Abstract
Xylanase gene isolated from Bacillus brevis was expressed in E. coli BL21. Sequencing of the gene (Gen Bank accession number: HQ179986) showed that it belongs to family 11 xylanases. The recombinant xylanase was predominantly secreted to culture medium and showed mesophilic nature (optimum activity at 55°C and pH 7.0). The cell free culture medium exhibited 30 IU/ml xylanse activity. The enzyme did not show any cellulose activity and was active under wide range of temperature (40°C to 80°C) and pH (4 to 9). The enzyme showed considerable thermo stability and regained over 90% of activity, when returned to 55°C after boiling for 5 min. These physiochemical properties of B. brevis xylanse show high potential of its applications in paper and pulp industry.
Similar content being viewed by others
Introduction
Xylan is a major component of hemicellulose of forest and agricultural materials such as hardwood, grain straw, corn cobs and grasses (Dekker and Richards 1976; Wilkie 1979). Xylan can be enzymatically degraded to useful products like xylose (Dekker and Richards 1976; Wang et al. 1980), xylitol, and ethanol (Jeifries 1981; Sung et al., 1993). Xylan is generally insoluble in nature; however, a number of microorganisms with the help of some of their enzymes can readily solubilize xylan. D-xylanase (15 kD to 30 kD) is one of the key enzymes required for the degradation of xylan (Dekker and Richards 1976; Domiano et al. 2003).
One of the exciting prospects for recombinant xylanase is its use in the paper and pulp bio-bleaching (Saleem et al. 2009; Singh et al. 2013) to reduce the requirement of organo-chemicals for bleaching process (Kuhad et al. 1998). However, for its use in paper and pulp industry for bio-bleaching, xylanase pretreatment has to take place at a high temperature and in alkaline conditions; hence thermostable xylanases (Chapla et al. 2012; Saleem et al. 2012) with high pH optimum are of great importance. Moreover, for industrial application xylanase should be celleulase free and require minimum downstream processing for its production. Therefore, this study was carried out to increase the xylanase production by heterologous expression of Bacillus brevis xylanase gene in E. coli and secreting it in the medium, so that it require minimum downstream processing for its applications in paper and pulp industry. Analysis of some of its biochemical characteristics was carried out.
Materials and methods
Bacterial strains and culture media
Bacillus brevis strains, obtained from Bacillus genetic stock center (BGSC Accession number ATCC8246T) was used in the present study. For xylanase production by B. brevis M-9 medium supplemented with 1% xylan was used.
Cloning of β-1, 4-endo-xylanase gene from Bacillus brevis
Total genomic DNA of the bacteria was isolated following the modified procedure of Ausubel et al. (1994) and used for template with PCR primers designed from Brevibacillus brevis endo-1, 4-beta-xylanase (xylB) gene (GenBank DQ100303). The PCR primers used were as follows; Forward 5′CGGGGTACC TAA ATGTTTAAGTTTAAAAAGAATTTCTTAGTTGG; Reverse 5′ CGGAATTC TTACCACACTGTTACGTTAGAACTTCC. The Kpn I and Eco RI restriction sites incorporated for cloning are underlined. The forward primer has a stop codon (Bold highlighted) just before the start codon. It was incorporated to remove N-terminal S.tag fusion sequence present in the pET29 vector and to create a mini-cistron before the xalanse coding sequence. The PCR was conducted in a GeneAmp PCR System 9700 (PE Biosystems) for 35 cycles (each cycle consisting of 1 min of denaturation at 93°C, 1 min annealing at 55°C, 1 min extension at 72°C, and a final 10 min extension at 72°C) using Taq Polymerase (Fermentas). Amplified PCR products were extracted from agarose gels by QIAGEN gel extraction kit and cloned in pUC vector. The sequence verified gene was further subcloned in protein expression vector pET29A using the Kpn I and Eco RI sites and mobilized to E. coli BL 21(DE3) for protein expression.
Enzyme assays
The xylanase activity was determined with 1% birchwood xylan in 50 mM phosphate buffer of pH 7 at 55°C using the method described by Bailey et al. (1992). The enzymatic reaction was carried out for 5 min and the reducing sugars were determined using the DNS method (Miller 1959). The xylanase activity was measured in terms of international units (IU). One IU of xylanase is defined as one μ mole of xylose produced by 1 ml undiluted enzyme in 1 min. The μ moles of xylose produced by xylanase were deduced from xylose standard plot.
SDS-PAGE and zymogram
SDS-PAGE (12.5%) was performed by the standard methods as described by Laemmli (1970). Protein bands visualized by incubating gels with gentle shaking for 30 min in 10% trichloroacetic acid, 4 h in Coomassie brilliant blue R250Coomassie blue (0.025%) in (45:45:10 water:ethanol:acetic acid), and overnight in destaining solution (67% water, 25% ethanol, 8% acetic acid). Modified gels for the detection of in-situ xylanase activity (zymograms) were prepared by substituting a boiled solution of birchwood xylan (0.5%) for water during the preparation of the separating layer of the gel. Following electrophoresis, these gels were incubated for 1 h with gentle agitation in 2.5% Triton X-100 and then for 30 min at 80°C in pre-heated buffer (12.5 mM bis-tris propane, pH 6.0, at 80°C). In- situ xylanase activity was detected by staining the gels for 30 min in 0.1% Congo red and then destaining them in 1 M NaCl. The activity gels were rinsed in a dilute acid solution (10 mM HCl) to increase the contrast between the hydrolyzed (clear) and non-hydrolyzed (black) xylan prior to photography.
For the zymogram analysis, the crude enzyme samples were electrophoresed as above on SDS-PAGE containing xylan (0.1%). After running, the gel was washed four times for 30 min in 100 mM phosphate buffer (pH 7.0); the first two washes containing 25% isopropyl alcohol, to remove SDS and renature protein in the gel. The gel was then incubated for 20 min at 37°C before soaking in Congo Red solution for 5 min at room temperature and washing with 1 M NaCl until excess dye was removed from the active band. The zymogram was prepared after soaking the gel in 0.5% acetic acid solution. The background turned dark blue, and clear zones were observed in the areas exposed to xylanase activity (Nakamura et al. 1994).
Homology modeling
B. brevis xylanse protein structural model was developed through homology modeling using I-Tasser server (Roy et al. 2010). Quality of predicted structural models were evaluated through stereochemical parameters of Ramachandran Plot and verify-3D (Laskoswki et al. 1993; Luthy et al. 1992).
Results
B. brevis xylanase gene
The xylanase sequence obtained in the present study was aligned with the published sequence of xylanase from B. brevis (GenBank DQ100303). The nucleotide sequence alignment shows that both the sequences align completely without any gap (Additional file 1: Figure S1). However, there is a difference of 33 nucleotides between these two xylanase sequences. Alignments of translated sequences of these two proteins reveal a difference in seven amino acids (Additional file 2: Figure S2). Whether this difference is reflected in 3D model or not was compared by developing the 3D model of both Xylanses followed by superimposion (Figure 1A and B). These two structures differ only marginally from each other. High-lighting of differences on the 3D structure show that most of the differences in amino acid residues are present in two clusters. One around the active site and second below the site (Figure 1B). To understand weather the clustering of difference in amino acid sequences present in 3D model is restricted to B. brevis or same pattern is also present in other species, xylanase sequences from different Bacillus species were retrieved from the database and their 3D structure developed and evaluated. Results summarized in Figure 1C show that this pattern of clustering of mutation around active site is present in other Bacillus xylanase also (Figure 1C and D). The B. Brevis Xylanase protein sequence was further analyzed for presence of secretary signal peptide. The sequence analysis shows that a 28 amino acid long secretary signal peptide is present in the B. brevis Xylanse (Additional file 3: Figure S3). In silico sequence analysis and stereochemical evaluation of modeled B. brevis xylanase protein 3D structure shows high quality of predicted structural model, without any residue in disallowed region (Figure 2).
Structural comparisons of B. brevis xylanase with other Bacillus Sp. (A). Surface view of Xylanase protein 3D structure, (B). Superimposition of two B. brevis structures, Green regions shows differences in the amino acid sequences of the two structures. Three differences are around active site while two are grouped at the distant region of the proteins. (C & D). Superimposition with B. licheniformis and B. amyloliquefaciens xylanse 3D structures respectively.
Heterologous expression of B. brevis xylanase
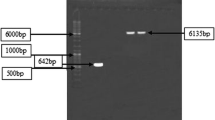
SDS-PAGE of IPTG induced Zymogram was developed that showed xylanase activity on the gel by clearing zone development on staining with Congo Red dye. Xylanase production was found more (approximately 1.5 times higher) in cloned expression host as compared to B. brevis (Figure 3). The optimization of temperature of both cloned and native (B. brevis) xylanases show mesophilic nature of B. brevis xylanase with maximum activity at 55°C Figure 4A. Similarly the optimum pH of the B. brevis xylanase was found at pH 7 (Figure 4B). After finding optimum temperature and pH, effect of high temperature on the enzyme activity was measured. Although enzyme showed considerable loss of enzyme activity at higher temperature and negligible activity was observed above 90°C, the enzyme regained more than 90% activity after boiling for 5 min and subsequent cooling at 37°C for 45 minutes. The enzyme was also evaluated for any cellulose activity and as expected, no detectable cellulase activity was observed.
SDS PAGE analysis for xylanase. (A). SDS-PAGE analysis of B. brevis xylanase expression in two clones (2&3). M: Protein ladder BL: BL21 (DE3) host cell transformed with pET29A vector (Control), 2&3 are two different colonies for pET-29-xylanse transformed in BL21 (DE3). Xylanse protein in dominantly visible only in medium, indicating that most of protein is getting secreted in the growth medium. (B). Xymogram analysis confirms tthat the 23 kDa protein band observed in SDS is a functional Xylanase enzyme.
Discussion
Many xylanase genes have been isolated from different microbial organisms and were expressed in E. coli, however, the xylanase expression level in E. coli was generally lower than the parent organism (Bernier et al. 1983; Gallardo et al. 2003; Paice et al. 1986; Panbangered et al. 1983, 1984, 1985; Sandhu and Kennedy 1984; Sipat et al. 1987) and confined mainly to the cytoplasmic or periplasmic fractions (Kulkarni et al. 1999). Hyperexpression of a B. circulans xylanase gene in E. coli was reported by (Yang et al. 1989) with a xylanase activity of 7 IU/ml found in intracellular fraction of E. coli. Extra-cellular activity of xylanase had also been reported earlier in recombinant E. coli for the bacterial xylanases from alkaliphilic Bacillus by Honda (1985), and alkaliphilic, and thermophilic Bacillus species by Shendye and Rao (1993). A synthetic gene encoding mature B. circulans xylanase has been designed to imitate the frequency of degenerate codons used in E. coli. This synthetic gene was then converted to B. subtilis xylanase gene via substitution of Thr147 Ser codon. Under the control of lac promoter, recombinant xylanase activity in solution form in cytoplasm was reported as high as 300 mg/ml (Sung et al. 1993).
In the present study we have isolated a xylanase gene from B. brevis and cloned it in E. coli. We understand that this is the first report on cloning and heterologous expression of B. brevis xylanase gene in BL21 host. The most significant part of this study is the expression of cloned xylanase gene in extracellular medium of E. coli which may be exploited for production of xylanase at commercial level. It is interesting that the xylanase activity in the E. coli clone is higher (2 times) over that of native B. brevis strain, possibly due to higher level of protein expression. The enzyme produced by E. coli is functionally active and capable of degrading birchwood xylan even in SDS-PAGE gel also. In silico sequence analysis and stereochemical evaluation of modeled B. brevis xylanase protein 3D structure shows high quality of predicted structural model, without any residue in disallowed region (Figure 2). Further comparison of xylanses from different bacterial sources reveals high level of structural similarity with point mutations. These point mutations might have evolved to provide adaptations to bacteria under different environmental conditions. Further studies on the DNA sequence of the cloned fragment containing xylanase gene should prove helpful in determining the regulatory sequences of the gene and the subunit structure of this enzyme.
Conclusion
B. brevis xylanase was efficiently expressed and secreted by E. coli (BL21). The xylanase activity was found more (2 times) in culture filtrate of BL21 as compared to B. brevis. Heterologous expression systems that produce large amounts of secreted proteins with in an organism that can be grown in industrial scale fermenters must be developed to facilitate higher enzyme production by using agri-waste as a carbon source for enzyme production. Since optimization of growth medium is a useful tool to attain high levels of enzyme activity at lower cost (Damaso et al. 2003) further studies on medium optimization might improve the yield of B. brevis xylanase production with the kind of BL21 expression systems described in this paper. An important further consideration is to develop higher-level enzyme producing integrant E. coli strains with a number of copies of B. brevis xylanase for growth in optimized mefdia.
References
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K: Current protocols in molecular biology. John Wiley and Sons, New York; 1994:2.0.1-2.14.8.
Bailey MJ, Biely P, Poutanen K: Interlaboratery testing of methods for assay of xylanase activity. J Biotechnol 1992, 23: 257-270. 10.1016/0168-1656(92)90074-J
Bernier R Jr, Driguez H, Desrochers M: Molecular cloning of a Bacillus subtilis xylanase gene in Escherichia coli . Gene 1983, 26: 59-65. 10.1016/0378-1119(83)90036-7
Chapla D, Patel H, Madamwar D, Shah A: Assessment of a thermostable xylanase from Paenibacillus sp.ASCD2 for application in prebleaching of eucalyptus kraft pulp. Waste Biomass Valor 2012, 3: 269-274. doi:10.1007/s12649-012-9112-z 10.1007/s12649-012-9112-z
Damaso MCT, Almeida MS, Kurtenbach E, Martins OB, Pereira N Jr, Andrade CMMC, Albano RM: Optimized expression of a thermostable xylanase from Thermomyces lanuginosus in Pichia pastoris . Appl Environ Microbiol 2003, 69: 6064-6072. 10.1128/AEM.69.10.6064-6072.2003
Dekker RFH, Richards GN: Hemicelluloses: their occurrence, purification, properties, and mode of action. Adv Carbohydr Chem Biochem 1976, 32: 276-352.
Domiano VB, Bocchins DA, Gomes E, Silva D: Applications of crude xylanase from Bacillus licheniformis 77–2 to the bleaching of eucalyptus kraft pulp. World J Microbiol Biotechnol 2003, 19: 139-144. 10.1023/A:1023244911314
Gallardo O, Diaz P, Pastor FI: Cloning and characterization of xylanase A from the strain Bacillus sp. BP-7: comparison with alkaline pI-low molecular weight xylanases of family 11. Curr Microbiol 2003, 48: 276-279.
Honda H, Kudo T, Horikoshi K: Molecular cloning and expression of the xylanase gene from alkalophilic Bacillus strain C-125 in Escherichia coli . J Bacteriol 1985, 161: 784-785.
Jeifries TW: Conversion of xylose to ethanol under aerobic conditions by Candida tropicalis . Biotechnol Lett 1981, 3: 213-218. 10.1007/BF00154647
Kuhad RC, Manchanda M, Singh A: Optimization of xylanase production by a hyper-xylanolytic mutant strain of Fusarium oxysporum . Process Biochem 1998, 33: 641-647. 10.1016/S0032-9592(98)00025-9
Kulkarni N, Shendye A, Rao M: Molecular and biotechnological aspects of xylanases. FEMS Microbiol Rev 1999, 23: 411-456. 10.1111/j.1574-6976.1999.tb00407.x
Laemmli UK: Cleavage of structural proteins during the assembly of the head of bacteriophage-T4. Nature 1970, 227: 680-685. 10.1038/227680a0
Laskoswki RA, MacArthur MW, Moss DS, Thorton JM: PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 1993, 26: 283-291. 10.1107/S0021889892009944
Luthy R, Bowie JU, Eisenberg D: Assessment of protein models with three-dimensional profiles. Nature 1992, 356: 83-85. 10.1038/356083a0
Miller GL: The use of dinitrosalicylic acid reagent for the determination of reducing sugars. Anal Chem 1959, 31: 426-428. 10.1021/ac60147a030
Nakamura S, Nakai R, Wajabatacgu K, Ishiguro Y, Aono R, Horikoshi K: Thermophilic alkaline xylanase from newly isolated alkaliphilic and thermophilic Baillus sp. strain TAR-I. Biosci Biotechnol Biochem 1994, 58: 78-81. 10.1271/bbb.58.78
Paice M, Bourbonnais R, Desrochers M, Jurasek L, Yaguchi M: A xylanase gene from Bacillus subtilis : nucleotide sequence and comparison with B. pumilus gene. Arch Microbiol 1986, 144: 201-206. 10.1007/BF00410947
Panbangered W, Shinmyo A, Kinoshita S, Okada H: Purification and properties of endoxylanase produced by Bacillus pumilus . Agric Biol Chem 1983, 47: 957-963. 10.1271/bbb1961.47.957
Panbangered W, Kawaguchi O, Tomita T, Shinmyo A, Okada H: Isolation of two B- xylosidase genes of Bacillus pumilus and comparisions of their gene products. Eur J Biochem 1984, 138: 267-273. 10.1111/j.1432-1033.1984.tb07911.x
Panbangered W, Fukusake E, Epifanio E, Shinmyo A, Okada H: Expression of a xylanase gene of Bacillus pumilus in Escherichia coli and Bacillus subtilis . Appl Microbiol Biotechnol 1985, 22: 259-264.
Roy A, Kucukurla A, Zhang Y: I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 2010, 5: 25-738.
Saleem M, Tabassum MR, Yasmin R, Imran M: Potential of xylanase from thermophilic Bacillus sp. XTR-10 in biobleaching of wood kraft pulp. Bioresource Technol 2009, 100(12):3069-3075. 10.1016/j.biortech.2009.01.020
Saleem M, Aslam F, Akhtar MS, Tariq M, Rajoka MI: Characterization of a thermostable and alkaline xylanase from Bacillus sp. and its bleaching impact on wheat straw pulp. World J Microbiol Biotechnol 2012, 28: 513-522. 10.1007/s11274-011-0842-z
Sandhu JS, Kennedy JF: Molecular cloning of Bacillus polymyxa (1–4)-P-D-xylanase gene in Escherichia coli . Enzyme Microb Technol 1984, 6: 271-274. 10.1016/0141-0229(84)90129-7
Shendye A, Rao M: Cloning and extracellular expression in Escherichia coli of xylanase from an alkalophilic thermophilic Bacillus sp. NCIM 59. FEMS Microbiol Lett 1993, 108: 297-302. 10.1111/j.1574-6968.1993.tb06119.x
Singh V, Pandey VC, Agrawal S: Potential of Laceyella sacchari strain B42 crude xylanase in biobleaching of kraft pulp. African J of Biotechnol 2013, 12(6):570-579.
Sipat A, Taylor KA, Lo RYC, Forsberg CW, Kreli PJ: Molecular cloning of a xylanase gene from Bacteroides succinogenes and its expression in Escherichia coli . Appl Environ Microbiol 1987, 53: 477-481.
Sung WL, Luk CK, Zahab DM: Overexpression and purification of the Bacillus subtilis and Bacillus circulans xylanases in Escherichia coli . Protein Express Purif 1993, 4: 200-206. 10.1006/prep.1993.1026
Wang PY, Johnson BF, Schneider H: Fermentation of D-xylose by yeasts using glucose isomerase in the medium to convert D-xylose to D-xylulose. Biotechnol Lett 1980, 2: 273-278.
Wilkie KCB: The hemicelluloses of grasses and cereals. Adv Carbohyr Chem Biochem 1979, 36: 215-262.
Yang RCA, MacKenzie CR, Billows D, Nearing SA: Identification of two distinct Bacillus circulars xylanases by molecular cloning of the genes and expression in Escherichia coli . Appl Environ Microbiol 1989, 55: 568-574.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
GKG - Carried out all research work and framed manuscript. MK - Participated in framing the manuscript. VN and CA - Participated in Bioinformatics work. BR - Participated in framing the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
40064_2013_1451_MOESM1_ESM.ppt
Additional file 1: Figure S1: Nucleotide sequence alignment of published B. brevis (B. brevis _o) and new B. brevis (B. brevis _G) xylanase sequences. identical sequences are highlighted in boxes. (PPT 111 KB)
40064_2013_1451_MOESM2_ESM.ppt
Additional file 2: Figure S2: Amino acid sequence alignment of published B. brevis (B. brevis _O) with the new B. brevis (B. brevis _G). Identical residues are highlighted in the box. (PPT 86 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Goswami, G.K., Krishnamohan, M., Nain, V. et al. Cloning and heterologous expression of cellulose free thermostable xylanase from Bacillus brevis. SpringerPlus 3, 20 (2014). https://doi.org/10.1186/2193-1801-3-20
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/2193-1801-3-20