/10.1038/nature16546
Linkous A, Balamatsias D, Snuderl M, Edwards L, Miyaguchi K, Milner T, Reich B, Cohen-Gould L, Storaska A, Nakayama Y et al (2019) Modeling patient-derived glioblastoma with cerebral organoids. Cell Rep 26:3203–3211. https://doi.org/10.1016/j.celrep.2019.02.063
Liu C, Sage JC, Miller MR, Verhaak RG, Hippenmeyer S, Vogel H, Foreman O, Bronson RT, Nishiyama A, Luo L et al (2011) Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell 146:209–221. https://doi.org/10.1016/j.cell.2011.06.014
Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM, Reifenberger G et al (2021) The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol 23:1231–1251. https://doi.org/10.1093/neuonc/noab106
Mahdi J, Shah AC, Sato A, Morris SM, McKinstry RC, Listernick R, Packer RJ, Fisher MJ, Gutmann DH (2017) A multi-institutional study of brainstem gliomas in children with neurofibromatosis type 1. Neurology 88:1584–1589. https://doi.org/10.1212/WNL.0000000000003881
McCormick F (1995) Ras signaling and NF1. Curr Opin Genet Dev 5:51–55. https://doi.org/10.1016/s0959-437x(95)90053-5
Ostrom QT, de Blank PM, Kruchko C, Petersen CM, Liao P, Finlay JL, Stearns DS, Wolff JE, Wolinsky Y, Letterio JJ et al (2015) Alex’s Lemonade Stand Foundation infant and childhood primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro Oncol 16(Suppl 10):x1–x36. https://doi.org/10.1093/neuonc/nou327
Pan Y, Bush EC, Toonen JA, Ma Y, Solga AC, Sims PA, Gutmann DH (2017) Whole tumor RNA-sequencing and deconvolution reveal a clinically-prognostic PTEN/PI3K-regulated glioma transcriptional signature. Oncotarget 8:52474–52487. https://doi.org/10.18632/oncotarget.17193
Pan Y, Hysinger JD, Barron T, Schindler NF, Cobb O, Guo X, Yalcin B, Anastasaki C, Mulinyawe SB, Ponnuswami A et al (2021) NF1 mutation drives neuronal activity-dependent initiation of optic glioma. Nature 594:277–282. https://doi.org/10.1038/s41586-021-03580-6
Pan Y, Xiong M, Chen R, Ma Y, Corman C, Maricos M, Kindler U, Semtner M, Chen YH, Dahiya S et al (2018) Athymic mice reveal a requirement for T-cell-microglia interactions in establishing a microenvironment supportive of Nf1 low-grade glioma growth. Genes Dev 32:491–496. https://doi.org/10.1101/gad.310797.117
Persson AI, Petritsch C, Swartling FJ, Itsara M, Sim FJ, Auvergne R, Goldenberg DD, Vandenberg SR, Nguyen KN, Yakovenko S et al (2010) Non-stem cell origin for oligodendroglioma. Cancer Cell 18:669–682. https://doi.org/10.1016/j.ccr.2010.10.033
Raabe EH, Lim KS, Kim JM, Meeker A, Mao XG, Nikkhah G, Maciaczyk J, Kahlert U, Jain D, Bar E et al (2011) BRAF activation induces transformation and then senescence in human neural stem cells: a pilocytic astrocytoma model. Clin Cancer Res 17:3590–3599. https://doi.org/10.1158/1078-0432.CCR-10-3349
Roggendorf W, Strupp S, Paulus W (1996) Distribution and characterization of microglia/macrophages in human brain tumors. Acta Neuropathol 92:288–293. https://doi.org/10.1007/s004010050520
Rossi ML, Hughes JT, Esiri MM, Coakham HB, Brownell DB (1987) Immunohistological study of mononuclear cell infiltrate in malignant gliomas. Acta Neuropathol 74:269–277. https://doi.org/10.1007/BF00688191
Ryall S, Tabori U, Hawkins C (2020) Pediatric low-grade glioma in the era of molecular diagnostics. Acta Neuropathol Commun 8:30. https://doi.org/10.1186/s40478-020-00902-z
Sancho-Martinez I, Nivet E, Xia Y, Hishida T, Aguirre A, Ocampo A, Ma L, Morey R, Krause MN, Zembrzycki A et al (2016) Establishment of human iPSC-based models for the study and targeting of glioma initiating cells. Nat Commun 7:10743. https://doi.org/10.1038/ncomms10743
Santoro C, Perrotta S, Picariello S, Scilipoti M, Cirillo M, Quaglietta L, Cinalli G, Cioffi D, Di Iorgi N, Maghnie M et al (2020) Pretreatment endocrine disorders due to optic pathway gliomas in pediatric neurofibromatosis type 1: multicenter study. J Clin Endocrinol Metab. https://doi.org/10.1210/clinem/dgaa138
Selt F, Hohloch J, Hielscher T, Sahm F, Capper D, Korshunov A, Usta D, Brabetz S, Ridinger J, Ecker J et al (2017) Establishment and application of a novel patient-derived KIAA1549:BRAF-driven pediatric pilocytic astrocytoma model for preclinical drug testing. Oncotarget 8:11460–11479. https://doi.org/10.18632/oncotarget.14004
Sexton-Oates A, Dodgshun A, Hovestadt V, Jones DTW, Ashley DM, Sullivan M, MacGregor D, Saffery R (2018) Methylation profiling of paediatric pilocytic astrocytoma reveals variants specifically associated with tumour location and predictive of recurrence. Mol Oncol 12:1219–1232. https://doi.org/10.1002/1878-0261.12062
Simmons GW, Pong WW, Emnett RJ, White CR, Gianino SM, Rodriguez FJ, Gutmann DH (2011) Neurofibromatosis-1 heterozygosity increases microglia in a spatially and temporally restricted pattern relevant to mouse optic glioma formation and growth. J Neuropathol Exp Neurol 70:51–62. https://doi.org/10.1097/NEN.0b013e3182032d37
Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB (2003) Identification of a cancer stem cell in human brain tumors. Cancer Res 63:5821–5828
Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB (2004) Identification of human brain tumour initiating cells. Nature 432:396–401. https://doi.org/10.1038/nature03128
Solga AC, Toonen JA, Pan Y, Cimino PJ, Ma Y, Castillon GA, Gianino SM, Ellisman MH, Lee DY, Gutmann DH (2017) The cell of origin dictates the temporal course of neurofibromatosis-1 (Nf1) low-grade glioma formation. Oncotarget 8:47206–47215. https://doi.org/10.18632/oncotarget.17589
Stokland T, Liu JF, Ironside JW, Ellison DW, Taylor R, Robinson KJ, Picton SV, Walker DA (2010) A multivariate analysis of factors determining tumor progression in childhood low-grade glioma: a population-based cohort study (CCLG CNS9702). Neuro Oncol 12:1257–1268. https://doi.org/10.1093/neuonc/noq092
Sun Y, Alberta JA, Pilarz C, Calligaris D, Chadwick EJ, Ramkissoon SH, Ramkissoon LA, Garcia VM, Mazzola E, Goumnerova L et al (2017) A brain-penetrant RAF dimer antagonist for the noncanonical BRAF oncoprotein of pediatric low-grade astrocytomas. Neuro Oncol 19:774–785. https://doi.org/10.1093/neuonc/now261
Susanto E, Marin Navarro A, Zhou L, Sundstrom A, van Bree N, Stantic M, Moslem M, Tailor J, Rietdijk J, Zubillaga V et al (2020) Modeling SHH-driven medulloblastoma with patient iPS cell-derived neural stem cells. Proc Natl Acad Sci U S A 117:20127–20138. https://doi.org/10.1073/pnas.1920521117
Tu A, Robison A, Melamed E, Buchanan I, Hariri O, Babu H, Szymanski L, Krieger M (2018) Proliferative index in pediatric pilocytic astrocytoma by region of origin and prediction of clinical behavior. Pediatr Neurosurg 53:395–400. https://doi.org/10.1159/000490466
Yuan M, White D, Resar L, Bar E, Groves M, Cohen A, Jackson E, Bynum J, Rubens J, Mumm J et al (2021) Conditional reprogramming culture conditions facilitate growth of lower-grade glioma models. Neuro Oncol 23:770–782. https://doi.org/10.1093/neuonc/noaa263
Zang J, Ye J, Zhang C, Sha M, Gao J (2019) Senescent hepatocytes enhance natural killer cell activity via the CXCL-10/CXCR3 axis. Exp Ther Med 18:3845–3852. https://doi.org/10.3892/etm.2019.8037
Zhang J, Wu G, Miller CP, Tatevossian RG, Dalton JD, Tang B, Orisme W, Punchihewa C, Parker M, Qaddoumi I et al (2013) Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet 45:602–612. https://doi.org/10.1038/ng.2611
Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N et al (2014) An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34:11929–11947. https://doi.org/10.1523/JNEUROSCI.1860-14.2014
Zhao M, Ma Q, Xu J, Fu S, Chen L, Wang B, Wu J, Yang L (2015) Combining CXCL10 gene therapy and radiotherapy improved therapeutic efficacy in cervical cancer HeLa cell xenograft tumor models. Oncol Lett 10:768–772. https://doi.org/10.3892/ol.2015.3281
Zhu Y, Harada T, Liu L, Lush ME, Guignard F, Harada C, Burns DK, Bajenaru ML, Gutmann DH, Parada LF (2005) Inactivation of NF1 in CNS causes increased glial progenitor proliferation and optic glioma formation. Development 132:5577–5588. https://doi.org/10.1242/dev.02162
Acknowledgements
This work was supported by a National Cancer Institute R50 Research Specialist Award (1-R50-CA233164-01 to C.A.), pre-pilot funding for small-animal MRI from the Siteman Cancer Center (to C.A.), grants from the Pediatric Brain Tumor Foundation (P20-00873 to D.H.G.) and Ian’s Friends Foundation (to D.H.G.), and a Research Program Award from the National Institutes of Neurological Disorders and Stroke (1-R35-NS07211-01 to D.H.G.). The GEiC Center at WUSM engineered the hiPSCs and is subsidized by NCI Cancer Center Support Grant P30-CA091842. The Washington University Ophthalmology Core facility is funded by National Eye Institute Grant P30EY002687.
Author information
Authors and Affiliations
Contributions
CA and DHG designed and analyzed the experiments. CA, JC, OC, SMS, AC, AFW, CMK, AHZ, GX, JRG conducted and/or interpreted the experiments. FR provided reagents and conducted the histopathological analyses. The manuscript was assembled by CA and DHG was responsible for the final production of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Anastasaki, C., Chatterjee, J., Cobb, O. et al. Human induced pluripotent stem cell engineering establishes a humanized mouse platform for pediatric low-grade glioma modeling. acta neuropathol commun 10, 120 (2022). https://doi.org/10.1186/s40478-022-01428-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40478-022-01428-2