Abstract
Background
Pollution due to the heavy metals is a problem that may have negative consequences on the hydrosphere. One of the best procedures in removing the toxic metals from the environment is using metal resistant bacteria.
Results
In the present study eight nickel resistant bacteria were isolated from industrial wastewaters. Three of them were selected as the most resistant based on their Maximum tolerable concentration (8, 16 and 24 mM Ni2+). Their identification was done according to morphological, biochemical characteristics and 16SrDNA gene sequencing and they were identified as Cupriavidus sp ATHA3, Klebsiella oxytoca ATHA6 and Methylobacterium sp ATHA7. The accession numbers assigned to ATHA3, ATHA6 and ATHA7 strains are JX120152, JX196648 and JX457333 respectively. The Growth rate of the most resistant isolate, Klebsiella oxytoca strain ATHA6, in the presence of Ni2+ and the reduction in Ni2+ concentration was revealed that K oxytoca ATHA6 could decrease 83 mg/mL of nickel from the medium after 3 days.
Conclusion
It can be concluded that the identified Ni resistant bacteria could be valuable for the bioremediation of Ni polluted waste water and sewage.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
The pollution of the environment with toxic heavy metals is increasing throughout the world along technological development. Copper, Chromium, Cadmium and Nickel are known to be the most commonly heavy metals used and the more wide spread contaminants of the environment [1, 2]. Wastewater contains significant concentration of heavy metals that are not degraded by the conventional process of wastewater treatment. The main source of heavy metals is the industrial activities such as metal processing, mining and electroplating, tanning, carpet washing and dying. Presence of high concentration of toxic heavy metals in waste water can cause severe problems to human health [3]. Bioremediation can be used to effectively reduce contaminant toxicity, mobility or volume to levels that are innocuous to human health and ecosystem [4]. Microorganisms possess mechanisms that regulate metal ion accumulation to avoid heavy metal toxicity and there are many reports about microbial resistance to heavy metals [5–7]. To survive under metal- stressed conditions, bacteria have evolved several types of mechanisms to tolerate the uptake of heavy metal ions. These mechanisms include the efflux of metal ions outside the cell, accumulation of the metal ions inside the cell and reduction of the heavy metal ions to a less toxic state [8].
Nickel is the 24th most abundant element in the earth crust [9]. Microorganisms have evolved in the presence of this metal, which is necessary in trace amounts for a variety of metabolic processes but in high concentration causing oxidative stress in the cell. The best known mechanisms of nickel resistance are mediated by efflux pumps such as cnr CBA (cobalt- nickel resistance) from Cupriavidus metallidurans CH34 (formerly Ralstonia metallidurans CH34), NccCBA (Nickel-cobalt-cadmium) and NreB (nickel resistance) from Achromobacter xylosoxidans 31A, CznABC (Cadmium-zinc-nickel) from Helicobacter pylori[10]. The aims of this study are isolation and characterization of nickel resistant bacteria (NiRB) from industrial wastewater, MTC (maximum tolerable concentration) and multiple metal tolerance determination of them.
Materials and methods
Sampling
Wastewater samples were collected in screw capped sterilized bottle from Factory’s steel- copper- smithy- sewage treatment plant of Isfahan (Iran). Some physicochemical parameters of wastewater viz., temperature (°C), pH, BOD, COD were measured [11].
Isolation and screening of Ni-resistant bacteria
Wastewater sample was spread on PHGII agar Plates containing 0.5 mM of Ni2+. PHGII agar plates were prepared by dissolving 2 g NaCl, 4 gr peptone and 1gr yeast extract in 1000 mL distilled water, pH was adjusted at 7.0 and then 15 gr agar was added in the 1000 mL Flasks. The medium was autoclaved at 121°C and 15 Lb pressure for 15 minutes, then supplemented with Ni2+ (0.5 mM/L). The growth of bacterial colonies was observed after 5 days of incubation at 30°C [12].
Identification of the bacterial isolate
Selected isolates were grown on LB (Luria- bertani) media. The shape and color of the colonies were examined under the Microscope after gram staining. Isolates were biochemically analyzed for the activities of oxidase, Catalase, VP-MR test, motility, indole production and citrate utilization according to Bergey’s manual of systematic Bacteriology [13].
Determination of maximum tolerable concentration and multi metal resistance
The Maximum Tolerable Concentration of heavy metal was selected as the highest concentration of heavy metal that allows growth after 2 days [14]. The increasing concentration of Ni (1, 2, 4, 8, 16, 24, 32 mM) on PHGII agar plates were used for testing the MTCs of NiRB. Then multi metal resistances of selected bacteria were also investigated [15].
16 SrDNA gene amplification and sequencing
Molecular identification of the most NiRB was performed by 16S rDNA PCR Sequencing [16]. Genomic DNA was extracted and the 16S rDNA gene was amplified by using the universal bacterial Primers of F27 (5′-CAGGGTACCAGAGTTTGA-3′) andR1492 (5′-CTCTCTGCAGTACGGCTAC-3′).
PCR was performed as a 25 μL reaction Mixture containing 2 μL of DNA extract as a template. Each primer at a concentration of 8 PM, 2 mM mgCl2 and dNTPS at a concentration of 0.2 mM, as well as 1.25 U of Taq polymerase and buffer were used. After the initial denaturation for 5 min at 95°C, there was 32 Cycle consisting of denaturation at 94°C for 1 min, annealing at 57°C for 1 min, extension at 72°C for 1 min and final extension at 72°C for 5 min. PCR was carried out in a gene AMP PCR system (Eppendorf). PCR products were analyzed by 1% (W/V) agarose gel electrophoresis in 0.5x TBE buffer with ethidium bromide (05 μg/ml). PCR products were then sequenced and compared with the National Center for Biotechnology Information (NCBI) database using the BLAST search available through the center’s website (http://www.ncbi.nlm.nih.gov/BLAST). The 16S rDNA sequences were then submitted to the Gene Bank using the BankIt service.
Nickel removal by isolated bacterium
Growth of selected isolate was carried out in PHGII broth supplemented with (12 mmol Ni2+/mL) and without Nickel (control).
The cultures were incubated at 30°C for 6 days and from each medium (control and treated) the amount of 5 ml was taken out under sterilized conditions after 24, 48, 72, 96, 144, 168 and 216 hours, respectively. The cultures were centrifuged at 3000 rpm for 5 minutes and supernatants were used for the estimation of Ni by atomic absorption Spectrophotometer (a Perkin-Elmer 800 atomic absorption spectrophotometer).
Results
The physicochemical characteristics of waste waters are shown in Table 1. From the three studied industrial wastewater, eight Ni resistant bacteria were isolated. The biochemical characteristics of isolated bacteria were shown in Table 2.
Maximum Tolerable Concentration of Ni- resistant bacteria were shown in Table 3 and three isolates were selected as the most resistant (S5, S7 and S8). Then the Multi metal resistant of the 3 selected isolates was determined and are shown in Table 4. Also, the gene sequence analysis were done to further identify the bacteria and to support the results from the biochemical analysis of the most resistant isolates. The phylogenetic distance trees of results are shown in Figure 1. The percentage of maximum similarity and GenBank accession number are shown in Table 5.
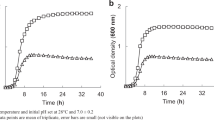
The Growth rate of the most resistant isolate, Klebsiella oxytoca strain ATHA6, in the presence of Ni2+ (704 mg/ml) and the reduction in Ni2+ concentration are shown in Figure 2. It is revealed that K oxytoca ATHA6 could decrease 83 mg/mL of nickel from the medium after 3 day.
Discussion
Heavy metal-resistant microorganisms are thought to naturally occur; primarily in metal-contaminated soils. Studies suggest that metals as well as other soil physicochemical parameters can contribute to bacterial resistance to metals [17]. Soil pH can influence the solubility of metals, [18]. The pH values of the soil samples revealed no significant differences at all localities and ranged from 7.4 to 8.4. Both the BOD and COD tests are a measure of the relative oxygen-depletion effect of a waste contaminant [19]. Both have been widely adopted as a measure of pollution effect. So according to results the studied waste water were belonged to weakly contaminant wastewaters.
Surviving microorganisms in a stressed environment especially in presence of heavy metals is dependent to development of tolerance mechanisms [20]. They also play important roles in the cycling of toxic metals in the biosphere. While toxic mechanism of all heavy metals is similar, multiple tolerances are common phenomena among heavy metal resistant bacteria. So, in this research efforts were made to isolate nickel resistant bacteria (NiRB) from industrial effluents.
Eight NiRB were isolated from studied industrial effluents. Most of them were Gram- negative cocci and bacilli; they were identified as Moraxella (Bovis), Acinetobacter (Lwoffi), Providencia (stuartii), Branhamella (Catarhalis), Cupriavidus sp ATHA3, S. aureus, Methylobacterium sp. ATHA7, Klebsiella oxytoca ATHA6.
Virender et al. [2] also isolated gram- negative NiRB isolates [2]. All the isolates of this research exhibited resistance with maximum tolerable concentration (MTC) for nickel ranging from 1 to 24 mM. Among them, three isolated strains of ATHA3, ATHA7 and ATHA 6 were found to have significant higher MTC than the other isolates (8, 16 and 24 mM respectively) and selected as the most Ni resistant isolates for further study. Sevgi et al. [21] isolated NiRB and their resistance was found to be in the range of 2.0-4.0 mM of NiCl2 while it has also been reported that nickel-resistant bacteria [21]. Alcaligenes xylosoxidans, isolated from a galvanization tank, tolerated relatively high concentrations of nickel (40 mM) [22].
Spatel et al. [6] also isolated Ni resistant Pseudomonas fragi from industrial effluent containing different concentrations of nickel sulfate. Well growth of isolate was observed in the medium containing up to 2.5 mmol/L of nickel [6]. Rajbanshi studied on heavy metal resistant bacteria in sewage treatment plant and could isolate nickel resistant Staphylococcus spp. and Bacillus spp., which showed MIC ranging from 150 μg/mL to 500 μg/ml [23]. Arundhati and Paul [24] also isolated the nickel-resistant bacterium Cupriavidus sp. which exhibited a typical inducible Ni resistance in Ni supplemented (1.0-10.0 mM) Tris-minimal medium. This strain could accumulate a maximum of 29.3 μM Ni/g protein after 48 h of growth in 5 mM Ni. They were stated that nickel resistance in Cupriavidus may be due to extracytoplasmic binding and accumulation coupled with expression of specific periplasmic proteins [24].
Multi Metal Tolerance Test of the three NiRB isolates indicated highest tolerance to Cd (8 mM) by ATHA3 and ATHA7 strains and lowest to Cu (1 mM) by ATHA6. Pb resistance in all cases was 2 mM. NiRB isolates of Sevgi et al. [21] study, showed multi metal resistance as follows: cobalt (20 mM), zinc (10 mM), cadmium (1 mM), and copper (1 mM). Malik also isolated a total of 70 bacterial isolates from industrial and agricultural soils and tested for their resistance against several heavy metals. They reported that the industrial isolates were more resistant to different metals than agricultural isolates. They showed that 88.8% of isolates from industrial soil were resistant to Ni, 80% to Cr, 82.8% Zn, and 71.4% to Cd. Multiple tolerances occur only to toxic compounds that have similar mechanisms underlying their toxicity [25]. Since all heavy metals are similar in their toxic mechanisms multiple tolerances are common phenomena among heavy metal resistant bacteria [26].
The effect of Ni concentration on the bio removal capacity and growth rate of ATHA6 is shown in the Figure 1. Another finding of metal presence in the culture medium was reduction in growth rate as compared with control. This has been explained that the exposed microorganism to metals stress deviate its energy from growth to maintenance of other functions as a greater demand of energy to resist metal toxicity [27]. Bio removal efficiency increased with time, and maximum efficiency was observed at 72 h of growth (11.78% reduction in Ni2+ concentration). Bio removal was negligible after this time. Specific surface properties and the physiological state of the microorganisms might have a role in metal uptake. High biomass production is also important for better bio removal.
For the phylogenetic analysis of the most resistant NiRB isolates, 16S rDNA genes were amplified as described before. About 1500 bp PCR products were used for DNA sequencing with the same forward and reverse primers as the PCR primers. The 16S rDNA gene sequences of ATHA3, ATHA6 and ATHA7 strains were submitted to GenBank. The accession numbers assigned to ATHA3, ATHA6 and ATHA7 strains are JX120152, JX196648 and JX457333 respectively.
Comparison of 16S rDNA gene sequences revealed that ATHA3 showed about 98% similarity to Cupriavidus metalidurans strain CH34. Cupriavidus metallidurans CH34 is a β-proteobacteria adapted to metal contaminated environments. It has AtmA, a chromosomal ABC transporter, causing resistance to nickel [28].
As a result of a BLAST search, 99% similarity was observed between the 16S rDNA gene sequence of ATHA6 and Klebsiella oxytoca. Stoppel and Schlegel [29] found chromosomally coded Nickel resistance genes of K. oxytoca that were not found to occur in combination with any of the other known nickel resistance determinants [29]. So, the nickel resistance genes of K. oxytoca may encode a second type of nickel resistance which appears independently from other nickel resistance determinants.
96% similarity of pink pigmented ATHA7 and Methylobacterium Variabile strain GR3 was observed. A 96% similarity in 16S rDNA gene sequences is a low value for identification at species level. Therefore, ATHA7 may be a member of a novel bacterial species. Various pink-pigmented Methylobacteria were also obtained from the rhizosphere and endosphere of hyper accumulating plant Thlaspi goesingense grown in Redschlag, Austria. These strains were found to exhibit different multiple heavy metal resistance characteristics to Ni, Cd, Co, Zn and Cr [30]. Aboudrar et al. [31] also confirmed that, the NiRB strains could reduce the soil bioavailability of Ni and also induce promoting effects on the growth of plant. So, they stated that NiRB strains could serve as an effective metal-immobilizing and growth-promoting bioinoculant for plants in Ni-stressed soils [31].
Conclusion
It can be concluded that there are several different heavy metal resistant bacteria in the contaminated environment that should be isolated and identified for further studies. The most NiRB isolated of this study are Cupriavidus sp ATHA3, Klebsiella oxytoca ATHA6 and Methylobacterium sp ATHA7. These identified Ni resistant bacteria could be valuable for the bioremediation of Ni polluted waste water and sewage.
References
Hussein H, Farag S, Moawad H: Isolation and characterization of pseudomonas resistance to heavy metals contaminants. Arab J Biotechnol 2003, 1(7):13–22.
Virender S, Chauhan PK, Kanta R: Isolation and characterization of pseudomonas resistant to heavy metals contaminants. Int J Pharm Sci Rev Res 2010, 3: 164–167.
Keramati P, Hoodaji M, Tahmourespour A: Multimetal resistance study of bacteria highly resistant to Mercury isolated from dental clinic effluent. Afr J Microbiol Res 2011, 5: 831–837.
Toroglu S, Dıncer S: Heavy metal resistances of Enterobacteriaceae from Aksu River (Turkey) polluted with different sources. Asian J Chem 2009, 21(1):411–420.
Fillali BK, Taoufik J, Dzairi FZ: Waste water bacterial isolates resistant to heavy metals and antibiotics. Currt Microbiol 2000, 41: 151–156.
Spatel J, Cpatel P, Kalia K: Isolation and characterization of Nickel Uptake by Nickel Resistant Bacterial Isolate (NiRBI). Biomed Environ Sci 2006, 19: 297–301.
Narasimhulu K, Sreenivasa P, Vinod A: Isolation and identification of bacterial strains and study of their resistance to heavy metals and antibiotics. J Microbial Biochem Technol 2010, 2: 74–76.
Laila MA, Wagdy KB, Thanaa HA: Heavy metal resistance and gene expression analysis of metal resistance gene in gram- positive and gram- negative bacteria present in Egyptian soils. J Appl Sci In Environ Sanit 2011, 6(2):201–211.
Iyaka AY: Nickel in soils: A review of its distribution and impacts. Sci Res Essays 2011, 6(33):6774–6777.
Salvador M, Carolina G, Jose E: Novel nickel resistance genes from the rhizosphere metagenome of plants adapted to acid mine drainage. Appl Environ Microbiol 2007, 73(19):6001–6011.
APHA - American public Health Association: Standard Methods for the Examination for Water and Wastewater. 20th edition. Washington: AWWA, WPCF; 1998.
Teitzel GM, Persek MR: Heavy metal resistance of biofilm and Planktonic Pseudomonas aeruginosa . Appl Environ Microbiol 2003, 69(4):2313–2320.
Claus D, Berkeley RCW, et al.: Genus pseudomonas. In Bergey's Manual of Systematic Bacteriology. Volume 2. 2nd edition. Edited by: Sneath PHA. Baltimore, MD: Williams & Willkins Co.; 1968:140–219.
Coral MN, Korkmaz H, Arikan B: Plasmid heavy metal mediated resistances in Enterobacter spp. Isolated from sofulu landfill. Ann Microbiol 2005, 55(3):175–179.
Hassen A, Saidi N, Cherif M: Resistance of environmental bacteria to heavy metals. Bio Resou Technol 1998, 64: 7–15.
Chen WP, Kuo TT: A simple and rapid method for the preparation of gram negative bacterial genomic DNA. Nucleic Acids Res 1993, 21: 2260.
Shi Z, Cao Z, Qin D, Zhu W, Wang Q, Li M, Wang G: Correlation models between environmental factors and bacterial resistance to antimony and copper. PLoS ONE 2013, 8(10):e78533. 10.1371/journal.pone.0078533
Klimek B: Effect of long-term zinc pollution on soil microbial community resistance to repeated contamination. B Environ Contam Tox 2012, 88(4):617–622.
Samudro G, Mangkoedihardjo S: Review on BOD, COD and BOD/COD ratio: a triangle zone for toxic, biodegradable and stable levels. Int J Academic Res 2010, 2(4):235–239.
Margesin R, Schinner F: Bioremediation (natural attenuation and biostimulation) of diesel-Oil-contaminated soil in an alpine glacier skiing area. Appl Environ Microbiol 2001, 67: 3127–3133.
Sevgi E, Coral G, Gizir AM: Investigation of heavy metal resistance in some bacterial strains isolated from industrial soils. Turk J Biol 2010, 34: 423–431.
Schmidt T, Schlegel HG: Combined nickel-cobalt-cadmium resistance encoded by the ncc locus of Alcaligenes xylosoxidans31A . J Bacteriol 1994, 176: 7045–7054.
Rajbanshi A: Study on heavy metal resistant bacteria is Guhewori sewage treatment plant. J our nature 2008, 6: 52–57.
Arundhati P, Paul AK: Nickel uptake and intracellular localization in Cupriavidus pauculus kps 201. Adv biosci biotechnol 2010, 1: 276–280.
Malik A, Khan IF, Alem A: Plasmid incidence in bacteria from agricultural and industrial soils. World J Microbiol Biotechnol 2002, 18: 827–833.
Dalsgarrd A, Guardbassi L Danish EPA Environment project No. 722. Occurrence and Fate of Antibiotics Resistant Bacteria in Sewage 2002.
Hernandz B, Dorian A: Zinc and lead biosorption by Delftia tsuruhatensis : a bacterial strain resistant to metals isolated from mine tailings. J water Resource Protec 2012, 4: 1–11.
Mikolay A, Nies DH: The ABC-Transporter AtmA is involved in nickel and cobalt resistance of Cupriavidus metallidurans strain CH34. Anotonic Van Leev wenhoek 2009, 96(2):183–191.
Stoppel RD, Schlegel HG: Nickel- resistant bacteria from anthropogenically nickel- Polluted and naturally nickel Percolated ecosystems. Appl Environ Microbiol 1995, 61(6):2276–2285.
Idris R, Kuffner M, Bodrossy L: Characterization of Ni-tolerant methylobacteria associated with the hyperaccumulating plant Thlaspi goesingense and description of Methylobacterium goesingense sp. nov. Syst Appl Microbiol 2006, 29(8):634–644.
Aboudrar W, Schwartz C, Morel J-L, Boularbah A: Effect of nickel-resistant rhizosphere bacteria on the uptake of nickel by the hyperaccumulator Noccaea caerulescens under controlled conditions. J Soils Sediments 2013, 13(3):501–507.
Acknowledgements
The authors are thankful to the Islamic Azad University of Khorasgan (Isfahan) branch, Isfahan, Iran, for providing laboratory facilities.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
The overall implementation of this study including experiments, data analysis, and manuscript preparation were the results of joint efforts by individuals who are listed as co-authors of this paper. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Alboghobeish, H., Tahmourespour, A. & Doudi, M. The study of Nickel Resistant Bacteria (NiRB) isolated from wastewaters polluted with different industrial sources. J Environ Health Sci Engineer 12, 44 (2014). https://doi.org/10.1186/2052-336X-12-44
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/2052-336X-12-44